Abstract
Interleukin (IL)-12 activates T helper (Th) 1 cells to produce interferon (IFN)-γ which inhibits atopic inflammation. IL-12 acts through interaction with its receptor, especially β2 subunit. In several studies, the low production of IFN-γ in peripheral mononuclear cells of atopic patients on response to IL-12 stimulation has been reported. Therefore we investigated the IL-12 receptor β2 (IL-12Rβ2) mRNA expression and RNA editing, nucleotide 2451 C-to-U conversion, to find the cause of low responsiveness to IL-12 in atopy. Quantitative real time PCR for mRNA expression and sequence analysis for RNA editing were performed in 80 atopic patients and 54 healthy controls. The expression of IL-12Rβ2 mRNA was significantly lower in atopic patients than healthy controls (p<0.05). In sequence analysis, RNA editing on nucleotide 2451 was not found from either atopic patients or healthy controls. In additional evaluation, there was no relationship between expression of IL-12Rβ2 mRNA and serum total IgE or blood eosinophil count. Reduced IL-12Rβ2 mRNA expression in atopic patients indicate the reduced capacity to respond to IL-12 which induce IFN-γ production and this may contribute to Th2-skewed immune response in atopy.
Keywords: interleukin-12 receptor; IL-12Rβ2; Gene Expression Profiling; Atopy; Asthma; Dermatitis, Atopic; RNA Editing
INTRODUCTION
Atopy is characterized by enhanced immunoglobulin E (IgE) response to common environmental antigens and leads to clinical disorders such as asthma, eczema and rhinitis (1). Allergic inflammation and IgE synthesis are known to be a T helper (Th) 2 phenomenon promoted by a distinctive allergen-specific Th2 cell subset (2).
Th cells can be divided into two subpopulations, Th1 and Th2, based on the cytokines they produce. Th2 cells produce interleukin (IL)-4, IL-5 and IL-13, which cause eosinophilia and promote immunoglobulin class switching to IgE (3). Th1 cells produce interferon (IFN)-γ, which inhibit Th cells to differentiate to Th2 cells and down-regulates the production of IgE.
IL-12 is the single most powerful factor inducing the synthesis of IFN-γ and acts by binding to its specific receptor. The receptor of IL-12 (IL-12R) is composed of two subunits, β1 and β2, that assemble to form a high affinity IL-12 receptor complex (4). While the β1 subunit is relatively constantly expressed, the β2 subunit expression is restricted to Th1 cells and highly regulated by several cytokines.
Several investigators have shown that peripheral blood mononuclear cells (PBMCs) from atopic patients produced less IFN-γ than those of healthy individuals when they were stimulated with IL-12 (5-9). The underlying mechanisms of the low responsiveness to IL-12 in atopic patients have not been known. In one study, reduced expression of IL-12Rβ2 mRNA was found to have relation with low production of IFN-γ in atopic patients (13). They explained that the decreased capacity of up-regulation of IL-12Rβ2 might decrease the responsiveness of Th1 cells to IL-12. In another study, RNA editing, especially, nucleotide 2451 C-to-U conversion of IL-12Rβ2, was reported in atopic patients (1). They suggested that above RNA editing might give rise to a conformational change in IL-12Rβ2 and cause impairment of Th1 cells to respond to IL-12.
The aim of this study was to investigate mRNA expression of IL-12Rβ2 as well as the RNA editing, nucleotide 2451 C-to-U conversion, of IL-12Rβ2 as a possible cause of low responsiveness to IL-12 in atopic patients. In additional evaluation, the correlation of the IL-12Rβ2 mRNA expression with serum total IgE concentration and blood eosinophil count were evaluated in atopic patients.
MATERIALS AND METHODS
Subjects
Eighty atopic patients (52 male, 28 female, 34.2±18.9 yr) with major allergic diseases such as bronchial asthma (10 male, 4 female, 30.7±22.1 yr), atopic dermatitis (26 male, 15 female, 36.5±18.5 yr) or allergic rhinitis (17 male, 8 female, 31.6±18.1 yr) were studied. The diagnosis of allergic diseases was established by specific allergic symptoms, positive results of allergen specific IgE test and/or increased serum total IgE concentration. Fifty four healthy controls (23 male, 31 female, 47.1±14.3 yr) who had no history of atopic disease and had normal serum total IgE concentration were studied.
