Abstract
Drosophila MSL complex binds the single male X chromosome to upregulate gene expression to equal that from the two female X chromosomes. However, it has been puzzling that ~25% of transcribed genes on the X do not stably recruit MSL complex. Here, we find that almost all active genes on the X are associated with robust H4 Lys16 acetylation (H4K16ac), the histone modification catalyzed by MSL complex. The distribution of H4K16ac is much broader than that of MSL complex, and our results favor the idea that chromosome-wide H4K16ac reflects transient association of MSL complex, occurring through spreading or chromosomal looping. Our results parallel those of localized Polycomb repressive complex and its more broadly distributed H3K27me3 chromatin mark, suggesting a common principle for the establishment of active and silenced chromatin domains.
Keywords: dosage compensation, histone acetylation, MOF
Eukaryotic gene expression is influenced by chromatin environment. The alteration of local and higher-order chromatin structure modulates the accessibility of DNA to the nuclear complexes that carry out processes such as transcription, DNA replication and DNA repair. A major class of chromatin regulators catalyzes the post-translational modification of histone proteins. Although their influence on gene expression through targeting to promoters is clearly established, we are only beginning to understand how histone modifications impact the formation and structure of chromatin domains1.
Dosage compensation, an essential process that equalizes the level of X-linked gene expression between males and females, is a model for studying chromosome-wide gene regulation. In Drosophila, dosage compensation is mediated by the MSL (male-specific lethal) complex, composed of at least five MSL proteins (MSL1, MSL2, MSL3, MOF and MLE) and two non-coding roX (RNA on X) RNAs2. MSL complex specifically recognizes hundreds of sites along the male X chromosome and upregulates transcription of X-linked genes approximately two-fold.
High resolution mapping studies support a two-step model for MSL targeting. In the first step, MSL complex distinguishes the X from autosomes by sequence-dependent recognition of a set of high affinity sites on the X, known as chromatin entry sites (CES)3,4. Subsequently, MSL complex spreads to the middle and 3′ ends of active genes on the X chromosome5,6. Whereas MSL1 and MSL2 are essential for the MSL complex to recognize any sites on the X chromosome, MSL3, MOF and MLE are required specifically for MSL spreading from CES7–9. Spreading involves the recognition of general features of active genes, including the transcription-coupled H3K36me3 mark, and can occur in the absence of X-specific sequence elements10–13. Although many transcribed genes clearly recruit MSL complex, it is difficult to define the total number of binding sites along chromatin. In three global analyses, a subset of transcribed genes did not stably recruit MSL complex5,6,14. One study estimated that ~25% of genes on the X chromosome bound by RNA polymerase II are not bound by MSL complex in embryos6. In our previous studies, we found that 73% of transcribed genes and 83% of H3K36me3-associated genes are bound by MSL complex, with an intermediate class that could not be unambiguously defined as bound or unbound5,11. Thus, if MSL complex is recruited to transcribed genes by recognizing features of active transcription, a key outstanding question is why ~25% of these genes do not appear to recruit MSL complex. This is particularly important to understand in light of recent evidence that nearly all genes on the X chromosome undergo dosage compensation15.
The localization of MSL complex to the bodies of active genes also suggests that it regulates transcription elongation or the recycling of polymerase to the promoter for reinitiation5,6,16. Although it is unknown how MSL complex upregulates transcription, the identification of MOF suggested a chromatin-based mechanism17. MOF, also known as KAT8, is a MYST family histone acetyltransferase (HAT) that acetylates histone H4 at Lys16 (refs. 17–20), a modification enriched on the male X chromosome21,22. The catalytic activity of MOF is required for MSL spreading from CES and for the preferential enrichment of H4K16ac along the polytene male X chromosome9.
Like many histone acetylation marks, H4K16ac stimulates transcription in vitro and in vivo19,23,24. However, H4 Lys16 is distinct from other acetylated residues on the H4 tail in that it regulates a specific subset of genes in S. cerevisiae, suggesting that the role of H4K16ac is not limited to charge neutralization of the N-terminal H4 tail redundantly with other acetylation marks25. One possibility is that H4K16ac also regulates the binding of chromatin-associated proteins. In favor of this idea, H4K16ac inhibits the binding of Sir3 and blocks the spread of heterochromatin in S. cerevisiae26,27. In addition, H4K16ac antagonizes the activity of the ISWI family of ATP-dependent chromatin remodelers in vitro and in vivo28–30. Another consequence of H4K16ac is the decondensation of higher order chromatin structure. H4K16ac inhibits the compaction of chromatin fibers and interfiber interactions to a similar extent as deletion of the entire H4 tail30,31. Consistent with this observation, enrichment of H4K16ac on the Drosophila male X chromosome is associated with the diffuse morphology of this chromosome21,22.
