Abstract
Posttranscriptional regulation of gene expression of mRNAs containing adenine-uridine rich elements (AREs) in their 3′ untranslated regions is mediated by a number of different proteins that interact with these elements to either stabilise or destabilise them. The present review concerns the TPA-inducible sequence 11 (TIS11) protein family, a small family of proteins, that appears to interact with ARE-containing mRNAs and promote their degradation. This family of proteins has been extensively studied in the past decade. Studies have focussed on determining their biochemical functions, identifying their target mRNAs, and determining their roles in cell functions and diseases.
1. Introduction
Mechanisms of posttranscriptional gene regulation by micro-RNA (miRNA) and short interfering RNA (siRNA) have been highlighted in recent years as having important roles in control of a variety of developmental and functional processes. The present review concerns a small family of proteins, the 12-O-tetradecanoylphorbol-13-acetate (TPA) inducible sequence 11 (TIS11) family, which also function in posttranscriptional gene regulation and whose functions may overlap and interact with miRNA and siRNA control mechanisms. The TIS11 family consists of four mammalian members and include TIS11 (ZFP36, TTP, Nup475, GOS24), TIS11b (Berg36, ERF-1, ZFP36L1, BRF-1), and TIS11d (ZFP36L2, ERF-2, BRF-2). The fourth family member described in rodents, Zfp36l3, was expressed in mouse placenta, but was not detected in human placenta or other human tissues [1]. TIS11-like proteins have also been identified in drosophila and yeast [2–4]. These proteins contain two tandemly repeated zinc finger motifs through which they bind to adenine uridine (AU) rich elements (AREs) in mRNA and mediate ARE-mediated mRNA decay [5]. Table 1 lists the three human TIS11 family members, their chromosomal locations as well as reported mRNA targets. This family of proteins have been reported to promote deadenylation, decapping, and finally degradation of mRNAs by either exosome (3′-5′ degradation) or XRN1 exonuclease (5′-3′ degradation) [6].
Table 1.
Human TIS11 family members and reported mRNA targets of the TIS11 family.
| Gene | Alternative names | Chromosomal location | Reported mRNA targets | reference | mechanism |
|---|---|---|---|---|---|
| TIS11 | ZFP36, TTP, Nup475, GOS24 | 19q13.1 | TNF* | [8] | mRNA stability |
| GM-CSF | [15] | mRNA stability | |||
| IL-3 | [16] | mRNA stability | |||
| IL-6 | [17] | mRNA stability | |||
| cyclooxygenase | [18] | mRNA stability | |||
| PAI type 2 | [19] | mRNA stability | |||
| Pitx2 | [20] | mRNA stability | |||
| TIS11 | [14] | mRNA stability | |||
| IL-2 | [21] | mRNA stability | |||
| 1,4galactosyltransferase | [22] | mRNA stability | |||
| IL-12 | [23] | ? | |||
| Ccl2 | [24] | mRNA stability | |||
| Ccl3 | [24] | mRNA stability | |||
| c-myc | [25] | mRNA stability | |||
| cyclin D1 | [25] | mRNA stability | |||
| Fos | [26] | mRNA stability | |||
| Ier3 | [27] | mRNA stability | |||
| Genome analysis 250 mRNAs | [27] | mRNA stability | |||
| MIP-2 | [23] | ? | |||
| p21 | [26] | mRNA stability | |||
| E47 | [28] | mRNA stability | |||
| VEGF | [29] | mRNA stability | |||
| IL-10 | [30] | mRNA stability | |||
| Genome analysis 137 mRNAs | [30] | mRNA stability | |||
| polo-like kinase 3 | [31] | mRNA stability | |||
| TIS11b | ZFP36L1, Berg36, ERF-1, BRF-1 | 14q22-24 | TNF | [32] | mRNA stability |
| GMCSF | [33] | mRNA stability | |||
| IL-3 | [34] | mRNA stability | |||
| VEGF | [35] | mRNA stability | |||
| c-IAP2 | [36] | mRNA stability | |||
| VEGF | [37] | translation | |||
| STAR | [38] | mRNA stability | |||
| TIS11d | ZFP36L2, ERF-2, BRF-2 | 2p22.3-p21 | TNF | [32] | mRNA stability |
| GM-CSF | [33] | mRNA stability | |||
| IL-3 | [33] | mRNA stability | |||
*Targets in bold confirmed in cells derived from knockout animals are so-called “physiological” targets.
AREs are found mostly at the 3′ end of many mRNAs encoding cytokines, transcription factors, cell cycle regulators, and apoptosis regulators. It is estimated that up to 8% of mRNAs may contain AREs [7]. AREs are subdivided into three classes: Class I, Class II, and Class III. Class II is further subdivided into Class IIA, IIB, IIC, IID, IIE [6]. Class I is characterised by the AUUUA motif and a U-rich region. Class III is characterised by a U-rich sequence found in, for example, the mRNA of c-jun. Class IIA contains five consecutive AUUUA pentamer sequences; Class IIB contains four consecutive AUUUA pentamers; Class IIC contains three consecutive AUUUA pentamers; Class IID contains two consecutive AUUUA pentamers, and finally Class IIE contains one AUUUA motif [6].
2. The TIS11 Family in Posttranscriptional Gene Regulation
One of the first indications that the prototype TIS11 protein functions as an AU binding protein came from work which showed that TIS11 inhibited TNFα production from macrophages by destabilising its messenger RNA (mRNA) and this appeared to be due to direct binding of TIS11 to the TNFα ARE [8]. It was later shown that the optimal and minimally required RNA sequence for TIS11 binding is UUAUUUAUU [9–12]. The adenine residues and the spacing between them are critical in ensuring a stable association between the TIS11 peptide and RNA, even though TIS11 was still able to strongly bind to an AUUUUA peptide, and intermediately to AUUA and AUUUUUA peptides [9, 10]. TIS11 interaction with the RNA sequence is of relatively high affinity [10]. It should be noted that AREs are found at the 3′ end of each of the TIS11 family mRNAs suggesting that they may regulate themselves by a negative feedback loop [13, 14].
Overexpression of human TIS11 in HEK293 cells caused significant reduction in the levels of an artificial reporter mRNA containing part of the TNF-α 3′ untranslated region (3′ UTR), and this was dependent on the amount of TIS11 plasmid transfected [32]. Similar effects were seen for rat TIS11b or Xenopus TIS11d, although TIS11d was less efficient in inducing TNF-α reporter mRNA decay than TIS11 and TIS11b [32]. Two zinc finger motifs in TIS11 are necessary and sufficient for binding to the AREs, and also mediate TNF-α mRNA decay [32]. Similarly, other studies reported that TIS11 and other members of the TIS11 family can also mediate decay of mRNA for GM-CSF and IL-3 [15, 33]. Critical residues in the human TIS11 family zinc finger domains for binding to the TNF-α ARE probe or GM-CSF ARE probes were Cys124, Cys147, His128, Cys162, and His166, and mutations of these residues to other amino acids completely abolished TIS11 binding [39]. In the same study it was shown that coexpression of wild type TIS11 and a Cys124R nonbinding mutant resulted in stabilisation of artificial reporter TNF-α mRNA, even though wild type TIS11 was able to bind reporter TNF-α mRNA [39]. This finding suggested that the presence of a nonbinding mutant acts as a dominant negative over TIS11 destabilising activity, possibly by interacting with proteins that regulate TIS11 destabilising function [39].