Methods
Venous blood was drawn from atopic patients and healthy controls. PBMCs were isolated by gradient centrifugation method using Ficoll-Paque™plus (Amersham Biosciences, Uppsala, Sweden) and were suspended in the RPMI 1640 (Invitrogen, Grand Island, NY, U.S.A.) culture medium in 1×106 density. Then the cells were cultured in the presence of 10 µg/mL PHA for 24 hr at 37℃ in a humidified atmosphere containing 5% CO2 (Forma CO2 incubator model 3039, Formal Scientific Inc, Ohio, U.S.A.).
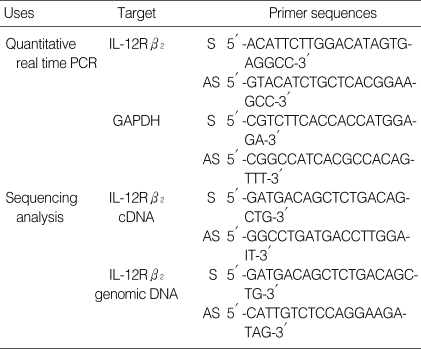
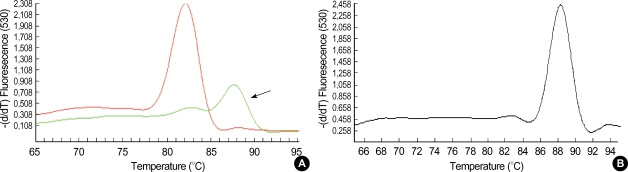
Total cellular RNA was extracted from cultured PBMCs using High Pure RNA isolation kit (Roche, Penzberg, Germany) and 5 µg of RNA was converted to complementary DNA (cDNA) by reverse transcription (RT) PCR in a total reaction mixture volume of 20 µL. The real time PCR reactions for quantification of IL-12Rβ2 cDNA and GAPDH (Glyceraldehyde-3-phosphate dehydrogenase) were performed, respectively. GAPDH was analyzed as an internal control. The master PCR reaction mixure was contained 2 µL of Light-Cycler® FastStart DNA master SYBR green I (Roche Molecular Biochemicals, Mannheim, Germany), 2.5 µL of MgCl2 in final concentration of 4 mM, each 10 pM of sense and antisense primers, 2 µL of cDNA and distilled water in final volume of 20 µL. The LightCycler® FastStart DNA master SYBR green I was composed of Taq DNA polymerase, SYBR green I dye, 10 mM MgCl2 and dNTPs (dATP, dCTP, dGTP, dUTP) in the reaction buffer. The PCR thermal conditions were as follows: 10 min at 95℃ and 40 cycles of 10 sec at 95℃, 5 sec at 57℃ and 4 sec at 72℃ for IL-12Rβ2 and 10 min at 95℃ and 40 cycles of 10 sec at 95℃, 5 sec at 55℃ and 15 sec at 72℃ for GAPDH. The primer sequences were summarized in Table 1. The quantitative real time PCR was performed using LightCycler® system (Roche Molecular Biochemical, Mannheim, Germany). During each cycle on the real time of reaction and after completion of primer extension at 72℃, fluorescence at 530 nm (F1 channel) was recorded. The amount of IL-12Rβ2 mRNA was calculated relative to the amount of GAPDH present in each sample and described as IL-12Rβ2/GAPDH ratio. All the analysis was performed in duplicate. Melting curve analysis was used to detect non-specific amplification which could be occurred during PCR procedures using SYBR green I (Fig. 1).
Table 1.
Primers for quantitative real time PCR and sequencing analysis
S, sense; AS, antisense.
Fig. 1.
Melting curves of amplification products of IL-12Rβ2 mRNA (A) and GAPDH (B). Solid arrow indicates non-specific primer-dimers product.