In order to investigate the consequences of MSL activity on the male X chromosome, we sought to map the distribution of H4K16ac at high resolution. Although H4K16ac colocalizes with MSL complex at the cytological level9,22, we have recently shown that higher resolution studies may reveal mechanistically important differences in binding patterns11,13. Our findings indicate that while the pattern of H4K16ac generally resembles that of MSL complex at the molecular level, there are three key differences. First, nearly all active genes on the X chromosome show high levels of H4K16ac. Secondly, enrichment for H4K16ac is also associated with the remainder of the X chromosome, providing evidence for a global role for MSL complex on the X. In both cases, our data suggest that H4K16ac deposition is the result of transient MSL complex association. Finally, we observe H4K16ac enrichment at the 5′ ends of active genes on the X and autosomes in males and females, as reported recently32–34. Taken together, our data indicate that MOF activity is highly specific for the male X chromosome, whereas the general 5′ H4K16ac may require the redundant action of multiple HAT enzymes.
Results
H4K16ac is broadly associated with the male X
We analyzed the distribution of H4K16ac in characteristically male SL2 cells by ChIP-chip using antibodies specific for H4K16ac and NimbleGen arrays tiled at 100 bp resolution along the entire X chromosome and the left arm of chromosome 2 (2L). As expected, we observed preferential enrichment of H4K16ac over the X chromosome (Fig. 1a). Genes bound by MSL complex in ChIP-chip experiments, as defined in ref. 11, are marked by high H4K16ac ChIP signal. However, we also observed substantial enrichment of H4K16ac along the X chromosome at sites lacking MSL binding. Though the levels of ChIP signal varied, the baseline level of H4K16ac is increased on the X relative to 2L, spanning both genes and intergenic regions. One recent study also noted the global enrichment of H4K16ac on the male X chromosome34, while another did not33, conceivably due to decreased sensitivity of the ChIP assay and restriction of the analysis to the top 15% of bound probes (Supplementary Fig. 1).
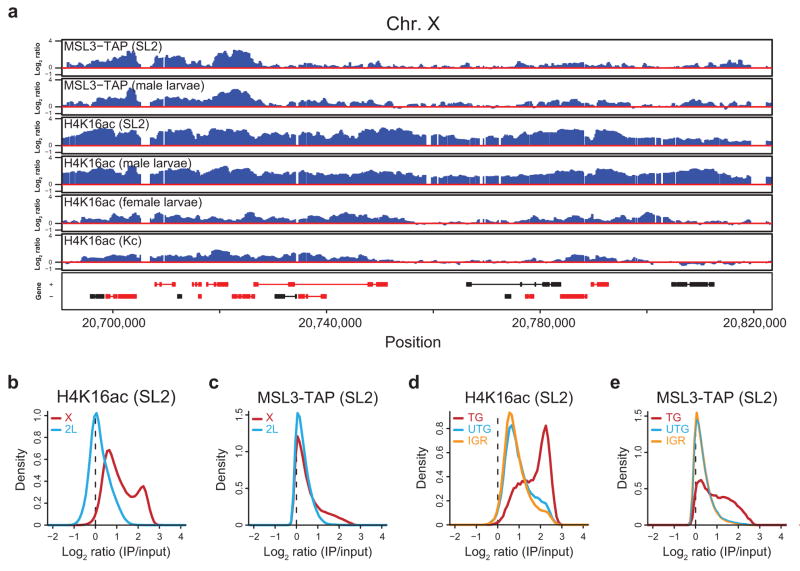
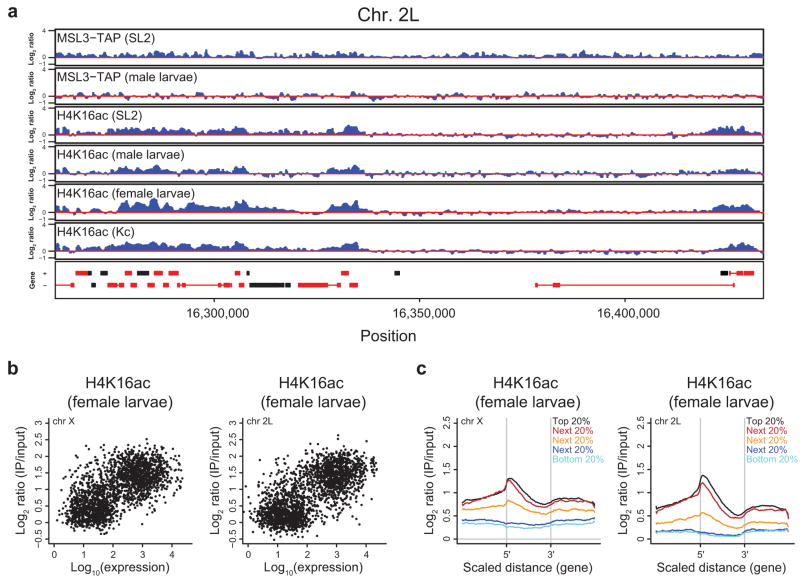
Figure 1.
