Abstract
Pharmacogenetics provides great opportunity for improving both the chance of therapeutic benefit and the ability to avoid adverse drug events. To date, the majority of pharmacogenetic studies have been performed using germline DNA. DNA collection in most clinical trials provides a wealth of samples from which pharmacogenetic studies can be launched. However, there is concern that the data from germline DNA pharmacogenetics might be of limited value for diseases, such as cancer, where germline variants may not adequately represent the genetic data obtained from the somatic DNA. In this perspective, we evaluate the literature that compares pharmacogenetic variants between germline DNA and matched somatic DNA. The analysis of these studies indicates that there is almost complete concordance between germline and somatic DNA in variants of pharmacogenetic genes. Although somatic variants are clinically significant and independently provide genetic information that cannot be gained from the germline, the use of germline DNA for pharmacogenetic studies is achievable and valuable. This use of germline DNA offers great opportunities for the implementation of individualized therapy.
Keywords: germline, pharmacogenetics, somatic
Pharmacogenetics provides great opportunity for improving both the chance of therapeutic benefit and the ability to avoid adverse drug events. To date, the majority of pharmacogenetic studies have been performed using germline DNA extracted from a patient’s blood or buccal specimen. This is largely due to the ease of collection of these specimens and the existing DNA extraction infrastructure preference for these matrices. At this point, many clinical trials, regardless of their disease focus or end point, request germline DNA collection (typically from blood or buccal specimens) upon study initiation. This provides a wealth of samples from which pharmacogenetic studies can be launched. However, there is concern that the data from germline DNA pharmacogenetics might be of limited value for diseases, such as cancer, where germline, or inherited, DNA variants may not adequately represent the genetic data obtained from the somatic, or acquired, DNA from the tumor tissue. The concern goes both ways, as often the only available patient tissue is formalin-fixed paraffin-embedded (FFPE) tumor tissue, raising the issue of suitability for toxicity predictor in germline tissues. Thankfully, there is data to address this important element of translational genomics. In this perspective, we summarize the literature that compares pharmacogenetic variants between germline DNA and matched somatic DNA. In addition, we address the predictive value of somatic DNA variants. Our analysis of the comparison studies indicates that there is almost complete concordance between variants in the pharmacogenetic genes examined in germline and somatic DNA (Figure 1). We conclude that while both germ-line and somatic DNA are of great importance in cancer, germline DNA provides a reliable and easily available resource and should continue to be the primary genetic source for pharmacogenetic studies.
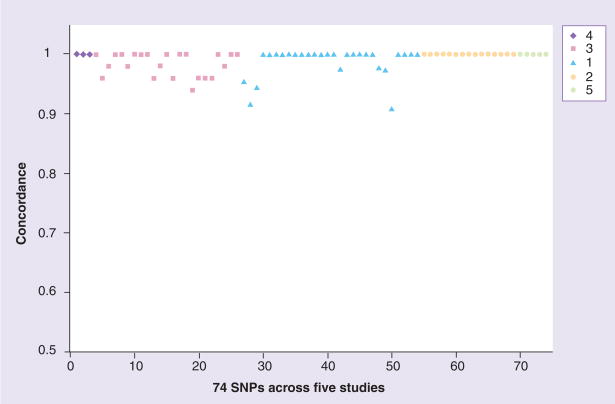
Figure 1. Concordance for 74 SNPs across five studies.
The legend indicates number of reference. The X-axis shows each individual SNP for each study, while the Y-axis describes the concordance between the two tissues compared in each specific study. The concordance value can be between 0 (no concordance) and 1 (complete concordance), but clearly cluster between 0.9 and 1.0 in the studies examined.
The studies reviewed below assess the concordance between variants from germline and tumor DNA as well as germline DNA from blood compared with germline DNA from FFPE normal adjacent tissue in not only genes with previously established roles in pharmacogenetics [1–3], but also genes shown to be involved in other physiological functions, including angiogenesis and breast cancer pathogenesis [4,5]. It has been shown that SNPs in an individual’s germline play a role in the development of cancers. Therefore, establishing the concordance between germline and somatic DNA variants may have value for cancer biology, variability in drug response, and possibly in development of cancers and metastasis [6]. Table 1 summarizes the findings of the discussed manuscripts, describes the tissues being compared, and most importantly the concordance between the germline and tumor genotypes. The studies are grouped into two categories based on the type of preparation used for the tissues genotyped, including fresh/frozen tissue or paraffin-embedded tissue.