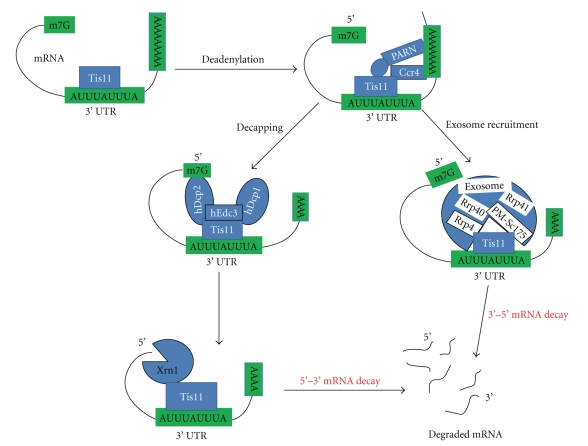
TIS11-dependent degradation of mRNA requires deadenylation [40]. Deadenylation is strongly induced by TIS11 when two nonamers (UUAUUUAUU) are present in a sequence, whereas deadenylation and mRNA degradation by TIS11 in probes containing only one nonamer is much weaker [12]. The ability of TIS11 to promote deadenylation was dependent on the presence of Mg+2 and it was suggested that there is involvement of PolyA specific ribonuclease (PARN) in the process. It was shown that TIS11 requires PARN to promote deadenylation of an ARE containing mRNA probe [40]. Association between TIS11 and PARN is indirect rather than direct and other proteins may form the bridge between them. Alternatively, the presence of TIS11 may displace an ARE stabilising factor that inhibits deadenylation caused by PARN [40]. Another study using immunoprecipitation demonstrated that TIS11 does not associate directly with PARN [41]. It was also shown that TIS11 co-immunoprecipitates with hDcp1 and hDcp2 (decapping enzymes), hXrn1 (5′-3′ exonuclease), hCcr4 (deadenylase), hRrp4 (a component of the exosome), or hEdc3 (enhances the activity of the decapping enzymes) through the N-terminal domain, whereas the zinc fingers and the C-terminal domain are not involved in these interactions [41, 42]. Additionally, deletion of the N-terminal domain of TIS11 partially abrogated induction of mRNA decay by TIS11. It was therefore proposed that TIS11 is involved in deadenylation and degradation of mRNA by recruiting enzymes such as hDcp1, hDcp2 and hCcr4 through the N-terminal domain [41]. Other transacting elements may also be involved in TIS11 function recruited through the C-terminal domain, since deletion of this domain in both TIS11 and TIS11b resulted in stabilisation of a β-globin mRNA containing AREs [41]. One such element may be the exosome, a multicomponent complex with 3′-5′ exonuclease activity, and it was reported that TIS11 requires and recruits the exosome specifically in ARE containing mRNA probes, to mediate degradation of a deadenylated mRNA [43]. It was also shown that the exosome consists of 14 different components, among which are hRrp4, hRrp40, hRrp41, hRrp42, hRrp43, hRrp46, hCsl4, and hMtr3 [43]. It seems likely that TIS11 mediated mRNA degradation involves multiple enzymes including PARN, decapping enzymes such as hDcp1, hDcp2, hEdc3, exonucleases including the exosome, and Xrn1 (5′-3′ exonuclease) as well, to preferentially degrade ARE containing mRNAs [42, 43]. Figure 1 shows the pathways and major components of ARE mediated mRNA decay.
Figure 1.
Pathways and major components of ARE-mediated mRNA decay by TIS11 proteins. TIS11 protein binds to the ARE sequence in the 3′ UTR and recruits deadenylases either directly (hCcr4) or indirectly (PARN). Deadenylated mRNA can be recruited by the exosome, a multiprotein structure containing proteins such as Rrp4, Rrp40, Rrp41, and PM-Scl75 that form the 3′ to 5′ exoribonuclease complex. Decapping of mRNA can follow deadenylation mediated by decapping enzymes such as Dcp1 and Dcp2 and degradation of mRNA can then be mediated by the 5′ to 3′ Xrn1 exonuclease.
Reported mRNA targets of the TIS11 family include TNF-α in many cell types [14, 24, 44–47], GM-CSF [15], IL-3 [16], IL-2 [21], IL-6 [17], IL-10 [30], Ccl2 and Ccl3 [24], 1,4-galactosyltransferase [22], vascular endothelial growth factor (VEGF) [29], cyclooxygenase-2 [18], plasminogen activator inhibitor type 2 [19], paired-like homeodomain 2 (Pitx2) [20], E47 [28], polo-like kinase 3 [31], and TIS11 itself [14]. VEGF mRNA was also reported to be a target of TIS11b [35] although in TIS11b deficient fibroblasts VEGF levels were shown to be elevated as a consequence of increased translation rather than an effect on VEGF mRNA [37]. More recently, TIS11b has been reported to target and degrade steroid acute regulatory (STAR) protein mRNA [38]. Utilisation of siRNA inhibition for TIS11 revealed that it targets FOS and p21 in the THP-1 cell line and primary human monocytes [26] and IL-12 and MIP-2 in macrophages [23]. Similarly, TIS11 is involved in the regulation of c-myc and cyclin-D following treatment of glioblastoma and prostate cancer cell lines with rapamycin [25]. This suggests that TIS11 also regulates certain transcription factors and cell cycle regulators. A global search for novel mRNA targets of TIS11 identified 250 potential targets and characterised immediate early response gene 3 (Ier3) mRNA as a novel target [27]. Notably, in this study Ier3 was clearly shown not to be a target for TIS11b [27] and in a later study polo-like kinase 3 was shown also to be a target for TIS11 but not TIS11b or TIS11d [31]. These examples illustrate that mRNA targets of TIS11 family members may be different for different family members. In fact, there is growing awareness in the field that many in vitro assays designed to measure mRNA interaction and decay may identify “nonphysiological targets” and that “physiological targets” of TTP family members can best be determined by analysis of mRNA degradation kinetics in knockout mice or cells derived from knockout mice [27]. Another more recent global analysis of TIS11 targets identified 137 potential TIS11 mRNA targets [30]. Interestingly, only a subset of the putative TIS11 targets contained the UUAUUUAUU sequence. This implies that either the RNA binding specificity of TIS11 proteins is not fully understood or that TIS11 proteins can be indirectly recruited to mRNA targets via other RNA-binding proteins, or that many false positives have been indentified in target screens. Table 1 summarises the reported mRNA targets for TIS11 family proteins and reported mechanisms.
A number of recent studies have reported links between TIS11 and miRNA functions and processes. Dicer, an enzyme required in the processing of miRNAs, was needed for TIS11 mediated degradation of TNFα ARE [48]. Furthermore, these studies revealed that miRNA16 can pair with eight bases in the TNFα ARE and this pairing is required for TIS11 mediated degradation of TNFα ARE [48]. TIS11 and miR16 do not bind directly to each other but interact through association with Argonaute/eiF2C family members [48]. Thus, TIS11 mediated mRNA degradation may require components of the miRNA processing pathway and indeed miRNA and TIS11 proteins may act codependently in mediating degradation. TIS11 was also identified as a component of the RNAi pathway by a genome wide screen in drosophila melanogaster [49]. TIS11 genes contain many predicted targets for miRNA suggesting another level of complexity in the interrelationship between TIS11 family members and miRNAs [50]. A picture is therefore emerging of potentially complex interplay of different mechanisms of posttranscriptional gene regulation which include the TIS11 family.
3. Identification of Human TIS11 Family Members
Human TIS11 is located on chromosome 19q13.1, and was isolated and cloned by several groups after stimulation of cells with serum, TPA, or insulin [51–53]. Human TIS11b is located on chromosome 14q22-24 and was isolated from chronic lymphocytic leukemia cells (CLL) after stimulation with TPA [54, 55], from human cDNA libraries probed with the rat cMG1 [13] or as a gene that reversed the phenotype of mutant H1080 cells which were unable to degrade IL-3 mRNA [34]. Finally human TIS11d is located on 2p22.3-p21 and was isolated from human cDNA libraries probed with the mouse TIS11d mRNA [56]. The sequence of each of the three human genes shows little variation between individuals, and most polymorphisms are detected in the introns, promoter, and 3′ UTR regions [57]. Thirteen polymorphisms were detected in the protein-coding regions of the three TIS11 family genes and six of these would result in amino acid changes. In TIS11b a polymorphism generating a dinucleotide substitution was detected that would prevent splicing of the single intron of TIS11b but to date the significance of this is unknown [57]. Further polymorphisms of TIS11 were identified in a more recent study and one, an C to T transition in the protein coding domain of TIS11, was significantly associated with rheumatoid arthritis in African-Americans [58].