H4K16ac is globally enriched along the male X. (a) The distribution of H4K16ac on the male X chromosome is broader than MSL complex. Active genes that lack stable MSL binding are associated with H4K16ac. ChIP-chip profiles for MSL3-TAP (from SL2 cells and male larvae5,11) and H4K16ac (from SL2 and Kc cells, male and female larvae), generated from NimbleGen tiling arrays, are shown for a representative region of the X chromosome. Genes are color-coded according to their transcription status (red, transcribed; black, untranscribed) as defined previously5. Genes on the top row are transcribed from left to right, and genes on the bottom row are transcribed from right to left. Numbers along the x-axis denote chromosomal position along the X in base pairs. The y-axis shows the ChIP signal expressed as the log2 ratio of IP/input. (b, c) H4K16ac is enriched along the majority of the X chromosome relative to 2L and shows a bimodal distribution on the X. In contrast, MSL3-TAP enrichment is restricted to a smaller subset of probes on the X. The distribution of log ratios at all probes on the X (red) and 2L (blue) is shown for H4K16ac (b) and MSL3-TAP (c) ChIP signal in SL2 cells. ChIP signal (x-axis) is expressed as the log2 ratio of IP DNA relative to input. (d, e) H4K16ac, like MSL3- TAP, is most highly enriched over actively transcribed genes on the X. The distribution of log ratios at all probes on the X chromosome, classified according to probes present in transcribed genes (TG, red), untranscribed genes (UTG, blue) and intergenic regions (IGR, orange), is shown for H4K16ac (d) and MSL3-TAP (e) in SL2 cells.
In order to examine the extent of H4K16ac on the X chromosome systematically, we compared its distribution over the X and 2L at the probe level. There is a clear shift in the baseline of H4K16ac on the X relative to 2L (Fig. 1b), and ~50% of the X chromosome is marked by H4K16ac (Supplementary Fig. 2a). In contrast, at the same cut-off, ~20% of probes on the X chromosome are enriched by MSL3-TAP, despite the broad pattern of MSL complex binding to the bodies of many transcribed genes on the X5,6. Furthermore, a shift in the peak of the distribution between the X and 2L was not observed for MSL3-TAP binding (Fig. 1c) and we did not detect an increased baseline of H3K4me2 and H3K36me3 on the X (ref. 11, data not shown), suggesting that this level of X chromosome enrichment is not a general feature of all histone modifications.
The distribution of H4K16ac on the X chromosome is bimodal (Fig. 1b), indicating that there are two categories of sites on the X that are enriched for H4K16ac. The X chromosome is generally characterized by an elevated baseline of H4K16ac; however, active genes show the highest H4K16ac ChIP signals, consistent with the localization of MSL complex (Fig. 1d,e, TG in red). It has been noted that ~25% of active genes on the X chromosome do not clearly recruit the complex (refs. 5,6,11,14, Supplementary Fig. 3a). Since MSL complex is not as broadly distributed as the histone modification that it deposits (Fig. 1 and Supplementary Fig. 2a), we asked whether H4K16ac generally marks active genes on the X chromosome. Indeed, almost all active genes on the X are associated with a high level of H4K16ac, above the elevated baseline of the X chromosome (Fig. 2a). Similar to MSL complex5,6, H4K16ac enrichment is biased towards the middle and 3′ ends of active genes on the X (Supplementary Fig. 3b). Genes that lack detectable MSL binding, as defined in ref. 11, also display this 3′ bias and are associated with a level of H4K16ac intermediate between that of untranscribed genes and MSL-bound genes (Supplementary Fig. 3c,d). Interestingly, a modest enrichment of MSL complex is observed at these genes, indicating a low level of MSL occupancy that is near the detection limit of the assay.
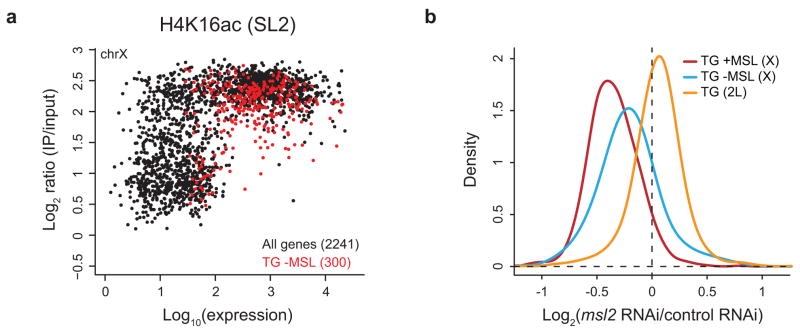
Figure 2.
High levels of H4K16ac are associated with transcribed genes on the male X. (a) Nearly all transcribed genes on the X are associated with high levels of H4K16ac. The maximum ChIP signal of H4K16ac for each gene in SL2 cells (y-axis) is plotted against its expression (in log scale) determined from Affymetrix microarrays (x-axis)5. The 2241 genes on the X chromosome are in black. The 300 genes on the X that are transcribed, but lack detectable MSL binding (TG -MSL) as defined in ref. 11, are overlaid in red. While all genes on the X are associated with a high baseline of H4K16ac characteristic of the male X chromosome, nearly all active genes are associated with even higher levels of H4K16ac. (b) MSL complex regulates the expression of transcribed genes on the X chromosome, including those that lack stable binding of the complex. Relative changes in gene expression following msl2 RNAi measured in ref. 36 are plotted for transcribed genes: (1) on the X chromosome that are bound by MSL3-TAP (834 genes, red); (2) on the X chromosome that lack detectable MSL binding (300 genes, blue); and (3) on 2L (1050 genes, orange), as classified in ref. 11.
