Abstract
In the past 21 years, a modest increase in the range of antimalarial drugs approved for clinical use has been complemented by a more impressive expansion in the analysis and understanding of the molecular mechanisms underlying resistance to these agents. Such resistance is a major factor in the increasing difficulty in controlling malaria, and important developments during this period are recounted here.
The drug-resistance problem
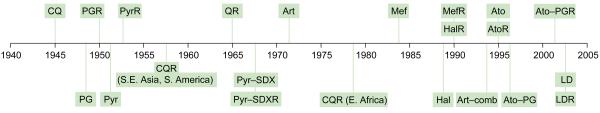
The first article in the newly emerged Parasitology Today to deal with drug-resistant malaria appeared in early 1986 and reported [1] that Plasmodium falciparum in Central and West Africa was becoming resistant ‘not only to chloroquine, but also to all existing alternative treatments except quinine’. The article concluded with the announcement that the new drugs mefloquine and halofantrine would become widely available for clinical use later that year, although mefloquine resistance had already been observed in earlier trials (Figure 1). At that time, the principal targets of the well-established antifolates (principally pyrimethamine and sulfadoxine) were known, thanks to a substantial body of earlier work on antifolate bacterial inhibitors, but those of other antimalarials were less clear. Moreover, the molecular basis of resistance to any of these drugs was completely unknown. In the past two decades, which have witnessed astounding progress in the molecular analysis and manipulation of Plasmodium, this situation has been substantially rectified and new classes of drugs such as the artemisinins and atovaquone have also entered clinical use (Figure 1). However, the problem of drug-resistant malaria is undoubtedly worse now than in 1986 [2], and the challenge of turning detailed molecular understanding into effective disease reduction remains stubbornly daunting.
Figure 1.
Approximate dates of introduction of antimalarial drugs as clinical agents, and observations of treatment failure. Quinine (Q) has been used in Europe since the 17th century and resistance to it was observed sporadically from the early 20th century. By 1965, consistent reports of treatment failure were appearing, mainly from Southeast Asia. Quinine is now reserved for use as a drug of last resort. CQR had spread to essentially all of sub-Saharan Africa by 1988. Despite a rapid spread of pyrimethamine (Pyr)-sulfadoxine (SDX) resistance in Southeast Asia from the mid-to-late 1960s onwards, this combination has been used widely and successfully, albeit with diminishing efficacy, to combat chloroquine (CQ)-resistant parasites in Africa since the early 1990s. Artemisinin was used in Chinese field trials as monotherapy in the 1970s. High rates of recrudescence were observed because of the very short half-life of the drug in human plasma (~4 h), although there are also reports of parasites from clinical cases with reduced sensitivity to artemisinins in vitro. Artemisinin derivatives are now used clinically only in combinations with other antimalarial drugs – no clinical resistance to these has yet been observed. Mefloquine (Mef) resistance had been reported in some field trials in the early 1980s. Atovaquone (Ato) is now used clinically only as atovaquone– proguanil (PG) combination (Malarone). LapDap (chlorproguanil–dapsone) can overcome many Pyr–SDX-resistant infections but it targets the same enzymes as this combination and is not 100% efficacious. Other abbreviations: Art, artemisinin and derivatives; comb, combinations; Hal, halofantrine; LD, LapDap; R, resistance.
The antifolates and the quinolines
Between 1988 and 1997, important insights into the genetic basis of resistance to the antifolate drugs emerged as extensive analyses were undertaken of the known target dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) genes in sensitive and resistant parasites. Correlations of sequence alterations with resistance to pyrimethamine and sulfadoxine were soon refined into proofs of a causal relationship, mainly by expression and purification of the various polymorphic recombinant enzymes and measurement of their altered drug-binding properties (Table 1). These studies were reinforced by the powerful methodology of parasite transfection, newly developed in 1995, which demonstrated that insertion of the resistant-type dhfr sequences into the genome of sensitive parasites resulted in predicted levels of pyrimethamine resistance, an approach later used to consolidate similar data for dhps mutations and mutations involved in resistance to the quinoline drugs [3-5].