TIS11, TIS11b, and TIS11d have all been associated with induction of mRNA decay (see Table 1), and even though redundancy in their function has been suggested, studies with knock-out mice and analysis of tissue distribution suggest that they may have cell specific regulation and functions. TIS11 knock-out mice appear normal at birth but later develop severe cachexia, autoimmunity, patchy alopecia, arthritis, and conjunctivitis, which can be reversed by administration of anti-TNF-α antibody [44]. TIS11b knock-out mice do not survive to birth but die in utero between days E8 and E13, due either to failure in chorioallontoic fusion [59], or to defective extraembryonic vasculogenesis [37]. Mice in which a truncated form of TIS11d was generated, which left the RNA binding domain and the nuclear export signal intact [60], appeared normal at birth, but females were infertile. Expression of the TIS11 family in mouse tissues has shown that the TIS11 protein is highly expressed in spleen, thymus, lung, large intestine, and liver, weakly expressed in brain and pancreas but is not expressed at all in the testis and uterus [61]. TIS11b is highly expressed in kidneys, liver, lung, pancreas, and heart, and weakly in skeletal muscle, colon, thymus, spleen, small intestine, brain, and peripheral blood leukocytes [62]. TIS11d is highly expressed in lung, liver, skeletal muscle, kidneys, pancreas, placenta, and less strongly in heart and brain [63].
4. Structure of TIS11 Family Members
The structures of the first zinc finger motif of TIS11 [64] and human TIS11d before and after binding to an ARE probe have been reported [65]. The spacing between the cysteines and histidine in the zinc fingers (CX8CX5CX3H) and the length of the linker between the zinc finger motifs are conserved between TIS11, TIS11b, and TIS11d. Because of the similarity of the zinc finger motifs between the three family members, it is expected that the model of mRNA binding to TIS11d will apply to all family members. The first zinc finger motif of TIS11 was found to be very different in comparison with other metal and zinc binding core domains, in terms of diameter and thickness [64]. For human TIS11d each of the two zinc fingers is folded independently into a compact domain connected by a linker sequence [65]. The structure is stabilised by binding of zinc to side chains of Cys159, Cys168, Cys174, His178 for the first zinc finger (ZF1) motif and Cys197, Cys206, Cys212, His216 for the second zinc finger (ZF2). Hydrogen bonds and van der Waals forces are involved in further stabilisation of this structure [65]. The orientation of the fingers is fixed, because the linker region has a relatively rigid structure stabilised by hydrogen bonds at each end and has little flexibility apart from the central region (residues 186-189) [65]. TNF-α ARE was used as a probe in this study, with the sequence 5′-UUAUUUAUU-3′. Zinc finger 1 (Arg153-Phe180) interacted with a single UAUU subsite on the 3′ end while zinc finger 2 (Lys191-Ala218) interacted with the same subsite on the 5′ end.
5. Regulation of the TIS11 Family at the mRNA Level
TIS11 family mRNAs are induced by TPA in a wide variety of cells including CLL cells, mouse fibroblasts, human epithelial cells, and PC phaeochromocytoma cells [53, 54]. Insulin can induce TIS11 in NIH3T3 mouse fibroblasts [66] but in RIE-1 cells only TIS11b is up regulated by insulin while the other two family members remain unaffected [67]. Lipopolysaccharide (LPS) induces TIS11 as early as 30 minutes poststimulation in murine macrophage RAW264.7 cells or human macrophages (THP-1) [14, 68, 69]. TIS11 was also induced by LPS in primary human white blood cells including lymphocytes, monocytes, and neutrophils [70]. TIS11 can be induced by GM-CSF in myeloid cells [71] and by TGF-β in a T-cell line [72] or a combination of TPA and ionomycin in T lymphocytes [73]. TIS11 can be induced by anisomycin (a p38 activator) and further induced by a combination of anisomycin plus IFN-γ or IFN-β as early as 1 hour poststimulation in mouse embryonic fibroblasts [24]. In the same study it was shown that addition of IFN-γ to LPS treated primary mouse bone marrow macrophages further increased induction of TIS11 mRNA in comparison with LPS treated cells [24]. Glucocorticoids have also been reported to induce TIS11 mRNA and protein in lung epithelial cells and this induction may well be important for glucocorticoid mediated control of inflammatory gene expression [74, 75]. The second family member, TIS11b, has been shown to be induced by TPA, bryostatin, and rituximab in CLL cells [54, 76, 77], calcium ionophore, anti-IgM and anti-CD20 in Burkitt's lymphoma cell lines [55, 78], insulin in RIE-1 cells [67] parathyroid hormone in human primary osteoblasts [79] or adrenocorticotropic hormone (ACTH) hormone in adrenocortical cells [80].
An important consideration is that the kinetics of induction of the mRNA for TIS11 genes appears to be dependent on the type of stimulus and cell type. For example, TIS11 mRNA is evident at 15 minutes after insulin or serum stimulation in mouse fibroblasts returning to almost baseline levels at two hours [66, 81]. In RAW264.7 cells, TIS11 mRNA induction by LPS is biphasic with the first peak at one hour and the second peak at five hours poststimulation [69]. It has been shown that both phases of the biphasic TIS11 mRNA induction after LPS treatment in RAW264.7 are regulated through the p38 pathway at the mRNA level [69] and p38 and ERK pathway at the protein level [82]. Similarly TIS11b peaks at two hours poststimulation and is clearly above basal levels at four hours after stimulation in insulin treated RIE-1 cells [67] but in ACTH treated human adrenocortical cells, peak induction of the gene occurs at three hours poststimulation and is above the basal levels for up to 24 hours poststimulation [80]. TPA stimulation of CLL cells induced TIS11b more strongly and earlier than bryostatin [76] and in the BL60-2 Burkitts lymphoma cell line, anti-IgM induced TIS11b more strongly than anti-CD20 stimulation [78]. H7 (a broad specificity inhibitor for the PKC and PKA pathways) did not have any effect on induction of TIS11b mRNA by TPA or bryostatin in CLL cells [76]. Similarly, induction of TIS11b in RIE-1 cell line by insulin was unaffected by PKC inhibition, but inhibition of PI3K pathway by wortmannin partially abrogated induction, whereas the same PI3K inhibitor had no effect on induction of TIS11b by TPA [67].
6. Regulation of the TIS11 Family at the Protein Level
The TIS11 family is also regulated at the protein level through either phosphorylation and/or localisation in different cellular compartments.
6.1. Phosphorylation
The TIS11 protein was suggested to be phosphorylated since it was noticed that stimulation of the NIH3T3 cell line with various stimuli such as serum, TPA, platelet-derived growth factor (PDGF), fibroblast growth factor (FGF) for 2 hours, induced TIS11 protein detected as 45 kDa protein and not as the predicted 33 kDa protein [83]. In this particular study, phosphorylation of TIS11 was shown to be partially regulated by the p42 mitogen activated protein kinase (MAPK) pathway, which phosphorylates Ser220 in mouse TIS11 [83]. In another study in LPS stimulated murine macrophages, TIS11 protein was evident at 75 minutes poststimulation as a 36 kDa protein, and increased in size over time up to 45 kDa at 240 minutes poststimulation [84]. TIS11 protein is a substrate for MAPK p38α and p38β isoforms which probably control multiple phosphorylation events [84]. TIS11 phosphorylation was also reported to be mediated by the p38-regulated MAPK activated protein kinase 2 (MK2) [68]. Ser52, Ser178, Thr249, Thr250, Ser264, Ser80/82 were suggested to be the sites of phosphorylation by MK2 in murine TIS11 and Ser264 and Thr250 were suggested to be regulated by the p38a pathway [85]. Addition of a p38 inhibitor to RAW 264.7 cells previously stimulated by LPS for 2 hours resulted in rapid TIS11 dephosphorylation and degradation [82]. TIS11 phosphorylation is therefore in equilibrium with dephosphorylation probably mediated by protein phosphatase 2A [86]. Major phosphorylation sites responsible for murine TIS11 stabilisation were found to be Ser52 and Ser178 which were phosphorylated by MK2 [82, 87].