Fragment of exon 13 including nucleotide 2451 of IL-12Rβ2 was amplified from mRNA and genomic DNA, respectively. The PCR thermal conditions were as follows: 5 min at 94℃ and 40 cycles of 20 sec at 94℃, 20 sec at 65℃ and 45 sec at 72℃ for mRNA and 10 min at 94℃ and 40 cycles of 1 min at 94℃, 1 min at 55℃ and 1 min at 72℃ for genomic DNA. The primer sequences were summarized in Table 1. The amplified products were purified and sequenced in automatic sequencer ABI prism® 3730xI (Applied Bisosystems, Foster City, CA, U.S.A.).
Serum total IgE concentration was measured using RIDA AllergyScreen test kit (R-biopharm AG, Darmstadt, Germany). Peripheral blood eosinophil count was measured using ADVIA120 (Bayer, Dublin, Ireland).
Statistical analysis
The comparison of IL-12Rβ2 mRNA expression between atopic patients including 3 disease groups and healthy controls was analyzed using independent t test. The comparison of IL-12Rβ2 mRNA expression among 3 disease groups was analyzed using one way ANOVA. The relationship between IL-12Rβ2 mRNA expression and serum total IgE were analyzed using one way ANOVA. The relationship between IL-12Rβ2 mRNA expression and blood eosinophil count was analyzed using Spearman's correlation test. All the statistical analysis was performed with SPSS program (version 12.0, SPSS Inc, Chicago, IL, U.S.A.) and the p value of <0.05 was considered as significant.
RESULTS
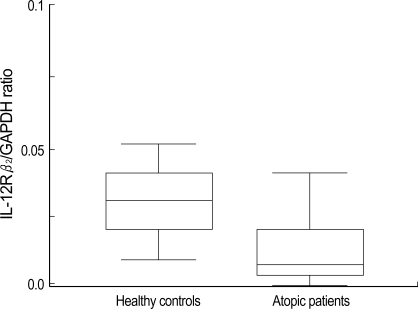
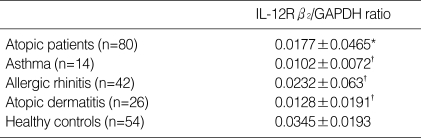
The PHA stimulated PBMCs from atopic patients had significantly lower expression of IL-12Rβ2 mRNA than the cells from healthy controls (p<0.05) (Fig. 2). The average IL-12Rβ2/GAPDH ratio was 0.035±0.02 in healthy controls and 0.018±0.047 in atopic patients. When patients were divided into 3 disease groups (asthma, atopic dermatitis, allergic rhinitis), each disease group showed lower IL-12Rβ2 mRNA expression than controls (p<0.05, respectively), also. But there was no significant difference of IL-12Rβ2 mRNA expression between 3 groups (p>0.05) (Table 2).
Fig. 2.
Comparison of IL-12Rβ2/GAPDH ratio between healthy controls (n=54) and atopic patients (n=80). The mRNA expression of IL-12Rβ2 is significantly lower in atopic patients than healthy controls (p<0.05).
Table 2.
Results of IL-12Rβ2 mRNA expression manifested by IL-12Rβ2/GAPDH ratio
*p<0.05 by independent t test compared with healthy controls. †p>0.05 by one way ANOVA compared among asthma, allergic rhinitis and atopic dermatitis. Values are mean±SD.
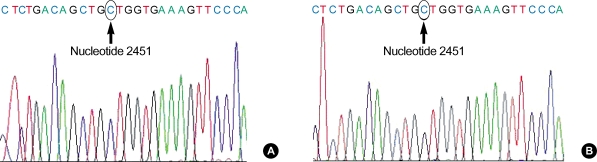
In the results of sequencing analysis, all the nucleotide 2451 was cytidine (C) in cDNA as well as genomic DNA from atopic patients and healthy controls (Fig. 3). Therefore, the RNA editing phenomenon, C-to-U conversion at nucleotide 2451 of IL-12Rβ2 was not found in present study.
Fig. 3.