Table 1.
Genetic changes in P. falciparum associated with resistance to current drugs in clinical use as antimalarials
| Drug | Gene(s)a,b encoding target(s) |
Principal amino acid changes associated with resistance levels in the fieldc |
Refsf |
|---|---|---|---|
| Sulfadoxine, dapsone | dhps | S436A/F, A437Gd, K540E, A581G, A613S/T | [3,5] |
| Pyrimethamine, trimethoprim | dhfr | N51I, C59R, S108Nd, I164L or C50R, N51I, S108Nd, I164L | [3,5] |
| Proguanil, chlorproguanil | dhfr | A16V, S108Td or N51I, C59R, S108Nd, I164L | [3,5] |
| Chloroquine | crt | C72S, M74I, N75D/E, K76Td, A220S, Q271E | [7,9,11-13,16] |
| Chloroquine | mdr1 | N86Yd, Y184F, S1034C, N1042D, D1246Y (subsidiary effect relative to crt mutations) |
[18,19] |
| Mefloquine, quinine, halofantrine, lumefantrine |
mdr1 | Copy number >1; wild-type N86 (can also be affected by mutations in crt to differing degrees in different strains) |
[18-20] |
| Tetracycline, doxycycline | (mt protein synthesis) | Not yet characterized | |
| Atovaquone | cytb | Y268S/Ne | [34-36] |
| Artemisinin and derivatives | atp6 | Clinical resistance to combinations not yet observed; atp6 likely to be eventual principal determinant, with L263 a key residue |
[26,27] |
| Artemisinin and derivatives | mdr1 | Clinical resistance to combinations not yet observed; status of mdr1 might affect precise sensitivity levels |
[18-20] |
Known targets (it cannot be excluded that there are as yet unidentified additional targets).
Abbreviations and gene products: atp, ATPase; crt, chloroquine resistance transporter; cytb, cytochrome b; dhfr, dihydrofolate reductase; dhps, dihydropteroate synthase; mdr, multidrug resistance; mt, mitochondrial.
The most commonly observed mutations associated with resistance in the field.
Key mutations; the relative importance of the other mutations is not assessed or indicated here. Some of these probably function as compensatory mutations to maintain function of the target protein, rather than directly affecting drug binding.
Observed in several of the very small number of clinical failures using Malarone reported to date.
Representative references only; numerous studies have reported many of the mutations listed.
Compared with the situation for the antifolates, unravelling the genetic basis of chloroquine resistance (CQR) was a much more substantial undertaking because the likely target genes were completely unknown. The bold approach taken by the Wellems group at the National Institutes of Health (http://www.nih.gov/) [6,7] began with a genetic cross of two cloned parasite populations (chloroquine-sensitive and chloroquine-resistant) during the sexual stage in the mosquito, and subsequent isolation of recombinant asexual progeny in a primate host. Phenotypic typing of progeny and mapping of their recombination loci pointed to a key gene residing within a 36-kb span on chromosome 7 [8]. Sequencing along this region identified several potential open reading frames (ORFs) that were analysed for polymorphisms correlating with the chloroquine-resistance (CQR) phenotype in P. falciparum strains from around the world. Some 15 years after the original cross, and following further detective work involving detailed transcriptional analysis of this locus, a highly fragmented ORF of 13 exons was eventually revealed, mutations in which showed complete linkage to the CQR phenotype in the 40 laboratory-adapted strains examined at that time [9]. Moreover, the genomic locus of this ORF exhibited the most pronounced sequence evolution during recent times, consistent with the geographically near-universal deployment of chloroquine since its introduction in the mid-1940s [10]. The gene involved, named pfcrt for ‘P. falciparum chloroquine resistance transporter’, is polymorphic in up to 15 nucleotide positions, as catalogued thus far, but only one amino acid change, K76T, has been found consistently in chloroquine-resistant parasites. However, a second alteration, A220S, also shows extremely strong linkage, possibly compensating for changes in the normal function of the CRT protein induced by K76T, and is likely to have a modulating effect on the level of resistance (Table 1). cDNA versions of pfcrt from resistant parasites transfected into sensitive parasites bestow the resistance phenotype, with the status of codon 76 confirmed as being especially important [7]. This is reflected in many recent field studies: for example, in West Africa where all chloroquine-resistant strains examined carried both the K76T and A220S changes, compared with ~40% prevalence in random pretreatment samples [11]. Indeed, no chloroquine-resistant isolate has yet been characterized that carries the wild-type lysine at position 76. However, it was observed recently that some sensitive strains can also carry the K76T mutation [12,13], emphasizing the need for still finer analysis of the complex mutational patterns seen in pfcrt.