The importance of Ser52 phosphorylation for the cytoplasmic localisation of murine TIS11 was demonstrated in a study which also suggested that Ser178 is important for this localisation [82]. Interestingly, Ser178 is also present in murine TIS11b and phosphorylation of this site creates a binding site for the 14-3-3 β isoform [85]. This is not the only isoform of 14-3-3 that the TIS11 family can bind to and it has been shown by a yeast two hybrid system that all TIS11 family members bind equally well to 14-3-3 η and β isoforms [88]. A study using MudPIT or MALDI/MS methods showed that the major phosphorylation sites in human TIS11 when purified from HEK293 cells were Ser66, Ser88, Thr92, Ser169, Ser186, Ser197, Ser218, Ser228, Ser276, Ser296 [89]. Ser197, Ser218, and Ser228 were predicted to be potential sites for protein kinase A, glycogen synthase kinase-3, and extracellular-signal-regulated kinase 1 (both Ser218 and Ser228), respectively [89].
Phosphorylation of TIS11 may alter its function as a destabilizing AU binding protein. One study showed that transfection of cells with TIS11 and MKK6, which activates p38, resulted in phosphorylation of TIS11 protein but this did not affect TIS11 ability to induce mRNA decay [69]. Another study using a series of serine or threonine mutants, alone or in combination, showed that these phosphorylation events did not alter the function of TIS11 in terms of TNF-α induced destabilisation or mRNA binding and deadenylation [47]. A further study showed that cotransfection of HEK293 cells with TIS11 and MKK6(E) dominant active MKK6 resulted in a slight increase in the half life of a TNF-α construct in comparison with cells transfected with TIS11 alone, suggesting some deactivation or inhibition of TIS11 function following phosphorylation [84]. Furthermore, it was reported that phosphorylation of murine TIS11 especially at Ser178 inhibited its binding ability to TNF-α ARE and thus that the unphosphorylated TIS11 is active in mRNA degradation [87]. It was proposed that reduced binding activity of phosphorylated murine TIS11 could be due to competitive binding with 14-3-3 proteins because phosphorylation of Ser52 and Ser178 has been reported to induce binding to 14-3-3 proteins [87]. More recently, it was reported that phosphorylation of human TIS11b did not affect its ability to bind to AREs and recruit RNA degradation enzymes but did nonetheless inhibit its ability to degrade ARE-containing mRNA [90]. Finally, another study involving human TIS11b, showed that phosphorylation of TIS11b by protein kinase B at Ser92 abrogated mRNA decay of an IL-3 ARE containing probe induced by overexpression of TIS11b [91].
6.2. Subcellular Localisation of TIS11 Family Members
In many cell types, TIS11 is expressed at very low/undetectable levels in the nucleus and cytoplasm of unstimulated cells and is almost entirely expressed in the cytoplasm of stimulated cells [82]. However, there was one early report showing that TIS11 is nuclear and remains nuclear after stimulation with serum [51]. Another exception appears to be human umbilical vein endothelial cells (HUVEC) in which TIS11 is cytoplasmic in resting HUVEC and becomes nuclear after stimulation with TNF-α [22]. Nuclear to cytoplasmic shuttling of murine Tis11 has been shown to be regulated through induction of phosphorylation at Ser52 and Ser178, primarily by the p38 pathway [82]. Hypophosphorylated forms of TIS11 were found to be nuclear and highly susceptible to proteasomal degradation [82].
Murine TIS11 has a nuclear export signal (NES) between amino acids 1–15 [92]. For murine TIS11b B the NES has been mapped between amino acids 305–313 and for murine TIS11d between amino acids 471–479 [92]. In a separate study, the rat TIS11 nuclear localisation signal (NLS) was mapped to the region containing the tandem repeats of the zinc finger motif (amino acids 88–161), and Arg127 and Arg131, which are located in the linker region between the two zinc finger motifs, were shown to be critical residues whereas Leu3, Isol16 and Leu10 were critical for nuclear export [93]. It has been shown by a yeast two hybrid system that TIS11 specifically associates with nucleoporin/Nup214 which is part of the nuclear pore on the cytoplasmic face of the pore complex [94] and this protein probably associates with the linker region between the zinc finger motifs to mediate nuclear localisation of TIS11. On the other hand nuclear export seems to be regulated through CRM1 since inhibition of CRM1 resulted in nuclear accumulation of TIS11 [93].
When cells are stimulated with a strong stress signal such as heat shock, TIS11 co-localises with TIA-1 in stress granules [95]. Localisation of TIS11 in stress granules also occurs when cells are treated with FCCP (p-trifluoromethoxy carbonyl cyanide phenyl hydrazone, a mitochondrial inhibitor), and oxidative stress but MK2 phosphorylation of TIS11 promotes its complex formation with 14-3-3 protein and excludes it from stress granules [96, 97]. Both zinc fingers of rat TIS11 are absolutely necessary for localisation in stress granules, especially amino acids Tyr105, Tyr113, Phe119, Gly109, and Gly114 [95]. On the contrary it was shown that arsenite treatment excludes TIS11 from stress granules through MK2 mediated or p38 and c-jun N-terminal kinase (JNK) mediated phosphorylation [47, 96]. TIS11 and TIS11b also localise to processing bodies (P-bodies), which are cytoplasmic foci of mRNAs and enzymes that have been proposed as sites of mRNA decay and translational silencing [42, 97, 98]. TIS11 and TIS11b appear to deliver ARE containing mRNAs to processing bodies for translational silencing and mRNA decay [98]. Overexpression of TIS11 and TIS11b resulted in stable association between stress granules and processing bodies suggesting that the TIS11 family plays an active role in tethering stress granules to processing bodies [97]. Stress granules are considered sites where mRNA is sorted for storage, reinitiation of translation or degradation [97, 99]. Interestingly, formation of processing bodies is absolutely necessary for RNA interference activity and translation arrest or mRNA degradation through siRNA and miRNA [100]. Thus, TIS11 family and miRNA posttranscriptional functions take place in the same subcellular location as well as possibly being interdependent and sharing mRNA degradation machinery [48].
7. TIS11 Family in Cell Functions and Disease
Overexpression of TIS11 family members induces apoptosis in a variety of cell lines including HeLa, U20S, SAOS2, and 3T3 [101, 102] and B-cell lymphoma cells [77] cells. Induction of apoptosis by all three TIS11 family members was completely abrogated in the presence of Bcl-2 or CrmA [101]. TIS11 synergistically induced apoptosis with TNF-α in 3T3 cells and the zinc fingers and the N-terminal domain of TIS11 are absolutely critical for this effect [102]. Mutant TIS11 lacking the zinc finger motifs failed to induce apoptosis and localised in the nucleus, whereas the wild type TIS11 localised in the cytoplasm [102]. The TIS11 family may regulate degradation of cell survival proteins such as, Bcl-2, directly. Bcl-2 mRNA contains a Class II ARE motif in the 3′UTR with at least one UUAUUUAUU nonamer which is conserved between different species and this sequence controls the stability of Bcl-2 [103]. RNA electrophoretic shift assays have confirmed an interaction between TIS11b and the Bcl-2 ARE (Murphy et al., unpublished observations). Because TIS11 also destabilises several mRNAs whose overexpression is related to malignant change, it has been suggested that they may form a class of tumour suppressor genes. In support of this hypothesis, it has been reported that injection of mast cells transformed with v-H-Ras resulted in tumour formation in mice as early as 6 weeks after injection due to production of autocrine IL-3 [104]. Transfection with v-H-Ras clones overexpressing TIS11 delayed tumour formation by at least 4 weeks [104]. Another study correlated response to cisplatin in head and neck squamous cell carcinoma cell lines with levels of TIS11b expression. Tumour cells expressing TIS11b underwent apoptosis following drug exposure, while tumour cells with low/absent levels of TIS11b were resistant to apoptosis, but became sensitive after transfection of cells with a TIS11b expressing plasmid [36]. Interestingly, significant downregulation of inhibitor of apoptosis protein 2 (cIAP2) but not Bcl-2 mRNA was seen in these cells even though both these mRNAs contain class II AREs [36].