Results of sequencing analysis of IL-12Rβ2 cDNA (A) and genomic DNA (B). Both bases of nucleotide 2451 of cDNA and genomic DNA are cytosines in atopic patients and healthy controls. Arrows indicate the base composition of nucleotide 2451 on exon 13.
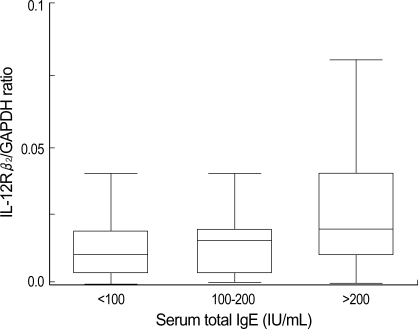
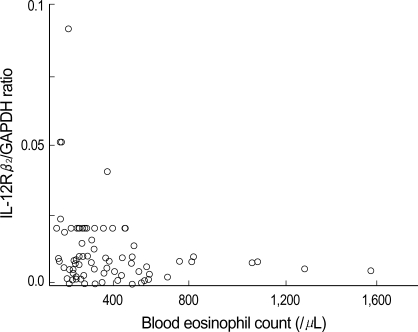
The atopic patients were divided into 3 groups according to the semiquantitative results of serum total IgE (<100 IU/mL, 100-200 mIU/mL and >200 IU/mL) and the average IL-12Rβ2/GAPDH ratio was 0.008±0.007, 0.008±0.007 and 0.028±0.064, respectively. When the expression of IL-12Rβ2 mRNA was compared, there was no significant differences among groups (p>0.05) (Fig. 4). The correlation between IL-12Rβ2 mRNA expression and blood eosinophil count was not found, either (p<0.05) (Fig. 5).
Fig. 4.
Comparison of IL-12Rβ2/GAPDH ratio among 3 groups divided by serum total IgE concentration. Significant difference is not found between 3 groups (one way ANOVA, p>0.05).
Fig. 5.
Relationship between IL-12Rβ2/GAPDH ratio and blood eosinophil count in atopic patients. Significant relationship between IL-12Rβ2 mRNA expression and blood eosinophil count is not found (Spearman's correlation test, p>0.05).
DISCUSSION
IL-12 is the single most powerful factor to induce Th1 immune response. It is produced by antigen present cells (APCs), including monocytes and dendritic cells. IL-12 is a heterodimeric cytokine composed of 35 and 40 kDa peptides and the genes are located on chromosome 3 and 5, respectively (11). By binding to its specific receptor complex, β1 and β2, IL-12 promotes the development of Th1 responses and induces production of IFN-γ. The β2 subunit is the signal transducing component of the receptor and highly regulated in contrast to the β1 subunit. IFN-γ inhibits Th cells to differentiate to Th2 cells and down-regulates the production of IgE in B lymphocytes. IFN-γ is also able to augment IL-12. This IL-12/IFN-γ loop helps the predominant Th1 cell differentiation and inhibits Th2 cell differentiation (12).
Since it was reported that PBMCs from atopic patients produced less IFN-γ than those of healthy individuals when they were stimulated with IL-12 (5-9), several investigations to find genetic mechanisms from interaction between IL-12 and IL-12R have been reported. Several IL-12 polymorphism or IL-12R β2 mutations such as truncated (2496 del 91) or missense (1577 A to G and 2799 A to G) mutation has been reported (13). Although these mutations were thought to lead to change in the conformational structure of β2 receptor, these are controversial, yet. In a recent study, the relationship between RNA editing of IL-12Rβ2 and atopy was reported by Kondo et al. (1). RNA editing is defined when there is any specific alteration in primary sequence of a RNA molecule. Such changes have marked effects on gene expression. They found that the patients who had nucleotide 2451 C-to-U conversion showed lower IFN-γ production and faint tyrosine phosphorylation of STAT-4 than those who did not have editing. This C-to-U conversion cause substitution of amino acid alanine to valine. Therefore, they proposed that the RNA editing might cause impairment of the IL-12 signal cascade and the subsequent reduction in IFN-γ production by disturbing conformational binding of IL-12 to IL-12R. Unfortunately, we could not find the C-to-U conversion at nucleotide 2451 from either atopic patients or healthy controls.