The identification of the CRT protein as the key player in CQR, although undoubtedly a tour de force of persistence and insight, marked a new beginning, rather than an end, to the story. Bioinformatics analyses indicate that CRT belongs to a previously undefined collection of proteins within a superfamily of drug and metabolite transporters [14,15]. It was proposed that the crucial K76T mutation would alter the selectivity of CRT such that chloroquine could efficiently exit the food vacuole in which it acts, a prediction that has received recent experimental support [13,16]. It now seems clear that the positive charge of K76 normally prevents efflux of the diprotonated form of chloroquine (the predominant form in the acidic vacuole), an effect that is lost on mutation to T or other nonbasic residues. However, mutations that restore a positive charge to the pore can compensate for the loss of that carried by K76 and, thus, reverse the CQR phenotype, as is seen in at least one of the apparently anomalous sensitive strains mentioned earlier, in which an S163R alteration accompanies K76T [13].
Cross-resistance
Early studies, particularly from Southeast Asia, had reported that cross-resistance was apparent among some chloroquine-resistant parasites to related antimalarials such as the arylaminoalcohols quinine, mefloquine and halofantrine, suggesting that these drugs were also acting on the parasite food vacuole. Another approach to investigating resistance involving these drugs was, therefore, to search for an ATP-binding cassette (ABC)-type membrane transporter that could alter the flux of drugs targeted to this vacuole. This was based on an analogy with mammalian tumour cells that exhibit multidrug resistance in which the P-glycoprotein is upregulated and mediates an ATP-dependent expulsion of a wide range of inhibitors. This can be reversed by Ca2+-channel blockers such as verapamil, and the fact that CQR levels in P. falciparum could also be diminished by verapamil lent further credence to this approach. One gene, pfmdr1, with significant similarity to the human multidrug resistance (mdr) gene exhibited polymorphisms that were apparently linked to CQR in some (but not all) studies [17,18]. Nevertheless, the localization of its gene product, P-glycoprotein homologue 1 (Pgh1), on the surface of the food vacuole indicated that it was appropriately placed to modulate intracellular concentrations of chloroquine and other inhibitors – a conclusion supported both by transfection studies [19] and recent field studies (e.g. Ref. [12]). Many such studies support the conclusion that there is a reciprocal relationship between CQR and resistance to the arylaminoalcohols [18]. Importantly, whereas pfmdr1 might have a significant but subsidiary role in CQR, its amplification to copy numbers of up to five seems to be a primary mechanism whereby parasites can become more resistant to mefloquine and other arylaminoalcohols [20]. However, changes in pfcrt alone can also lead to similar patterns of resistance to these drugs [13] and seem to be the primary determinant in the reversal of CQR by verapamil [16]. It is also now apparent that putative parasite transporters other than pfcrt and pfmdr1 can influence the precise level of sensitivity to these antimalarial drugs [21].