TIS11 (but not TIS11b) specifically interacts with the Tax protein from either bovine leukaemia virus or human T lymphotropic virus 1 (HTLV-1) through the C-terminal domain [46]. In the presence of TIS11, Tax was less oncogenic probably due to inhibition of Tax transactivation activity [46]. Interestingly TIS11 was able to abrogate TNF-α induction by Tax in unstimulated RAW264.7 cells [46]. TIS11b expression is upregulated in human HTLV-1-infected cells by Tax transactivator protein which binds to two TIS11b upstream control elements [105].
TIS11b was found to be one of the genes that are over expressed in cell lines and primary cells expressing the AML1-ETO fusion protein, that is, found in 40% of Acute Myeloid Leukaemia subtype M2 [62]. When ectopically expressed TIS11b promoted myeloid progenitor L-G cell proliferation and inhibited differentiation [62]. Intriguingly, siRNA mediated TIS11b knockdown in murine embryonic stem cells promoted their differentiation to cardiomyocytes [106]. Increased expression of TIS11b was reported in primary breast tumours [107] and hepatocellular carcinomas [108]. In addition, TIS11, TIS11b, and TIS11d have been reported to be over-expressed in a variety of the NCI 60 panel of human cancer cell lines [109].
The involvement of the TIS11 family in inflammatory disease was one of the earliest reported disease associations of this family. TIS11 knockout mice developed a generalised autoimmune-like disease that was largely controlled by the addition of neutralising anti-TNF antibodies [44]. TIS11, in particular, has been reported to mediate degradation of inflammatory cytokine mRNAs (see Table 1 and references therein). Recently, targeting TIS11 has been shown to have therapeutic potential in an animal model of inflammatory bone loss [110].
8. Conclusion
The TIS11 family of proteins mediate posttranscriptional gene regulation, which appears to be important in a range of physiological and pathological processes. A number of important gaps in our knowledge still exist and require further study. We have limited knowledge of mRNA targets of TIS11 family members and whether individual TIS11 family members target distinct, overlapping, or identical targets to other family members. A few studies have suggested inhibition of translation may also be mediated by the TIS11 family as well as mRNA degradation and this requires further elucidation. A most interesting line of enquiry concerns the relationship of TIS11 family mediated posttranscriptional regulation to miRNA and siRNA mediated posttranscriptional regulation. Finally, their role in cell functions and disease have already provided evidence that they may be useful therapeutic targets in inflammatory diseases and cancer. Further studies are likely to identify other important regulatory mechanisms for this family in health and disease.
Abbreviations
- ARE:
Adenine uridine rich elements
- AU:
Adenine uridine
- CLL:
Chronic lymphocytic leukemia
- LPS:
Lipopolysaccharide
- MAPK:
Mitogen activated protein kinase
- miRNA:
Micro RNA
- MK2:
p38-regulated MAPK activated protein kinase 2
- PARN:
PolyA specific ribonuclease
- siRNA:
Short interfering RNA
- TPA:
12-O-tetradecanoylphorbol-13-acetate
References
- 1.Blackshear PJ, Phillips RS, Ghosh S, Ramos SVB, Richfield EK, Lai WS. Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the tristetraprolin family of CCCH tandem zinc finger proteins. Biology of Reproduction. 2005;73(2):297–307. doi: 10.1095/biolreprod.105.040527. [DOI] [PubMed] [Google Scholar]
- 2.Ma Q, Wadleigh D, Chi T, Herschman H. The Drosophila TIS11 homologue encodes a developmentally controlled gene. Oncogene. 1994;9(11):3329–3334. [PubMed] [Google Scholar]
- 3.Ma Q, Herschman HR. The yeast homologue YTIS11, of the mammalian TIS11 gene family is a non-essential, glucose repressible gene. Oncogene. 1995;10(3):487–494. [PubMed] [Google Scholar]
- 4.Thompson MJ, Lai WS, Taylor GA, Blackshear PJ. Cloning and characterization of two yeast genes encoding members of the CCCH class of zinc finger proteins: zinc finger-mediated impairment of cell growth. Gene. 1996;174(2):225–233. doi: 10.1016/0378-1119(96)00084-4. [DOI] [PubMed] [Google Scholar]
- 5.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochemical Society Transactions. 2002;30(6):945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 6.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nature Reviews Molecular Cell Biology. 2001;2(4):237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 7.Bakheet T, Williams BRG, Khabar KSA. ARED 2.0: an update of AU-rich element mRNA database. Nucleic Acids Research. 2003;31(1):421–423. doi: 10.1093/nar/gkg023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281(5379):1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 9.Worthington MT, Pelo JW, Sachedina MA, Applegate JL, Arseneau KO, Pizarro TT. RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/tristetraprolin protein. The Journal of Biological Chemistry. 2002;277(50):48558–48564. doi: 10.1074/jbc.M206505200. [DOI] [PubMed] [Google Scholar]
- 10.Blackshear PJ, Lai WS, Kennington EA, et al. Characteristics of the interaction of a synthetic human tristetraprolin tandem zinc finger peptide with AU-rich element-containing RNA substrates. The Journal of Biological Chemistry. 2003;278(22):19947–19955. doi: 10.1074/jbc.M301290200. [DOI] [PubMed] [Google Scholar]
- 11.Brewer BY, Malicka J, Blackshear PJ, Wilson GM. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of AU-rich mRNA-destabilizing motifs. The Journal of Biological Chemistry. 2004;279(27):27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- 12.Lai WS, Carrick DM, Blackshear PJ. Influence of nonameric AU-rich tristetraprolin-binding sites on mRNA deadenylation and turnover. The Journal of Biological Chemistry. 2005;280(40):34365–34377. doi: 10.1074/jbc.M506757200. [DOI] [PubMed] [Google Scholar]
- 13.Bustin SA, Nie X-F, Barnard RC, et al. Cloning and characterization of ERF-1, a human member of the Tis11 family of early-response genes. DNA and Cell Biology. 1994;13(5):449–459. doi: 10.1089/dna.1994.13.449. [DOI] [PubMed] [Google Scholar]
- 14.Brooks SA, Connoly JE, Rigby WFC. The role of mRNA turnover in the regulation of tritetraprolin expression: evidence for an extracellular signal-regulated kinase-specific, AU-rich element-dependent, autoregulatory pathway. The Journal of Immunology. 2004;172(12):7263–7271. doi: 10.4049/jimmunol.172.12.7263. [DOI] [PubMed] [Google Scholar]
- 15.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95(6):1891–1899. [PubMed] [Google Scholar]
- 16.Stoecklin G, Ming X-F, Looser R, Moroni C. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Molecular and Cellular Biology. 2000;20(11):3753–3763. doi: 10.1128/mcb.20.11.3753-3763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoecklin G, Stoeckle P, Lu M, Muehlemann O, Moroni C. Cellular mutants define a common mRNA degradation pathway targeting cytokine AU-rich elements. RNA. 2001;7(11):1578–1588. [PMC free article] [PubMed] [Google Scholar]
- 18.Sawaoka H, Dixon DA, Oates JA, Boutaud O. Tristetraprolin binds to the 3′-untranslated region of cyclooxygenase-2 mRNA: a polyadenylation variant in a cancer cell line lacks the binding site. The Journal of Biological Chemistry. 2003;278(16):13928–13935. doi: 10.1074/jbc.M300016200. [DOI] [PubMed] [Google Scholar]
- 19.Yu H, Stasinopoulos S, Leedman P, Medcalf RL. Inherent instability of plasminogen activator inhibitor type 2 mRNA is regulated by tristetraprolin. The Journal of Biological Chemistry. 2003;278(16):13912–13918. doi: 10.1074/jbc.M213027200. [DOI] [PubMed] [Google Scholar]