Table 1.
Comparison of germline and somatic genotype.
| Tissue type | Tissue preparation | No. patients genotyped | No. SNPs genotyped | Range of concordance | Average concordance | Ref. |
|---|---|---|---|---|---|---|
| Colorectal cancer versus paired normal mucosa | Fresh/frozen | 41 | 28 | 0.91–1.0 | 0.987 | [1] |
| Tumor cell lines versus paraffin-embedded cells | Paraffin-embedded | 8 | 4 | 1.0–1.0 | – | [2] |
| AML bone marrow versus germline buccal DNA | Fresh/frozen | 77 | 23 | 0.94–1.0 | 0.968 | [3] |
| Breast cancer versus lymph node | Paraffin-embedded | 21 | 3 | 1.0–1.0 | 1.0 | [4] |
| Blood DNA versus breast cancer adjacent normal | Paraffin-embedded | 106 | 5 | 1.0–1.0 | 1.0 | [5] |
The table includes the reference number, type of tissue and method of preparation, number of samples genotyped, and the range and average concordance for each study. AML: Acute myeloid leukemia.
Genotype comparison using fresh/frozen tumor & germline tissue samples
Prospective correlative science studies most often obtain fresh/frozen tissue for analysis. The sources from which DNA is extracted are typically blood samples for germline DNA and surgical resection or bone marrow biopsy samples for somatic/tumor DNA. Weiss et al. examined common pharmacogenetic gene polymorphisms in acute myeloid leukemia [3]. Because leukemia has frequent cytogenetic abnormalities, discordance between host and tumor DNA genotypes may be relevant for pharmacogenetic studies. The authors genotyped 21 SNPs and two gene deletions in 17 genes encoding proteins involved in drug metabolism, protection from oxidative stress, drug transport and DNA repair. The authors sought to examine the concordance between genotypes from paired pretreatment bone marrow samples (somatic/tumor) and nondiseased tissue from buccal cell samples (germline) from 80 adult acute myeloid leukemia patients. The results of these genetic comparisons indicated that 90% of the samples had complete concordance for all SNPs examined, while that value increased to 97% of individuals having one or less discrepant SNP (<4%, see Table 1) between the buccal and bone marrow DNA [3].
Marsh et al. performed a similar comparison in colorectal cancer [1]. The colorectal tumor and normal colon (germline) tissue were immediately frozen following surgical resection. This was followed by extraction of the genetic material and genotyping for 28 SNPs and one tandem repeat. The results of this genotype comparison showed that 41 out of 44 (93%) patient samples had one or less discordant genotypes between the germline and tumor DNA. Overall, there were 13 discordant genotypes of the 1139 comparisons (1.1%). In addition, because microsatellite instability is a hallmark of colorectal cancer, the authors sought to examine whether microsatellite-instability status corresponded with discordance. They found that although eight cases (20%) had microsatellite instability, this was not statistically significantly associated with genotype discordance (p = 0.67) [1]. Together, the studies of Weiss et al. and Marsh and colleagues establish that germline DNA genotype is highly conserved in DNA from fresh/frozen tumor tissue.
Genotype comparison using paraffin-embedded tumor samples & germline samples
For many large clinical trials the only available tissue for any correlative studies is FFPE tissue from either biopsy or resection. Schneider et al. addressed the issue of germline versus somatic DNA genotype by examining three polymorphisms, two in eNOS and one in VEGF. Because of the therapeutic and prognostic role of angiogenesis in breast cancer, 53 breast cancer tissue specimens from 21 patients were collected for this study. These included: 17 primary breast cancer, 17 involved lymph node and 19 uninvolved lymph node specimens. DNA samples were extracted from paraffin-embedded tissue. The authors analyzed well-established polymorphisms with high frequencies and potential functional consequence, including a the −786T>C polymorphism in the promoter and 894T>G (Glu298Asp) SNP in exon 7 of eNOS, and the 936C>T SNP in VEGF. The 21 different individuals with primary tumor and paired lymph node had 100% concordance between primary tumors and either involved or uninvolved lymph nodes. In addition to this finding, because the authors used concordance, it can be seen regardless of the part of tumor region that the DNA was extracted from in the paraffin block. This is an additional level of confidence in the concordance between germline and somatic DNA [4].