New drugs and drug combinations
Just as the addition of sulfadoxine to pyrimethamine in the early 1960s extended the useful life of pyrimethamine by decades, it is now clear that judicious combination of inhibitors can not only prolong efficacy but also give rise to highly potent treatments. A new combination of the old antifolate drugs chlorproguanil and dapsone (LapDap) has recently been introduced into Africa as a cheap treatment for parasites resistant to sulfadoxine–pyrimethamine [22]. The endoperoxide artemisinin, in the form of artesunate, was originally deployed in areas of multidrug resistance from 1994 onwards in Southeast Asia, where it has been used successfully in combination with mefloquine. Resistance to combination therapies based on artemisinin has yet to be seen in the field, and the rapid manner of killing by artemisinin means that the probability of resistant parasites emerging is much lower than for slower-acting drugs [23]. Nevertheless, resistance to artemisinin derivatives has been seen in the laboratory and it is important to know how this class of compound functions because such knowledge forms a crucial basis for dealing with resistance [24]. Heterologous expression and fluorescence localization studies indicate that artemisinins inhibit the Ca2+-dependent ATPase PfATP6 on membranous structures in the cytoplasm of the parasite and around the food vacuole (but not within it) after activation by iron (as Fe2+) [25]. However, this might not be the only target, given the propensity of these compounds to form promiscuous free radicals that can alkylate proteins and other molecules [25,26]. Importantly, it has recently been shown in heterologous in vivo expression studies that mutations in the key residue L263 in the inhibitor-binding site of PfATP6 can abolish sensitivity to artemisinins. This not only supports the notion that this protein is the principal drug target but also provides insight into the likely mechanism of eventual clinical resistance, in addition to pointers as to how derivatives of this family of inhibitors might be adapted most effectively in future [26,27].
In addition to mefloquine, the short-half-life artemisinin derivatives are being combined with several other antimalarials such as amodiaquine, sulfadoxine–pyrimethamine, lumefantrine and piperaquine, all of which operate on longer time scales [28,29], to eradicate parasites that escape the rapidly metabolized artemisinin and to minimize the likelihood of drug-resistant parasite forms emerging [30]. However, the fact that the Pgh1 transporter influences sensitivity to artemisinins and several of these other drugs is a potential cause for concern [18].
The 2-hydroxynaphthoquinone derivative atovaquone is another recently introduced inhibitor licensed for use as a clinical antimalarial in combination with the biguanide proguanil (as Malarone). Atovaquone functions as a structural analogue of coenzyme Q in the mitochondrial electron-transport chain and can collapse the organellar membrane potential. This arises from inhibition of cytochrome b in complex III of the chain, without affecting human mitochondria [31]. Studies of atovaquone used as monotherapy revealed rapid emergence of resistance [32], and point mutations in the cytochrome b gene are associated with considerable reductions in parasite susceptibility to the drug both in vitro and in vivo [33,34]. However, in combination with proguanil, there is clearly protection against early onset of resistance, and Malarone has been used successfully for several years, especially by travellers to Southeast Asia confronted with multidrug-resistant parasites, although the expense of this drug combination mitigates against its wide-scale use. Proguanil is metabolized in most people to the triazine cycloguanil, an anti-DHFR inhibitor that would not be expected to be efficacious against most Southeast Asian parasites, and although the basis of the synergy between the two components of Malarone is not yet clear it seems to involve the parental biguanide compound rather than its metabolic product. Although apparently still completely effective as a prophylactic, some cases of resistance to Malarone treatment have been reported [34-36], not all of which could be associated with a mutated cytochrome b gene [37], indicative of possible alternative routes to resistance.
Monitoring the situation in the field
Another important experiment that the passing of two decades has made possible is assessing the effect of long-term drug withdrawal in a given area. The recent discovery of the key K76T codon change in pfcrt has enabled stored samples to be retrospectively monitored for the frequency of this change. It was found that, after cessation of chloroquine use in Malawi in 1993, prevalence of the resistant genotype decreased from 85% in 1992 to 13% in 2000 [38], with a comparable reduction seen in China in 2001 following withdrawal in 1979 [39]. A similar reversion to wild type has been observed in the dhfr gene following the introduction of bednets and, thus, reduced usage of pyrimethamine–sulfadoxine in an area of Tanzania [40]. This re-emergence of sensitivity to the older drugs offers another encouraging strategy for deploying current antimalarials more effectively and, thus, restraining costs considerably.