These findings suggest that these genes might be previously unidentified MSL targets that transiently recruit the complex. In this case, we would expect the loss of MSL complex to affect their dosage compensation. Depletion of msl2 by RNA interference (RNAi) results in the decreased expression of known MSL target genes on the X chromosome (Fig. 2b, red) relative to genes on 2L (Fig. 2b, orange). The expression of genes on the X that do not stably bind the complex is also reduced (Fig. 2b, blue) although this effect is not as strong as for MSL-bound genes. Taken together, these results suggest that recruitment of MSL activity may be a general property of transcribed genes on the X chromosome. Lower levels of H4K16ac along the X chromosome suggest that MSL complex may function globally along the X, thereby utilizing a chromosome-wide mechanism to fine-tune transcription.
MSL complex mediates global acetylation of the male X
We next tested whether MSL complex is responsible for H4K16ac at sites on the X lacking detectable MSL ChIP signal. We performed RNAi in SL2 cells targeting the core MSL2 subunit and the MOF HAT. msl2 RNAi resulted in ~60–70% depletion of MSL2 and mof RNAi reduced MOF levels over 90%, compared to a non-specific GFP RNAi control, (Supplementary Fig. 4). Despite efficient knock-down of MSL2 and MOF, no reduction in cell viability was observed following msl2 and mof RNAi, consistent with previous observations (refs. 35,36, data not shown).
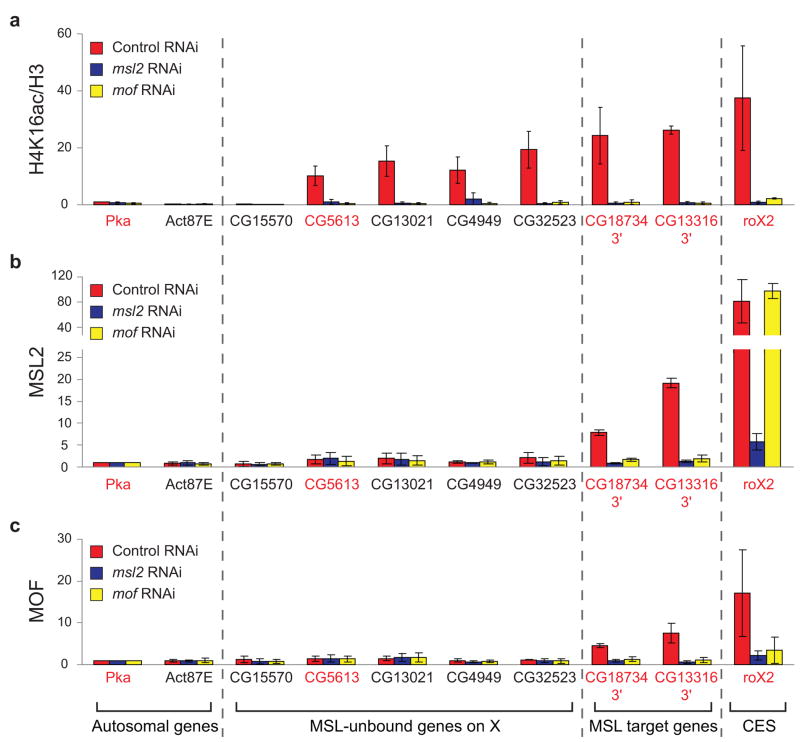
MOF, MSL2 and H4K16ac ChIP were performed in RNAi-treated SL2 cells, and samples were analyzed by quantitative PCR (qPCR). From the ChIP-chip data sets, we selected four genes on the X chromosome that are highly enriched by H4K16ac, but are clearly unbound by MSL complex (CG5613, CG13021, CG4949, and CG32523). Consistent with the microarray data, these loci were significantly enriched by H4K16ac but not by MSL2 or MOF in the control (Fig. 3a–c, red bars). For comparison, known MSL target genes show significant levels of MSL2, MOF and H4K16ac.