- 20.Briata P, Ilengo C, Corte G, et al. The Wnt/β-catenin → Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Molecular Cell. 2003;12(5):1201–1211. doi: 10.1016/s1097-2765(03)00407-6. [DOI] [PubMed] [Google Scholar]
- 21.Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. The Journal of Immunology. 2005;174(2):953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 22.Gringhuis SI, García-Vallejo JJ, van Het Hof B, van Dijk W. Convergent actions of IκB kinase β and protein kinase C δ modulate mRNA stability through phosphorylation of 14-3-3 β complexed with tristetraprolin. Molecular and Cellular Biology. 2005;25(15):6454–6463. doi: 10.1128/MCB.25.15.6454-6463.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jalonen U, Nieminen R, Vuolteenaho K, Kankaanranta H, Moilanen E. Down-regulation of tristetraprolin expression results in enhanced IL-12 and MIP-2 production and reduced MIP-3α synthesis in activated macrophages. Mediators of Inflammation. 2006;2006:8 pages. doi: 10.1155/MI/2006/40691. Article ID 40691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauer I, Schaljo B, Vogl C, et al. Interferons limit inflammatory responses by induction of tristetraprolin. Blood. 2006;107(12):4790–4797. doi: 10.1182/blood-2005-07-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marderosian M, Sharma A, Funk AP, et al. Tristetraprolin regulates cyclin D1 and c-Myc mRNA stability in response to rapamycin in an Akt-dependent manner via p38 MAPK signaling. Oncogene. 2006;25(47):6277–6290. doi: 10.1038/sj.onc.1209645. [DOI] [PubMed] [Google Scholar]
- 26.Patino WD, Kang J-G, Matoba S, Mian OY, Gochuico BR, Hwang PM. Atherosclerotic plaque macrophage transcriptional regulators are expressed in blood and modulated by tristetraprolin. Circulation Research. 2006;98(10):1282–1289. doi: 10.1161/01.RES.0000222284.48288.28. [DOI] [PubMed] [Google Scholar]
- 27.Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Molecular and Cellular Biology. 2006;26(24):9196–9208. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frasca D, Landin AM, Alvarez JP, Blackshear PJ, Riley RL, Blomberg BB. Tristetraprolin, a negative regulator of mRNA stability, is increased in old B cells and is involved in the degradation of E47 mRNA. The Journal of Immunology. 2007;179(2):918–927. doi: 10.4049/jimmunol.179.2.918. [DOI] [PubMed] [Google Scholar]
- 29.Essafi-Benkhadir K, Onesto C, Stebe E, Moroni C, Pagès G. Tristetraprolin inhibits ras-dependent tumor vascularization by inducing vascular endothelial growth factor mRNA degradation. Molecular Biology of the Cell. 2007;18(11):4648–4658. doi: 10.1091/mbc.E07-06-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoecklin G, Tenenbaum SA, Mayo T, et al. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. The Journal of Biological Chemistry. 2008;283(17):11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horner TJ, Lai WS, Stumpo DJ, Blackshear PJ. Stimulation of polo-like kinase 3 mRNA decay by tristetraprolin. Molecular and Cellular Biology. 2009;29(8):1999–2010. doi: 10.1128/MCB.00982-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. The Journal of Biological Chemistry. 2000;275(23):17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 33.Lai WS, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Tristetraprolin-mediated AU-rich element-dependent mRNA degradation can occur in the absence of a poly(A) tail. The Journal of Biological Chemistry. 2001;276(25):23144–23154. doi: 10.1074/jbc.M100680200. [DOI] [PubMed] [Google Scholar]
- 34.Stoecklin G, Colombi M, Raineri I, et al. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. The EMBO Journal. 2002;21(17):4709–4718. doi: 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciais D, Cherradi N, Bailly S, et al. Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene. 2004;23(53):8673–8680. doi: 10.1038/sj.onc.1207939. [DOI] [PubMed] [Google Scholar]
- 36.Lee SK, Kim SB, Kim JS, et al. Butyrate response factor 1 enhances cisplatin sensitivity in human head and neck squamous cell carcinoma cell lines. International Journal of Cancer. 2005;117(1):32–40. doi: 10.1002/ijc.21133. [DOI] [PubMed] [Google Scholar]
- 37.Bell SE, Sanchez MJ, Spasic-Boskovic O, et al. The RNA binding protein Zfp36l1 is required for normal vascularisation and post-transcriptionally regulates VEGF expression. Developmental Dynamics. 2006;235(11):3144–3155. doi: 10.1002/dvdy.20949. [DOI] [PubMed] [Google Scholar]
- 38.Duan H, Cherradi N, Feige J-J, Jefcoate C. cAMP-dependent posttranscriptional regulation of steroidogenic acute regulatory (STAR) protein by the zinc finger protein ZFP36L1/TIS11b. Molecular Endocrinology. 2009;23(4):497–509. doi: 10.1210/me.2008-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai WS, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. The Journal of Biological Chemistry. 2002;277(11):9606–9613. doi: 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- 40.Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Molecular and Cellular Biology. 2003;23(11):3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes and Development. 2005;19(3):351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Molecular Cell. 2005;20(6):905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 43.Chen C-Y, Gherzi R, Ong S-E, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107(4):451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 44.Taylor GA, Carballo E, Lee DM, et al. A pathogenetic role for TNFα in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4(5):445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki K, Nakajima H, Ikeda K, et al. IL-4-Stat6 signaling induces tristetraprolin expression and inhibits TNF-α production in mast cells. Journal of Experimental Medicine. 2003;198(11):1717–1727. doi: 10.1084/jem.20031701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Twizere J-C, Kruys V, Lefèbvre L, et al. Interaction of retroviral tax oncoproteins with tristetraprolin and regulation of tumor necrosis factor-α expression. Journal of the National Cancer Institute. 2003;95(24):1846–1859. doi: 10.1093/jnci/djg118. [DOI] [PubMed] [Google Scholar]
- 47.Rigby WFC, Roy K, Collins J, et al. Structure/function analysis of tristetraprolin (TTP): p38 stress-activated protein kinase and lipopolysaccharide stimulation do not alter TTP function. The Journal of Immunology. 2005;174(12):7883–7893. doi: 10.4049/jimmunol.174.12.7883. [DOI] [PubMed] [Google Scholar]
- 48.Jing Q, Huang S, Guth S, et al. Involvement of microRNA in AU-Rich element-mediated mRNA instability. Cell. 2005;120(5):623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 49.Dorner S, Lum L, Kim M, Paro R, Beachy PA, Green R. A genomewide screen for components of the RNAi pathway in Drosophila cultured cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(32):11880–11885. doi: 10.1073/pnas.0605210103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Molecular Immunology. 2008;45(7):1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DuBois RN, McLane MW, Ryder K, Lau LF, Nathans D. A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. The Journal of Biological Chemistry. 1990;265(31):19185–19191. [PubMed] [Google Scholar]