The extensive studies of gene polymorphisms associated with drug resistance have led to a battery of PCR-based tests for parasite resistance to chloroquine, the antifolates and other antimalarials [41-43]. However, typing of molecular markers gives only a likely, rather than certain, outcome of a treatment regime because the latter is also strongly dependent on the level of host immunity and other factors such as patient compliance and nutritional status. Nevertheless, the availability of such markers to monitor resistant parasite populations provides a powerful tool to rationalize or change the deployment of a given drug regime [17,42]. Analysis of variable sequences flanking the coding regions of key genes also provides considerable insight into the evolution and spread of drug-resistant strains [44,45]. However, the process of converting this knowledge into modified, licensed drugs that can overcome resistant forms will be a much longer-term process but it should provide an important complementary approach to the eventual deployment of entirely new classes of drugs that are targeted, for example, to recently discovered metabolic pathways in the apicoplast [46] or the mitochondrion [47]. Nor should it be forgotten that, in many situations, particularly in Africa, financial circumstances can override scientific considerations to a large degree, adversely affecting the likelihood that successful drug combinations will be efficiently deployed [48]. There is little doubt that malaria drug resistance will continue to be a topic of major importance during the next 21 years and that Trends in Parasitology will have plenty more to report as those years pass by.
Acknowledgements
The restricted number of references permitted for this article has meant citing reviews to a large extent, rather than original articles. To the authors of the latter, I proffer apologies and seek understanding.
References
- 1.Ambroise-Thomas P, Rossignol JF. Chemotherapy of resistant falciparum malaria. Parasitol. Today. 1986;2:79–80. doi: 10.1016/0169-4758(86)90163-8. [DOI] [PubMed] [Google Scholar]
- 2.Wongsrichanalai C, et al. Epidemiology of drug-resistant malaria. Lancet Infect. Dis. 2002;2:209–218. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 3.Hyde JE. Mechanisms of resistance of Plasmodium falciparum to antimalarial drugs. Microbes Infect. 2002;4:165–174. doi: 10.1016/s1286-4579(01)01524-6. [DOI] [PubMed] [Google Scholar]
- 4.Crabb BS. Transfection technology and the study of drug resistance in the malaria parasite Plasmodium falciparum. Drug Resist. Updat. 2002;5:126–130. doi: 10.1016/s1368-7646(02)00085-7. [DOI] [PubMed] [Google Scholar]
- 5.Gregson A, Plowe CV. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol. Rev. 2005;57:117–145. doi: 10.1124/pr.57.1.4. [DOI] [PubMed] [Google Scholar]
- 6.Carlton JMR, et al. Conservation of a novel vacuolar transporter in Plasmodium species and its central role in chloroquine resistance of P. falciparum. Curr. Opin. Microbiol. 2001;4:415–420. doi: 10.1016/s1369-5274(00)00228-9. [DOI] [PubMed] [Google Scholar]
- 7.Bray PG, et al. Defining the role of PfCRT in Plasmodium falciparum chloroquine resistance. Mol. Microbiol. 2005;56:323–333. doi: 10.1111/j.1365-2958.2005.04556.x. [DOI] [PubMed] [Google Scholar]