Rae et al. performed a similar study to examine the feasibility and accuracy of pharmacogenetic genotyping in a number of sample preparation techniques. They compared ten DNA samples isolated from whole blood with DNA isolated from the matched paraffin-embedded tumor sample. When genotyping a CYP2C8 SNP, two CYP2D6 SNPs and an ABCB1 SNP, they found 100% concordance between germline and somatic genotypes [2]. Therefore, the authors concluded, using a multi-tiered approach, that by using a high-yield DNA extraction method, one can reliably obtain concordant genotypes between these paraffin-embedded cells and their matched blood DNA sample.
Xie et al. also performed genotyping on paraffin-embedded normal tissue adjacent to breast cancer compared with germline blood DNA. Often, the adjacent tissue will exhibit loss of heterozygosity (LOH) in the stromal tissue surrounding the tumor [7]. It has been proposed that this LOH could alter the genotyping results if the ‘normal’ tissue was extracted in the field surrounding the tumor. SNPs in five genes that can be altered in breast cancer were evaluated; MTHFR, hOGG1, DBH, DRD2 and NQO1. For each of the five genes, between 95–99% of the 106 blood/tissue pairs produced genotyping results. One SNP in each gene was genotyped. In all five genes, 100% of the sample pairs that were genotyped produced concordant genotypes [5]. Although these results may reflect that the cells being genotyped were primarily stromal cells that do not harbor LOH, the numbers are convincing that the paraffin-embedded adjacent tissue can be used as ‘normal’ for genetic analysis. For this particular study, further genotyping on pharmacogenetic specific genes would need to be performed to assess whether this conclusion could be extrapolated from the breast cancer genes to pharmacogenetics as well.
The above described studies cover key issues for the use of a tumor sample, including DNA extraction methods, paraffin-embedding and tissue staining. They compare the genotypes as compared with their germline (either blood or buccal originated) counterparts. The studies encompass colorectal cancer, breast cancer, acute myeloid leukemia and general tumors across pharmacogenetic and other cancer genes. The results consistently show that concordance between germline and tumor DNA genotypes is virtually 100% (Figure 1). Additionally, there is almost 100% concordance between fresh tumor cells and paraffin-embedded cells. Further, Xie et al. concluded that paraffin-embedded normal adjacent tissue to a breast cancer tumor is 100% concordant with germline DNA from blood [5]. A number of preliminary conclusions can be drawn. Paraffin-embedded adjacent normal tissue can be used for normal tissue genotyping. This fixation method has only been applied to pharmacogenetic studies since 2003 and represents a source of genetic material that is accurate and useful [2,8]. In the studies reviewed above, paraffin-embedded tissue or cells can be genotyped with the same confidence as fresh tissue or cells, at least using the methods applied in the reviewed studies. Also, germline DNA from either blood or buccal cells can be genotyped for pharmacogenetic genes and the concordance with tumor DNA is virtually 100%.
There is predictive value in both germline DNA variants as well as somatic DNA variants. Somatic variants in many cancer-related genes are clinically significant and the effects of these mutations have implications on therapy. Many therapeutic agents have been designed to target the somatic change through the disruption of essential signaling pathways. Because of their role in tumorigenesis, kinases are the initial targets of many clinically-approved oncology drugs [9]. Somatic mutations in the kinase domain of EGFR are seen in the tumor tissue of non-small-cell lung cancer patients and are associated with increased response to gefitinib [10,11]. These mutations are present in the tumor prior to therapy. Imatinib targets the breakpoint of BCR-ABL translocation in the treatment of chronic myeloid leukemia [12]. Interestingly, the patient will typically become resistant to imatinib, most commonly due to BCR-ABL acquiring a somatic point mutation [13]. These somatic variants in chronic myeloid leukemia patients resistant to imatinib provide powerful information that will affect the course of the treatment. Somatic mutations in KIT have been reported in mast cell diseases, gastrointestinal stromal tumors, and familial testicular germ-cell tumors [14]. Because imatinib also has specificity for KIT, clinical trials have shown an increase in response [15].