- 52.Taylor GA, Lai WS, Oakey RJ, et al. The human TTP protein: sequence, alignment with related proteins, and chromosomal localization of the mouse and human genes. Nucleic Acids Research. 1991;19(12):p. 3454. doi: 10.1093/nar/19.12.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varnum BC, Lim RW, Sukhatme VP, Herschman HR. Nucleotide sequence of a cDNA encoding TIS11, a message induced in Swiss 3T3 cells by the tumor promoter tetradecanoyl phorbol acetate. Oncogene. 1989;4(1):119–120. [PubMed] [Google Scholar]
- 54.Murphy JJ, Norton JD. Cell-type-specific early response gene expression during plasmacytoid differentiation of human B lymphocytic leukemia cells. Biochimica et Biophysica Acta. 1990;1049(3):261–271. doi: 10.1016/0167-4781(90)90096-k. [DOI] [PubMed] [Google Scholar]
- 55.Ning Z-Q, Norton JD, Li J, Murphy JJ. Distinct mechanisms for rescue from apoptosis in Ramos human B cells by signaling through CD40 and interleukin-4 receptor: role for inhibition of an early response gene, Berg36. European Journal of Immunology. 1996;26(10):2356–2363. doi: 10.1002/eji.1830261013. [DOI] [PubMed] [Google Scholar]
- 56.Nie X-F, Maclean KN, Kumar V, McKay IA, Bustin SA. ERF-2, the human homologue of the murine TIS11d early response gene. Gene. 1995;152(2):285–286. doi: 10.1016/0378-1119(94)00696-p. [DOI] [PubMed] [Google Scholar]
- 57.Blackshear PJ, Phillips RS, Vazquez-Matias J, Mohrenweiser H. Polymorphisms in the genes encoding members of the tristetraprolin family of human tandem CCCH zinc finger proteins. Progress in Nucleic Acid Research and Molecular Biology. 2003;75:43–68. doi: 10.1016/s0079-6603(03)75002-8. [DOI] [PubMed] [Google Scholar]
- 58.Carrick DM, Chulada P, Donn R, et al. Genetic variations in ZFP36 and their possible relationship to autoimmune diseases. Journal of Autoimmunity. 2006;26(3):182–196. doi: 10.1016/j.jaut.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 59.Stumpo DJ, Byrd NA, Phillips RS, et al. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the tristetraprolin family. Molecular and Cellular Biology. 2004;24(14):6445–6455. doi: 10.1128/MCB.24.14.6445-6455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramos SBV, Stumpo DJ, Kennington EA, et al. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development. 2004;131(19):4883–4893. doi: 10.1242/dev.01336. [DOI] [PubMed] [Google Scholar]
- 61.Cao H, Tuttle JS, Blackshear PJ. Immunological characterization of tristetraprolin as a low abundance, inducible, stable cytosolic protein. The Journal of Biological Chemistry. 2004;279(20):21489–21499. doi: 10.1074/jbc.M400900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shimada H, Ichikawa H, Nakamura S, et al. Analysis of genes under the downstream control of the t(8;21) fusion protein AML1-MTG8: overexpression of the TIS11b (ERF-1, cMG1) gene induces myeloid cell proliferation in response to G-CSF. Blood. 2000;96(2):655–663. [PubMed] [Google Scholar]
- 63.Ino T, Yasui H, Hirano M, Kurosawa Y. Identification of a member of the TIS11 early response gene family at the insertion point of a DNA fragment containing a gene for the T-cell receptor β chain in an acute T-cell leukemia. Oncogene. 1995;11(12):2705–2710. [PubMed] [Google Scholar]
- 64.Amann BT, Worthington MT, Berg JM. A Cys3His zinc-binding domain from Nup475/tristetraprolin: a novel fold with a disklike structure. Biochemistry. 2003;42(1):217–221. doi: 10.1021/bi026988m. [DOI] [PubMed] [Google Scholar]
- 65.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nature Structural and Molecular Biology. 2004;11(3):257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 66.Lai WS, Stumpo DJ, Blackshear PJ. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. The Journal of Biological Chemistry. 1990;265(27):16556–16563. [PubMed] [Google Scholar]
- 67.Corps AN, Brown KD. Insulin and insulin-like growth factor I stimulate expression of the primary response gene cMG1/TIS11b by a wortmannin-sensitive pathway in RIE-1 cells. FEBS Letters. 1995;368(1):160–164. doi: 10.1016/0014-5793(95)00635-m. [DOI] [PubMed] [Google Scholar]
- 68.Mahtani KR, Brook M, Dean JLE, Sully G, Saklatvala J, Clark AR. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Molecular and Cellular Biology. 2001;21(19):6461–6469. doi: 10.1128/MCB.21.9.6461-6469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tchen CR, Brook M, Saklatvala J, Clark AR. The stability of tristetraprolin mRNA is regulated by mitogen-activated protein kinase p38 and by tristetraprolin itself. The Journal of Biological Chemistry. 2004;279(31):32393–32400. doi: 10.1074/jbc.M402059200. [DOI] [PubMed] [Google Scholar]
- 70.Fairhurst AM, Connolly JE, Hintz KA, et al. Regulation and localization of endogenous human tristetraprolin. Arthritis Research & Therapy. 2003;5(4):R214–R225. doi: 10.1186/ar778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Varnum BC, Lim RW, Kujubu DA, et al. Granulocyte-macrophage colony-stimulating factor and tetradecanoyl phorbol acetate induce a distinct, restricted subset of primary-response TIS genes in both proliferating and terminally differentiated myeloid cells. Molecular and Cellular Biology. 1989;9(8):3580–3583. doi: 10.1128/mcb.9.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ogawa K, Chen F, Kim Y-J, Chen Y. Transcriptional regulation of tristetraprolin by transforming growth factor-β in human T cells. The Journal of Biological Chemistry. 2003;278(32):30373–30381. doi: 10.1074/jbc.M304856200. [DOI] [PubMed] [Google Scholar]
- 73.Raghavan A, Robison RL, McNabb J, Miller CR, Williams DA, Bohjanen PR. HuA and tristetraprolin are induced following T cell activation and display distinct but overlapping RNA binding specificities. The Journal of Biological Chemistry. 2001;276(51):47958–47965. doi: 10.1074/jbc.M109511200. [DOI] [PubMed] [Google Scholar]
- 74.Smoak K, Cidlowski JA. Glucocorticoids regulate tristetraprolin synthesis and posttranscriptionally regulate tumor necrosis factor alpha inflammatory signaling. Molecular and Cellular Biology. 2006;26(23):9126–9135. doi: 10.1128/MCB.00679-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ishmael FT, Fang X, Galdiero MR, et al. Role of the RNA-binding protein tristetraprolin in glucocorticoid-mediated gene regulation. The Journal of Immunology. 2008;180(12):8342–8353. doi: 10.4049/jimmunol.180.12.8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ning Z-Q, Hirose T, Deed R, Newton J, Murphy JJ, Norton JD. Early response gene signalling in bryostatin-stimulated primary B chronic lymphocytic leukaemia cells in vitro. Biochemical Journal. 1996;319, part 1:59–65. doi: 10.1042/bj3190059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baou M, Jewell A, Muthurania A, et al. Involvement of TIS11b, an AU-rich binding protein, in induction of apoptosis by rituximab in B cell chronic lymphocytic leukemia cells. Leukemia. 2009;23:986–989. doi: 10.1038/leu.2008.340. [DOI] [PubMed] [Google Scholar]
- 78.Mathas S, Rickers A, Bommert K, Dörken B, Mapara MY. Anti-CD20-and B-cell receptor-mediated apoptosis: evidence for shared intracellular signaling pathways. Cancer Research. 2000;60(24):7170–7176. [PubMed] [Google Scholar]
- 79.Reppe S, Olstad OK, Rian E, Gautvik VT, Gautvik KM, Jemtland R. Butyrate response factor 1 is regulated by parathyroid hormone and bone morphogenetic protein-2 in osteoblastic cells. Biochemical and Biophysical Research Communications. 2004;324(1):218–223. doi: 10.1016/j.bbrc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 80.Chinn AM, Ciais D, Bailly S, Chambaz E, LaMarre J, Feige J-J. Identification of two novel ACTH-responsive genes encoding manganese-dependent superoxide dismutase (SOD2) and the zinc finger protein TIS11b [tetradecanoyl phorbol acetate (TPA)-inducible sequence 11b] Molecular Endocrinology. 2002;16(6):1417–1427. doi: 10.1210/mend.16.6.0844. [DOI] [PubMed] [Google Scholar]