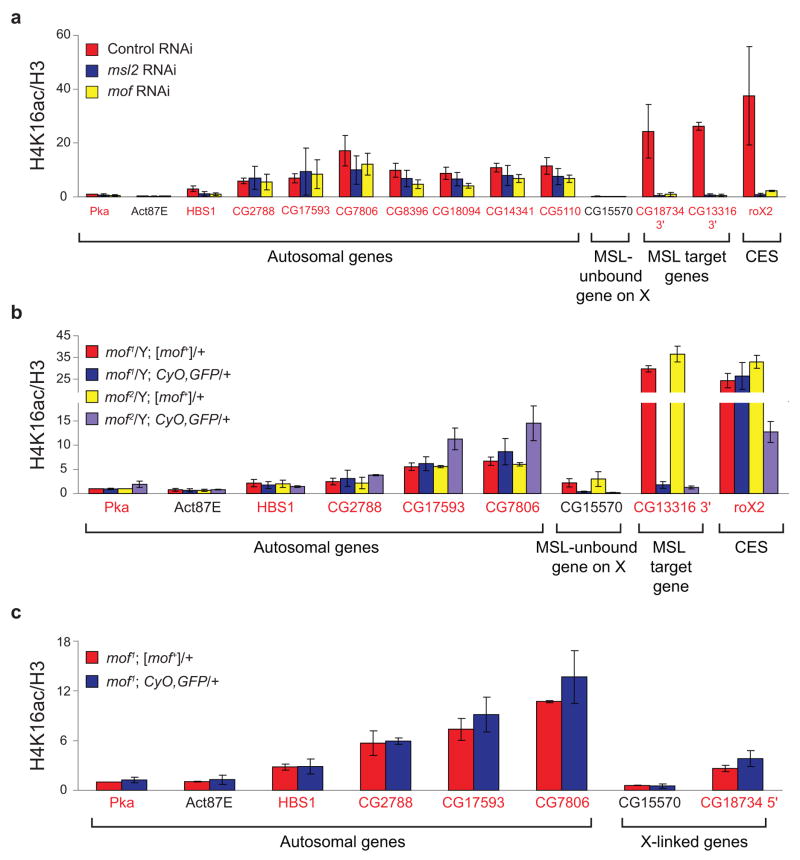
Figure 3.
MSL complex is required for broad H4K16ac on the male X. MSL2 and MOF are required for H4K16ac at sites on the X that lack detectable MSL binding. ChIP was performed following RNAi in SL2 cells for GFP as a negative control (red), msl2 (blue) or mof (yellow). Known MSL targets (3′ ends of MSL-bound genes and the roX2 CES (chromatin entry site)) and autosomal genes served as positive and negative controls, respectively. Five genes on the X that clearly lack detectable MSL binding11 were assayed. With the exception of CG15570, these genes were enriched by H4K16ac in SL2 ChIP-chip experiments, and primers were designed within the region of maximum ChIP signal. Genes were classified as transcribed (gene names in red) or untranscribed (gene names in black) based on Affymetrix analysis in SL2 cells5. H4K16ac ChIP signal (a) was quantified as percent IP normalized to input and H3 levels. The ChIP signals were normalized to the ChIP signal of the autosomal gene, Pka, in the control RNAi sample, and the replicates were averaged (see Supplementary Methods). Average MSL2 (b) and MOF (c) ChIP signals are presented as percent IP normalized to input and Pka levels. Error bars are defined as the standard deviation of three replicate experiments.
MSL2 is required for MSL complex to associate with the X chromosome by cytology7. Similarly, depletion of msl2 results in the loss of MSL2, MOF and H4K16ac at known targets to background levels (Fig. 3a–c, blue bars). In the absence of MOF, a core MSL complex is localized to CES, but cannot spread beyond these sites9. As with msl2, depletion of mof reduces MSL2, MOF and H4K16ac ChIP at target genes (Fig. 3a–c, yellow bars). Although MSL2 remains at CES as predicted by cytology, H4K16ac is reduced ~17-fold at roX2 following mof depletion. Importantly, MSL2 and MOF are both required for H4K16ac at sites on the X that are not stably associated with MSL complex, demonstrating that MSL activity on the X is not restricted to the active genes where it is detectably bound. Instead, global H4K16ac on the X might reflect transient action of MSL complex. Although stable binding of MSL complex is not required, we propose that H4K16ac is reinforced by intermittent association of MSL complex with these sites, and therefore is sensitive to depletion of MSL complex by RNAi. However, we cannot exclude the possibility that a second HAT which requires an intact MSL complex deposits H4K16ac at these sites.
Chromosome-wide acetylation of the male X in vivo
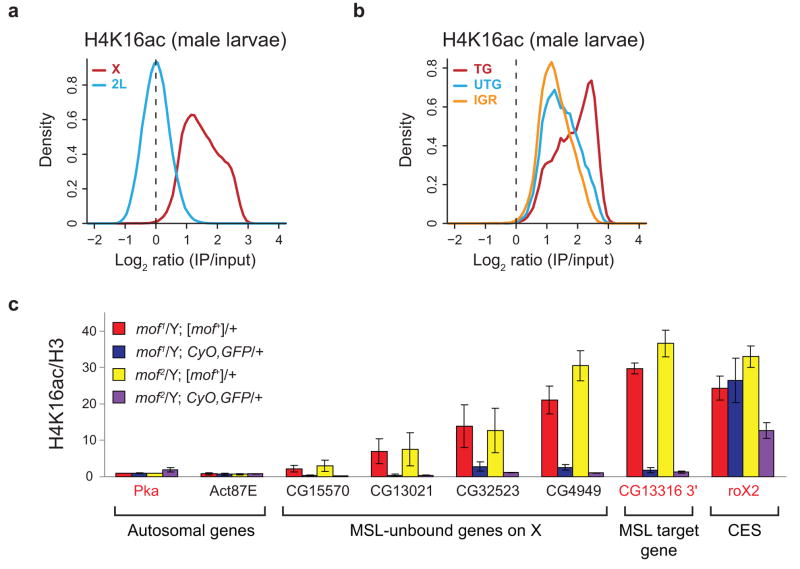
In order to determine if chromosome-wide acetylation of H4 Lys16 is a general property of the male X, we performed ChIP-chip analysis of H4K16ac in third instar male larvae. As observed in SL2 cells, H4K16ac is more broadly associated with the male X chromosome than MSL complex, assayed previously11, and exhibits a significant baseline shift relative to 2L (Figs. 1a and 4a and Supplementary Figs. 2b and 5a). In contrast, this was not observed in female larvae or “female” Kc cells, which do not express MSL complex (Supplementary Fig. 6a). Further characterization of H4K16ac on the X in male larvae revealed similar properties as were observed in SL2 cells. Active genes show the highest levels of H4K16ac enrichment (Fig. 4b and Supplementary Fig. 5b), and H4K16ac is biased towards the middle and 3′ ends of these genes (Supplementary Fig. 5c).