Loss-of-heterozygosity is commonly seen in cancers. LOH occurs when an individual has inherited a gene that has one allele inactivated and subsequently the second allele becomes inactivated by a mutation in the tumor, resulting in loss-of-function. Tumor-suppressor genes often become inactivated in oncology, resulting in tumorigenesis. In the presence of LOH, germline DNA would have a higher frequency of heterozygotes than the corresponding somatic DNA. To our knowledge, this has not presented any complications in genotyping with the exception of one study. In a study examining the tandem repeat sequence in the pharmacogenetic gene TYMS, Marsh et al. noted that in one out of 45 colorectal cancer patients the genotype was not identical between paired samples of colorectal tumor and normal tissue [16].
In conclusion, the use of germline DNA for pharmacogenetic studies is achievable and valuable. There is almost complete concordance between variants in pharmacogenetic genes between germline and somatic DNA as described above. Germline DNA is easily obtained and more readily available than somatic DNA. Somatic variants also provide significant predictive value. In some cases, somatic variants can be specifically targeted resulting in increased response. However, in pharmacogenetic studies, the availability and accuracy of germline DNA offers great opportunities for the implementation of individualized therapy in a timely fashion.
Future perspective
Pharmacogenetics is at an early point in its evolution – currently there are only a few genotypes that have shown a significant clinical impact on the individual patient. As the cost of whole-genome sequencing decreases over the next few years, targeted therapy for cancer will become more achievable. However, the amount of genetic information available for a given patient will far exceed current ability to translate it into better care.
Many current clinical trials and likely all future clinical trials will require the archiving of many sources of the patient’s molecular information. This will include germline DNA (from blood or buccal cells) or somatic DNA from the tissue specimens. To date, there have been only five publications directly addressing the issue of concordance between germline and somatic DNA in pharmacogenetics [1–5]. Because of the current limitation of available tissue in practice-changing clinical trials, the near future will bring an even greater need for understanding how DNA from fresh/frozen somatic/tumor DNA, germline DNA from blood/buccal cells, FFPE somatic/tumor or FFPE germline DNA from adjacent normal tissue can be used to better select populations of patients with the greatest risk–benefit relationship. These large, comprehensive studies will provide the concrete evidence that many researchers will require before relying on the use of only germline DNA for cancer studies. It is possible that the increased availability of both tumor and germline DNA from a patient will eliminate the need to rely exclusively on one or the other. However, in order to fully understand the disease and treat the patient in the most comprehensive manner, both the germline and somatic variants will require precise examination. Each has unique predictive value for different reasons. This fact will be consistent throughout time.
Executive summary
Concordance between matched germline (blood/buccal) and somatic (tumor-fresh/frozen) DNA is 93–100% for pharmacogenetic genotypes.
Concordance between matched formalin-fixed paraffin embedded (FFPE) germline versus FFPE somatic DNA and between matched FFPE adjacent tissue versus blood is 100% for pharmacogenetic genotypes.
FFPE tissue provides an accurate source of DNA for pharmacogenetic studies.
Germline DNA is sufficient for genetic analysis of pharmacogenetic genes.
Somatic DNA provides unique information that can have significant implications when the therapy has a molecular target in the tumor.