- 81.Taylor GA, Thompson MJ, Lai WS, Blackshear PJ. Mitogens stimulate the rapid nuclear to cytosolic translocation of tristetraprolin, a potential zinc-finger transcription factor. Molecular Endocrinology. 1996;10(2):140–146. doi: 10.1210/mend.10.2.8825554. [DOI] [PubMed] [Google Scholar]
- 82.Brook M, Tchen CR, Santalucia T, et al. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Molecular and Cellular Biology. 2006;26(6):2408–2418. doi: 10.1128/MCB.26.6.2408-2418.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Taylor GA, Thompson MJ, Lai WS, Blackshear PJ. Phosphorylation of tristetraprolin, a potential zinc finger transcription factor, by mitogen stimulation in intact cells and by mitogen-activated protein kinase in vitro. The Journal of Biological Chemistry. 1995;270(22):13341–13347. doi: 10.1074/jbc.270.22.13341. [DOI] [PubMed] [Google Scholar]
- 84.Zhu W, Brauchle MA, Di Padova F, et al. Gene suppression by tristetraprolin and release by the p38 pathway. American Journal of Physiology. 2001;281(2):L499–L508. doi: 10.1152/ajplung.2001.281.2.L499. [DOI] [PubMed] [Google Scholar]
- 85.Chrestensen CA, Schroeder MJ, Shabanowitz J, et al. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. The Journal of Biological Chemistry. 2004;279(11):10176–10184. doi: 10.1074/jbc.M310486200. [DOI] [PubMed] [Google Scholar]
- 86.Sun L, Stoecklin G, Van Way S, et al. Tristetraprolin (TTP)-14-3-3 complex formation protects TTP from dephosphorylation by protein phosphatase 2a and stabilizes tumor necrosis factor-α mRNA. The Journal of Biological Chemistry. 2007;282(6):3766–3777. doi: 10.1074/jbc.M607347200. [DOI] [PubMed] [Google Scholar]
- 87.Hitti E, Iakovleva T, Brook M, et al. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Molecular and Cellular Biology. 2006;26(6):2399–2407. doi: 10.1128/MCB.26.6.2399-2407.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnson BA, Stehn JR, Yaffe MB, Blackwell TK. Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and -independent mechanisms. The Journal of Biological Chemistry. 2002;277(20):18029–18036. doi: 10.1074/jbc.M110465200. [DOI] [PubMed] [Google Scholar]
- 89.Cao H, Deterding LJ, Venable JD, et al. Identification of the anti-inflammatory protein tristetraprolin as a hyperphosphorylated protein by mass spectrometry and site-directed mutagenesis. Biochemical Journal. 2006;394(1):285–297. doi: 10.1042/BJ20051316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maitra S, Chou C-F, Luber CA, Lee K-Y, Mann M, Chen C-Y. The AU-rich element mRNA decay-promoting activity of BRF1 is regulated by mitogen-activated protein kinase-activated protein kinase 2. RNA. 2008;14(5):950–959. doi: 10.1261/rna.983708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schmidlin M, Lu M, Leuenberger SA, et al. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. The EMBO Journal. 2004;23(24):4760–4769. doi: 10.1038/sj.emboj.7600477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Phillips RS, Ramos SBV, Blackshear PJ. Members of the tristetraprolin family of tandem CCCH zinc finger proteins exhibit CRM1-dependent nucleocytoplasmic shuttling. The Journal of Biological Chemistry. 2002;277(13):11606–11613. doi: 10.1074/jbc.M111457200. [DOI] [PubMed] [Google Scholar]
- 93.Murata T, Yoshino Y, Morita N, Kaneda N. Identification of nuclear import and export signals within the structure of the zinc finger protein TIS11. Biochemical and Biophysical Research Communications. 2002;293(4):1242–1247. doi: 10.1016/S0006-291X(02)00363-7. [DOI] [PubMed] [Google Scholar]
- 94.Carman JA, Nadler SG. Direct association of tristetraprolin with the nucleoporin CAN/Nup214. Biochemical and Biophysical Research Communications. 2004;315(2):445–449. doi: 10.1016/j.bbrc.2004.01.080. [DOI] [PubMed] [Google Scholar]
- 95.Murata T, Morita N, Hikita K, Kiuchi K, Kiuchi K, Kaneda N. Recruitment of mRNA-destabilizing protein TIS11 to stress granules is mediated by its zinc finger domain. Experimental Cell Research. 2005;303(2):287–299. doi: 10.1016/j.yexcr.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 96.Stoecklin G, Stubbs T, Kedersha N, et al. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. The EMBO Journal. 2004;23(6):1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kedersha N, Stoecklin G, Ayodele M, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. Journal of Cell Biology. 2005;169(6):871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes and Development. 2007;21(6):719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cougot N, Babajko S, Séraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. Journal of Cell Biology. 2004;165(1):31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jakymiw A, Lian S, Eystathioy T, et al. Disruption of GW bodies impairs mammalian RNA interference. Nature Cell Biology. 2005;7(12):1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 101.Johnson BA, Geha M, Blackwell KT. Similar but distinct effects of the tristetraprolin/TIS11 immediate-early proteins on cell survival. Oncogene. 2000;19(13):1657–1664. doi: 10.1038/sj.onc.1203474. [DOI] [PubMed] [Google Scholar]
- 102.Johnson BA, Blackwell TK. Multiple tristetraprolin sequence domains required to induce apoptosis and modulate responses to TNFα through distinct pathways. Oncogene. 2002;21(27):4237–4246. doi: 10.1038/sj.onc.1205526. [DOI] [PubMed] [Google Scholar]
- 103.Schiavone N, Rosini P, Quattrone A, et al. A conserved AU-rich element in the 3′ untranslated region of bcl-2 mRNA is endowed with a destabilizing function that is involved in bcl-2 down-regulation during apoptosis. The FASEB Journal. 2000;14(1):174–184. doi: 10.1096/fasebj.14.1.174. [DOI] [PubMed] [Google Scholar]
- 104.Stoecklin G, Gross B, Ming X-F, Moroni C. A novel mechanism of tumor suppression by destabilizing AU-rich growth factor mRNA. Oncogene. 2003;22(23):3554–3561. doi: 10.1038/sj.onc.1206418. [DOI] [PubMed] [Google Scholar]
- 105.Li B, Fink T, Ebbesen P, Liu XD, Zachar V. Expression of butyrate response factor 1 in HTLV-1-transformed cells and its transactivation by tax protein. Archives of Virology. 2003;148(9):1787–1804. doi: 10.1007/s00705-003-0114-9. [DOI] [PubMed] [Google Scholar]
- 106.Wegmüller D, Raineri I, Gross B, Oakeley EJ, Moroni C. A cassette system to study embryonic stem cell differentiation by inducible RNA interference. Stem Cells. 2007;25(5):1178–1185. doi: 10.1634/stemcells.2006-0106. [DOI] [PubMed] [Google Scholar]
- 107.Abba MC, Sun H, Hawkins KA, et al. Breast cancer molecular signatures as determined by SAGE: correlation with lymph node status. Molecular Cancer Research. 2007;5(9):881–890. doi: 10.1158/1541-7786.MCR-07-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zindy PJ, L'Helgoualc'h A, Bonnier D, et al. Upregulation of the tumor suppressor gene menin in hepatocellular carcinomas and its significance in fibrogenesis. Hepatology. 2006;44(5):1296–1307. doi: 10.1002/hep.21367. [DOI] [PubMed] [Google Scholar]
- 109.Carrick DM, Blackshear PJ. Comparative expression of tristetraprolin (TTP) family member transcripts in normal human tissues and cancer cell lines. Archives of Biochemistry and Biophysics. 2007;462(2):278–285. doi: 10.1016/j.abb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 110.Patil CS, Liu M, Zhao W, et al. Targeting mRNA stability arrests inflammatory bone loss. Molecular Therapy. 2008;16(10):1657–1664. doi: 10.1038/mt.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]