Figure 4.
Global H4K16ac on the male X in vivo. (a) H4K16ac is enriched along the majority of the X chromosome relative to 2L in male larvae. The distribution of log ratios at all probes on the X (red) and 2L (blue) is plotted for H4K16ac in male larvae. (b) As in SL2 cells, transcribed genes show the highest levels of H4K16ac in male larvae. The distribution of log ratios at all probes on the X chromosome, classified according to probes present in transcribed genes (TG, red), untranscribed genes (UTG, blue) and intergenic regions (IGR, orange), is shown. (c) MOF is required for H4K16ac in male larvae at sites on the X that lack detectable MSL3-TAP binding, as determined by ChIP-chip11. mof mutant male larvae (blue, purple) and their wild-type brothers (red, yellow) were generated from mof+ mothers. Known MSL targets (the roX2 CES (chromatin entry site) and CG13316 3′ end) and autosomal genes served as positive and negative controls, respectively. We assayed four genes on the X that lack detectable MSL binding11 but are enriched by H4K16ac in male larval ChIP-chip experiments (CG15570, CG13021, CG32523 and CG4949). Genes were classified as transcribed (gene names in red) or untranscribed (gene names in black) based on Affymetrix analysis in SL2 cells5. The H4K16ac ChIP signals were quantified as percent IP normalized to input and H3 levels. The ChIP signals for the two mof1 and the two mof2 genotypes were then normalized to Pka from mof1/Y; [mof+]/+ and mof2/Y; [mof+]/+ males respectively, and the replicates were averaged (see Supplementary Methods). The error bars represent the range from two independent experiments.
We next validated the enrichment of H4K16ac at sites lacking MSL complex and tested its dependence on MOF. H4K16ac ChIP was performed in male larvae harboring the catalytically-inactive mof1 or the truncated mof2 allele in the presence or absence of a 6.8 kb genomic mof+ transgene that rescues the viability of adult males (refs. 9,17, Supplementary Table 1). H4K16ac is enriched at the MSL target gene, CG13316, in the presence of the mof+ rescue transgene, but is reduced to background levels in mof mutant males (Fig. 4c, compare red and yellow vs. blue and purple bars). While residual activity of the mof1 allele is sufficient for H4K16ac at the roX2 CES (Fig. 4c, blue bar), acetylation at this site is reduced 2.5-fold in males harboring the stronger mof2 allele (Fig. 4c, purple bar). H4K16ac is also detected at genes on the X that do not stably bind MSL complex and is lost in mof1 and mof2 males. Taken together with our observations in SL2 cells, these results further demonstrate that MSL complex has two classes of targets on the male X chromosome; in addition to its known target genes, MSL complex functions more generally to globally acetylate the male X chromosome.
Limited role for MOF in 5′ H4K16ac
In agreement with recent reports32–34, we also detected a category of H4K16ac, not restricted by sex or chromosome (Figs. 1a and 5a). This modification is enriched over expressed genes on the X and 2L in female larvae and Kc cells with a bias towards the 5′ ends (Fig. 5b,c and Supplementary Fig. 6b,c). Autosomal 5′ H4K16ac is also detected in SL2 cells and male larvae (Fig. 5a and Supplementary Fig. 7). This pattern is reminiscent of the distribution of H4K16ac in human CD4+ T-cells37 and is distinct from H4K16ac in S. cerevisiae, which is generally excluded from the 5′ ends of genes38.
Figure 5.
5′ H4K16ac on X and 2L in females. (a) H4K16ac on active autosomal genes in males and females. ChIP-chip profiles for MSL3-TAP (from SL2 cells and male larvae5,11) and H4K16ac (from SL2 and Kc cells, male and female larvae) are shown for a representative region of chromosome 2L as in Figure 1a. (b) Transcribed genes on the X and 2L are enriched for H4K16ac in female larvae. The maximum H4K16ac ChIP signal for each gene in female larvae (y-axis) on the X (left) and 2L (right) is plotted against its expression (in log scale) in SL2 cells5 (x-axis). (c) H4K16ac is biased towards the 5′ ends of active genes in female larvae. A scaled average gene profile was generated for H4K16ac ChIP in female larvae over genes on the X (left) and 2L (right) ranked by expression quantiles from SL2 cells5.