Acknowledgments
Financial & competing interests disclosure
The authors are supported in part by NIH grants U01 GM63340, P50 CA106991 and P30 CA016086. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Marsh S, Mallon MA, Goodfellow P, McLeod HL. Concordance of pharmacogenetic markers in germline and colorectal tumor DNA. Pharmacogenomics. 2005;6(8):873–877. doi: 10.2217/14622416.6.8.873. [DOI] [PubMed] [Google Scholar]
- 2▪.Rae JM, Cordero KE, Scheys JO, Lippman ME, Flockhart DA, Johnson MD. Genotyping for polymorphic drug metabolizing enzymes from paraffin-embedded and immunohistochemically stained tumor samples. Pharmacogenetics. 2003;13(8):501–507. doi: 10.1097/00008571-200308000-00008. Study was one of the first to use formalin-fixed paraffin-embedded DNA for pharmacogenetic analyses. [DOI] [PubMed] [Google Scholar]
- 3.Weiss JR, Baer MR, Ambrosone CB, et al. Concordance of pharmacogenetic polymorphisms in tumor and germ line DNA in adult patients with acute myeloid leukemia. Cancer Epidemiol Biomarkers Prev. 2007;16(5):1038–1041. doi: 10.1158/1055-9965.EPI-06-0964. [DOI] [PubMed] [Google Scholar]
- 4.Schneider BP, Radovich M, Sledge GW, et al. Association of polymorphisms of angiogenesis genes with breast cancer. Breast Cancer Res Treat. 2007;111(1):157–163. doi: 10.1007/s10549-007-9755-9. [DOI] [PubMed] [Google Scholar]
- 5.Xie B, Freudenheim JL, Cummings SS, et al. Accurate genotyping from paraffin-embedded normal tissue adjacent to breast cancer. Carcinogenesis. 2006;27(2):307–310. doi: 10.1093/carcin/bgi215. [DOI] [PubMed] [Google Scholar]
- 6.Evans WE, McLeod HL. Pharmacogenomics – drug disposition, drug targets, and side effects. N Engl J Med. 2003;348(6):538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 7.Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science. 1996;274(5295):2057–2059. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- 8▪.Tayeb MT, Clark C, Haites NE, Sharp L, Murray GI, McLeod HL. CYP3A4 and VDR gene polymorphisms and the risk of prostate cancer in men with benign prostate hyperplasia. Br J Cancer. 2003;88(6):928–932. doi: 10.1038/sj.bjc.6600825. Study was one of the first to use formalin-fixed paraffin-embedded DNA for pharmacogenetic analyses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebolt-Leopold JS, English JM. Mechanisms of drug inhibition of signalling molecules. Nature. 2006;441(7092):457–462. doi: 10.1038/nature04874. [DOI] [PubMed] [Google Scholar]
- 10.Irmer D, Funk JO, Blaukat A. EGFR kinase domain mutations – functional impact and relevance for lung cancer therapy. Oncogene. 2007;26(39):5693–5701. doi: 10.1038/sj.onc.1210383. [DOI] [PubMed] [Google Scholar]
- 11.Moutinho C, Mateus AR, Milanezi F, Carneiro F, Seruca R, Suriano G. Epidermal growth factor receptor structural alterations in gastric cancer. BMC Cancer. 2008;8:10. doi: 10.1186/1471-2407-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12▪.Ikediobi ON. Somatic pharmacogenomics in cancer. Pharmacogenomics J. 2008;8(5):305–314. doi: 10.1038/tpj.2008.8. Provides a current review of the role of somatic variants in cancer therapy. [DOI] [PubMed] [Google Scholar]
- 13▪.Gorre ME, Mohammed M, Ellwood K, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293(5531):876–880. doi: 10.1126/science.1062538. Study is the first to establish the role of BCR-ABL in drug resistance. [DOI] [PubMed] [Google Scholar]
- 14.Rapley EA, Hockley S, Warren W, et al. Somatic mutations of KIT in familial testicular germ cell tumours. Br J Cancer. 2004;90(12):2397–2401. doi: 10.1038/sj.bjc.6601880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demetri GD. Identification and treatment of chemoresistant inoperable or metastatic GIST: experience with the selective tyrosine kinase inhibitor imatinib mesylate (STI571) Eur J Cancer. 2002;38(Suppl 5):S52–S59. doi: 10.1016/s0959-8049(02)80603-7. [DOI] [PubMed] [Google Scholar]
- 16.Marsh S, McKay JA, Cassidy J, McLeod HL. Polymorphism in the thymidylate synthase promoter enhancer region in colorectal cancer. Int J Oncol. 2001;19(2):383–386. doi: 10.3892/ijo.19.2.383. [DOI] [PubMed] [Google Scholar]