In other studies, the expression of IL-12Rβ2 mRNA was studied in atopy (10, 14). PBMCs from children with atopic airway symptoms showed lower mRNA expression of IL-12Rβ2 than non-atopic children and the production of IFN-γ after stimulation with IL-12 was also lower in atopic children. In present study, we also found that PBMCs from atopic patients expressed lower IL-12Rβ2 mRNA than those of healthy controls. This low expression findings of IL-12Rβ2 mRNA was found in all 3 atopic disease groups, but significant differences was not found between groups. These results correspond with previous studies and support their hypothesis that reduced capacity of up-regulation of IL-12Rβ2 may contribute to the Th2-skewed immune response in atopic patients.
The underlying mechanism of low mRNA expression of IL-12Rβ2 in atopic patients cannot be known yet. But there was a suggestion to introduce association of CD2 defects and low responsiveness to IL-12. In previous and present studies, the PBMCs were stimulated with phytohaemagglutinin, which activates lymphocytes via CD2. CD2 is an accessory molecule of T cell and has been thought to synergize the IL-12 action by up-regulation of the β2 chain (15, 16). Besides, the lower proportion of CD2+ lymphocytes was detected in atopic children (17). But further studies will be required whether there is connection between CD2 and atopy.
In additional evaluation, we also analyzed the relationship between IL-12Rβ2 mRNA expression and serum total IgE concentration or eosinophil count. There were not any significant associations between them. This result indicates that the IL-12Rβ2 mRNA expression would not affect production of IgE or eosinophils, directly. However, we thought that this analysis would be more reliable if we analyzed serum IgE concentration with quantitative method. In one study, a negative correlation between IL-2-induced IL-12Rβ2 mRNA expression and serum IgE concentration had been reported (10).
In conclusion, RNA editing which might cause the conformational changes in IL-12Rβ2 was not found, but less IL-12Rβ2 mRNA on response to PHA stimulation was found in atopic patients. This low expression of IL-12Rβ2 did not have relationship with atopic disease category, serum IgE concentration or blood eosinophil count. We guess low expression of IL-12Rβ2 may contribute to low responsiveness to IL-12, low production of IFN-γ and Th2-skewed immunity in atopic patients, subsequently.
References
- 1.Kondo N, Matsui E, Kaneko H, Aoki M, Kato Z, Fukao T, Kasahara K, Morimoto N. RNA editing of interleukin-12 receptor beta2, 2451 C-to-U (Ala 604 Val) conversion, associated with atopy. Clin Exp Allergy. 2004;34:363–368. doi: 10.1111/j.1365-2222.2004.01901.x. [DOI] [PubMed] [Google Scholar]
- 2.Romagnani S. Immunologic influences on allergy and the TH1/TH2 balance. J Allergy Clin Immunol. 2004;113:395–400. doi: 10.1016/j.jaci.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 3.Romagnani S. The Th1/Th2 paradigm. Immunol Today. 1997;18:263–266. doi: 10.1016/s0167-5699(97)80019-9. [DOI] [PubMed] [Google Scholar]
- 4.Presky DH, Yang H, Minetti LJ, Chua AO, Nabavi N, Wu CY, Gately MK, Gubler U. A functional interleukin 12 receptor complex is composed of two beta-type cytokine receptor subunits. Proc Natl Acad Sci USA. 1996;93:14002–14007. doi: 10.1073/pnas.93.24.14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilkens CM, Messer G, Tesselaar K, van Rietschoten AG, Kapsenberg ML, Wierenga EA. Lack of IL-12 signaling in human allergen-specific Th2 cells. J Immunol. 1996;157:4316–4321. [PubMed] [Google Scholar]