Since 5′ H4K16ac is not restricted to one sex, it is not expected to require the male-specific MSL complex. However, MOF is expressed in both males and females17, and in addition to its association with MSL complex, MOF co-purifies with the NSL (non-specific lethal) complex that is conserved between Drosophila and humans39. Therefore, one possibility is that MOF might mediate 5′ acetylation as a part of the NSL complex. Although we did not observe reproducible enrichment of MOF at the 5′ ends of transcribed autosomal genes by ChIP in SL2 cells (Supplementary Fig. 8a), this might result from a lower sensitivity of our MOF ChIP assay as compared to Kind et al.33.
To investigate the role of MOF in 5′ H4K16ac, we tested the consequences of MOF depletion in RNAi experiments in SL2 cells. Whereas H4K16ac at MSL-bound genes on the X was reduced to background levels following mof depletion (27- to 47-fold), autosomal H4K16ac was only modestly affected (1.1- to 3.1-fold) (Fig. 6a and Supplementary Fig. 8b, red vs. yellow bars). Unexpectedly, similar effects at the autosomal genes were observed upon depletion of msl2 (Fig. 6a, blue bars). Furthermore, H4K16ac remained at the 5′ ends of MSL target genes on the X chromosome after msl2 and mof RNAi, despite strong depletion at the 3′ ends (Supplementary Fig. 8b). The ChIP assay using an independent H4K16ac antibody yielded comparable results (Supplementary Fig. 8c). Therefore, we conclude that MOF cannot be the only HAT that acetylates H4 Lys16 at the 5′ ends of these genes.
Figure 6.
Limited role for MOF in 5′ H4K16ac. The 3′ ends of known MSL target genes and the roX2 CES (chromatin entry site) served as positive controls in SL2 cells and male larvae. Primers were designed in the 5′ ends of seven active genes on chromosome 2L that exhibited 5′-biased H4K16ac enrichment in ChIP-chip experiments. The HBS1 (chromosome 3L) P2 primer set was utilized to examine 5′ H4K16ac in a previous study33. Genes were classified as transcribed (gene names in red) or untranscribed (gene names in black) based on Affymetrix analysis in SL2 cells5. (a) MOF is not solely responsible for H4K16ac at the 5′ ends of autosomal genes in SL2 cells. RNAi was performed in SL2 cells for the negative GFP control (red), msl2 (blue) or mof (yellow). Average H4K16ac ChIP signal is quantified as in Figure 3a. (b) MOF is not required for H4K16ac at the 5′ ends of autosomal genes in male larvae. The average H4K16ac ChIP signals from mof mutant male larvae (blue, purple) and their wild-type brothers (red, yellow) are quantified as in Figure 4c. (c) MOF is not required for H4K16ac at the 5′ ends of genes on X or autosomes in female larvae. The H4K16ac ChIP signals from mof1 mutant females (blue) and their wild-type sisters (red) are presented as percent IP normalized to input and H3 levels. The ChIP signals were normalized to Pka from mof1; [mof+]/+ females before averaging the replicates (see Supplementary Methods). Error bars represent the range from two independent experiments.
The modest effects of mof depletion on 5′ H4K16ac in SL2 cells were not consistent with the ChIP results of Kind et al.33. Therefore, we turned to genetic mof mutants to address this question in vivo. We assayed H4K16ac at the 5′ ends of autosomal genes in mof1 and mof2 mutant male larvae. Whereas 3′-biased H4K16ac at MSL target genes on the X was greatly reduced in mof mutant males, autosomal sites of 5′ H4K16ac persisted (Fig. 6b, red and yellow vs. blue and purple bars), suggesting that MOF is not required for 5′ H4K16ac at these loci in vivo.
Although the HAT domain is absent from the truncated MOF2 protein, residual H4K16ac at the roX2 CES in mof2 mutant males may reflect the activity of maternally-deposited wild-type MOF. Therefore, we sought to perform ChIP assays in mof1 and mof2 mutant larvae lacking any maternal contribution of MOF. For this purpose, we needed homozygous mof mutant females to generate the desired progeny. We found that mof mutant females are viable, in contrast to their brothers, as expected (refs. 9,17, Supplementary Table 1). However, mof2 females were developmentally delayed, a phenotype that was rescued by a wild-type mof+ transgene (Supplementary Fig. 9). Furthermore, mof2 mutant females have significantly decreased fertility (data not shown), and we were not able to generate enough progeny for ChIP assays. These results suggest that MOF has a functional role(s) in females or that the truncated MOF2 protein has dominant-negative effects. mof1 male larvae lacking maternal MOF were also difficult to obtain, but we were able to assay H4K16ac in mof1 females in the absence of wild-type maternal MOF. Significant enrichment of H4K16ac was detected at the 5′ ends of genes on the X and autosomes in wild-type females (Fig. 6c, red bars). We observed comparable levels of H4K16ac in mof1 females, suggesting that MOF is not required for 5′ H4K16ac at these genes in vivo (Fig. 6c, blue bars). Taken together, our results suggest that MOF has, at the most, only modest effects on 5′ H4K16ac at the genes tested, in contrast to its highly specialized role on the male X. In humans, 5′-biased H4K16ac is one mark of many present in a complex signature over the promoters and bodies of genes37. Given the density of enzymes that are recruited to create these patterns, we favor a model in which MOF functions redundantly with other HATs with overlapping substrate specificities to acetylate H4 Lys16 at the 5′ ends of active genes.