- 8.Su X, et al. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- 9.Fidock DA, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wootton JC, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 11.Djimde A, et al. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 12.Durrand V, et al. Variations in the sequence and expression of the Plasmodium falciparum chloroquine resistance transporter (Pfcrt) and their relationship to chloroquine resistance in vitro. Mol. Biochem. Parasitol. 2004;136:273–285. doi: 10.1016/j.molbiopara.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Johnson DJ, et al. Evidence for a central role for PfCRT in conferring Plasmodium falciparum resistance to diverse antimalarial agents. Mol. Cell. 2004;15:867–877. doi: 10.1016/j.molcel.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran CV, Saier MH. The principal chloroquine resistance protein of Plasmodium falciparum is a member of the drug/metabolite transporter superfamily. Microbiology. 2004;150:1–3. doi: 10.1099/mic.0.26818-0. [DOI] [PubMed] [Google Scholar]
- 15.Martin RE, Kirk K. The malaria parasite's chloroquine resistance transporter is a member of the drug/metabolite transporter superfamily. Mol. Biol. Evol. 2004;21:1938–1949. doi: 10.1093/molbev/msh205. [DOI] [PubMed] [Google Scholar]
- 16.Lakshmanan V, et al. A critical role for PfCRT K76T in Plasmodium falciparum verapamil-reversible chloroquine resistance. EMBO J. 2005;24:2294–2305. doi: 10.1038/sj.emboj.7600681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wernsdorfer WH, Noedl H. Molecular markers for drug resistance in malaria: use in treatment, diagnosis and epidemiology. Curr. Opin. Infect. Dis. 2003;16:553–558. doi: 10.1097/00001432-200312000-00007. [DOI] [PubMed] [Google Scholar]
- 18.Duraisingh MT, Cowman AF. Contribution of the pfmdr1 gene to antimalarial drug resistance. Acta Trop. 2005;94:181–190. doi: 10.1016/j.actatropica.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Reed MB, et al. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 20.Price RN, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mu J, et al. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol. Microbiol. 2003;49:977–989. doi: 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- 22.Alloueche A, et al. Comparison of chlorproguanil–dapsone with sulfadoxine–pyrimethamine for the treatment of uncomplicated falciparum malaria in young African children: double-blind randomised controlled trial. Lancet. 2004;363:1843–1848. doi: 10.1016/S0140-6736(04)16350-2. [DOI] [PubMed] [Google Scholar]
- 23.White NJ, Pongtavornpinyo W. The de novo selection of drug-resistant malaria parasites. Proc. R. Soc. Lond. Ser. B. Biol. Sci. 2003;270:545–554. doi: 10.1098/rspb.2002.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meshnick SR. Artemisinin: mechanisms of action, resistance and toxicity. Int. J. Parasitol. 2002;32:1655–1660. doi: 10.1016/s0020-7519(02)00194-7. [DOI] [PubMed] [Google Scholar]
- 25.Eckstein-Ludwig U, et al. Artemisinins target the SERCA of Plasmodium falciparum. Nature. 2003;424:957–961. doi: 10.1038/nature01813. [DOI] [PubMed] [Google Scholar]
- 26.Krishna S, et al. Artemisinins: mechanisms of action and potential for resistance. Drug Resist. Updat. 2004;7:233–244. doi: 10.1016/j.drup.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Uhlemann AC, et al. A single amino acid residue can determine the sensitivity of SERCAs to artemisinins. Nat. Struct. Mol. Biol. 2005;12:628–629. doi: 10.1038/nsmb947. [DOI] [PubMed] [Google Scholar]
- 28.Olliaro PL, Taylor WRJ. Antimalarial compounds: from bench to bedside. J. Exp. Biol. 2003;206:3753–3759. doi: 10.1242/jeb.00653. [DOI] [PubMed] [Google Scholar]
- 29.Davis TME, et al. Piperaquine – a resurgent antimalarial drug. Drugs. 2005;65:75–87. doi: 10.2165/00003495-200565010-00004. [DOI] [PubMed] [Google Scholar]
- 30.White NJ. Antimalarial drug resistance. J. Clin. Invest. 2004;113:1084–1092. doi: 10.1172/JCI21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srivastava IK, et al. Resistance mutations reveal the atovaquone-binding domain of cytochrome b in malaria parasites. Mol. Microbiol. 1999;33:704–711. doi: 10.1046/j.1365-2958.1999.01515.x. [DOI] [PubMed] [Google Scholar]