- 6.Kondo N, Matsui E, Kaneko H, Fukao T, Teramoto T, Inoue R, Watanabe M, Kasahara K, Morimoto N. Reduced interferon-gamma production and mutations of the interleukin-12 receptor beta(2) chain gene in atopic subjects. Int Arch Allergy Immunol. 2001;124:117–120. doi: 10.1159/000053687. [DOI] [PubMed] [Google Scholar]
- 7.Matsui E, Kaneko H, Teramoto T, Fukao T, Inoue R, Kasahara K, Takemura M, Seishima M, Kondo N. Reduced IFN gamma production in response to IL-12 stimulation and/or reduced IL-12 production in atopic patients. Clin Exp Allergy. 2000;30:1250–1256. doi: 10.1046/j.1365-2222.2000.00931.x. [DOI] [PubMed] [Google Scholar]
- 8.Shikano H, Kato Z, Kaneko H, Watanabe M, Inoue R, Kasahara K, Takemura M, Kondo N. IFN-gamma production in response to IL-18 or IL-12 stimulation by peripheral blood mononuclear cells of atopic patients. Clin Exp Allergy. 2001;31:1263–1270. doi: 10.1046/j.1365-2222.2001.01141.x. [DOI] [PubMed] [Google Scholar]
- 9.Van der Pouw Kraan TC, Boeije LC, de Groot ER, Stapel SO, Snijders A, Kapsenberg ML, van der Zee JS, Aarden LA. Reduced production of IL-12 and IL-12-dependent IFN-gamma release in patients with allergic asthma. J Immunol. 1997;158:5560–5565. [PubMed] [Google Scholar]
- 10.Aniansson Zdolsek H, Janefjord CK, Fälth-Magnusson K, Jenmalm MC. Reduced IL-2 induced IL-12 responsiveness in atopic children. Pediatr Allergy Immunol. 2003;14:351–357. doi: 10.1034/j.1399-3038.2003.00075.x. [DOI] [PubMed] [Google Scholar]
- 11.Gubler U, Chua AO, Schoenhaut DS, Dwyer CM, McComas W, Motyka R, Nabavi N, Wolitzky AG, Quinn PM, Familletti PC. Coexpression of two distinct genes is required to generate secreted bioactive cytotoxic lymphocyte maturation factor. Proc Natl Acad Sci USA. 1991;88:4143–4147. doi: 10.1073/pnas.88.10.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manetti R, Parronchi P, Giudizi MG, Piccinni MP, Maggi E, Trinchieri G, Romagnani S. Natural killer cell stimulatory factor (interleukin 12 [IL-12]) induces T helper type 1 (Th1)-specific immune responses and inhibits the development of IL-4-producing Th cells. J Exp Med. 1993;177:1199–1204. doi: 10.1084/jem.177.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsui E, Kaneko H, Fukao T, Teramoto T, Inoue R, Watanabe M, Kasahara K, Kondo N. Mutations of the IL-12 receptor beta2 chain gene in atopic subjects. Biochem Biophys Res Commun. 1999;266:551–555. doi: 10.1006/bbrc.1999.1859. [DOI] [PubMed] [Google Scholar]
- 14.Janefjord CK, Jenmalm MC. PHA-induced IL-12R beta(2) mRNA expression in atopic and non-atopic children. Clin Exp Allergy. 2001;31:1493–1500. doi: 10.1046/j.1365-2222.2001.01206.x. [DOI] [PubMed] [Google Scholar]
- 15.O'Flynn K, Krensky AM, Beverly PC, Burakoff SJ, Linch DC. Phytohaemagglutinin activation of T cells through the sheep red cell receptor. Nature. 1985;313:686–687. doi: 10.1038/313686a0. [DOI] [PubMed] [Google Scholar]
- 16.Gollob JA, Li J, Reinherz EL, Ritz J. CD2 regulates responsiveness of activated T cells to interleukin 12. J Exp Med. 1995;182:721–731. doi: 10.1084/jem.182.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenmalm MC, Aniansson Zdolsek H, Holt PG, Björkstén B. Expression of and responses to CD2 and CD3 in 18-month-old children with and without atopic dermatitis. Pediatr Allergy Immunol. 2000;11:175–182. doi: 10.1034/j.1399-3038.2000.00083.x. [DOI] [PubMed] [Google Scholar]