- 32.Looareesuwan S, et al. Clinical studies of atovaquone, alone or in combination with other antimalarial drugs, for treatment of acute uncomplicated malaria in Thailand. Am. J. Trop. Med. Hyg. 1996;54:62–66. doi: 10.4269/ajtmh.1996.54.62. [DOI] [PubMed] [Google Scholar]
- 33.Vaidya AB, Mather MW. Atovaquone resistance in malaria parasites. Drug Resist. Updat. 2000;3:283–287. doi: 10.1054/drup.2000.0157. [DOI] [PubMed] [Google Scholar]
- 34.Korsinczky M, et al. Mutations in Plasmodium falciparum cytochrome b that are associated with atovaquone resistance are located at a putative drug-binding site. Antimicrob. Agents Chemother. 2000;44:2100–2108. doi: 10.1128/aac.44.8.2100-2108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fivelman QL, et al. Malarone treatment failure and in vitro confirmation of resistance of Plasmodium falciparum isolate from Lagos, Nigeria. Malar. J. 2002;1:1. doi: 10.1186/1475-2875-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wichmann O, et al. Screening for mutations related to atovaquone/proguanil resistance in treatment failures and other imported isolates of Plasmodium falciparum in Europe. J. Infect. Dis. 2004;190:1541–1546. doi: 10.1086/424469. [DOI] [PubMed] [Google Scholar]
- 37.Wichmann O, et al. Malarone treatment failure not associated with previously described mutations in the cytochrome b gene. Malar. J. 2004;3:14. doi: 10.1186/1475-2875-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kublin JG, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J. Infect. Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, et al. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People's Republic of China. Am. J. Trop. Med. Hyg. 2005;72:410–414. [PubMed] [Google Scholar]
- 40.Alifrangis M, et al. Increasing prevalence of wildtypes in the dihydrofolate reductase gene of Plasmodium falciparum in an area with high levels of sulfadoxine/pyrimethamine resistance after introduction of treated bed nets. Am. J. Trop. Med. Hyg. 2003;69:238–243. [PubMed] [Google Scholar]
- 41.Sangster N, et al. Resistance to antiparasitic drugs: the role of molecular diagnosis. Int. J. Parasitol. 2002;32:637–653. doi: 10.1016/s0020-7519(01)00365-4. [DOI] [PubMed] [Google Scholar]
- 42.Plowe CV. Monitoring antimalarial drug resistance: making the most of the tools at hand. J. Exp. Biol. 2003;206:3745–3752. doi: 10.1242/jeb.00658. [DOI] [PubMed] [Google Scholar]
- 43.Alifrangis M, et al. A simple, high-throughput method to detect Plasmodium falciparum single nucleotide polymorphisms in the dihydrofolate reductase, dihydropteroate synthase, and P. falciparum chloroquine resistance transporter genes using polymerase chain reaction and enzyme-linked immunosorbent assay-based technology. Am. J. Trop. Med. Hyg. 2005;72:155–162. [PubMed] [Google Scholar]
- 44.Cortese JF, et al. Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J. Infect. Dis. 2002;186:999–1006. doi: 10.1086/342946. [DOI] [PubMed] [Google Scholar]
- 45.Roper C, et al. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet. 2003;361:1174–1181. doi: 10.1016/S0140-6736(03)12951-0. [DOI] [PubMed] [Google Scholar]
- 46.Ralph SA, et al. Metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- 47.Biagini GA, et al. Antimalarial chemotherapy: young guns or back to the future? Trends Parasitol. 2003;19:479–487. doi: 10.1016/j.pt.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 48.Winstanley PA, et al. Clinical status and implications of antimalarial drug resistance. Microbes Infect. 2002;4:157–164. doi: 10.1016/s1286-4579(01)01523-4. [DOI] [PubMed] [Google Scholar]