Discussion
Nearly all genes on the X chromosome in the soma of the adult fly are dosage compensated15. Therefore, it has been puzzling that ~25% of transcribed genes are not clearly bound by MSL complex5,6,11,14. Here, our results provide an explanation. We find that almost all active genes on the X chromosome are associated with robust H4K16ac. The distribution of H4K16ac on the X chromosome is broader than MSL binding, and MSL complex is clearly required for this expanded H4K16ac domain. Therefore, MSL localization studies underestimate the number of MSL target genes on the X chromosome. Our results strengthen a model in which MSL complex targets genes by recognizing features of active transcription11. In addition, our results support a model in which MSL complex accounts for the majority of somatic dosage compensation in Drosophila.
These results begin to provide a molecular basis for previous cytological observations that the whole X chromosome has a distinct character from the autosomes. What is the impact of the strikingly elevated baseline levels of H4K16ac on the male X chromosome? H4K16ac may promote an open chromatin structure by directly inhibiting the compaction of chromatin fibers30,31. In addition, H4K16ac is known to antagonize the activity of the ISWI chromatin remodeling enzymes28–30, and counteracting ISWI activity may increase chromatin accessibility, at least in part, through lowering incorporation of linker histone H1 (ref. 40).
Consistent with a chromosome-wide increase in accessibility, the structure of the polytene male X is particularly sensitive to perturbation, since mutation of numerous chromatin regulators causes this chromosome to take on a short and bloated appearance (see refs. 41–43 for examples). Overexpression of MSL1 and MSL2 in males results in a similar X chromosome morphology44. Increased accessibility across the male X might facilitate MSL spreading to active genes and promote the recruitment of additional trans-acting factors involved in dosage compensation. For example, high-resolution mapping of the histone variant H3.3 in SL2 cells and the heterochromatin protein HP1 in adult flies has revealed preferential enrichment on the male X45,46. This may reflect a role for these factors in fine-tuning expression of X-linked genes, or alternatively may simply be a manifestation of the increased accessibility of this chromosome. A future goal will be to discriminate the primary causes of dosage compensation from potential downstream effects that might result from a chromosome-wide increase in accessibility.
Previously, a redistribution model was proposed to explain cytological evidence that MOF and H4K16ac staining on all chromosomes in females appears brighter than on autosomes in males47. According to this model, MSL complex specifically sequesters MOF to the X chromosome in males to decrease autosomal gene expression. The possibility that MOF regulates 5′ H4K16ac throughout the genome might be compatible with such a model33. However, our findings support a limited role for MOF in autosomal H4K16ac, suggesting that the transcriptional consequences of MOF redistribution would be limited. On the other hand, we discovered that females carrying a premature stop codon in the mof gene suffer developmental delay and poor fertility. While these phenotypes could be due to dominant-negative effects of the truncated MOF2 protein, they might reflect a non-sex-specific role for MOF. Although we cannot rule out the possibility that MOF is essential for 5′ H4K16ac at a subset of loci, it also could be required to regulate chromatin processes outside of transcription or to acetylate non-histone targets in Drosophila. MOF is essential for embryogenesis in mice48,49 and has been implicated in cell cycle progression50,51 and the DNA damage response52 in mammals, with evidence for some H4K16ac-independent functions51,53. Therefore, the potential effects of competition between various roles that MOF may play in Drosophila will be the subject of continuing debate.
The principle question raised by our work is: how does the MSL complex deposit H4K16ac at sites on the X where it is not stably bound? The detection of domains of histone modification that are broader than the localization of the modifying enzyme itself has been reported previously54–58. We favor the idea that the modification reflects transient association of MSL complex, for example by rapid scanning or looping59. A similar “hit-and-run” mechanism has been proposed to explain the activity of the histone methyltransferase E(z) in Drosophila, as E(z) is bound specifically at PREs, but the H3K27me3 modification that it deposits is found in broad domains54–57. Our results analyzing MSL deposition of the H4K16ac activation mark strongly parallel those of the Polycomb repressive complex, suggesting that similar principles govern chromosome spreading and domain organization in active and repressed regions of the genome.
Methods
Drosophila Stocks and Crosses
We raised flies on standard cornmeal-molasses medium at 25°C.
We generated mof1 mutant female larvae that lack a maternal contribution of wild-type MOF by crossing w cv mof1; +/CyO females to w cv mof1/Y; P[mof t6.8]18H1/CyO, twi>GAL4, UAS-2xEGFP males. The [w+mC, mof t6.8]18H1 transgene contains a 6.88 kb HindIII-HindIII fragment of the mof genomic region, previously shown to rescue mof1 mutant males17. We collected mof1 mutant (GFP+) and wild-type control (carrying the mof t6.8 genomic transgene, GFP-) female larvae.
A maternal contribution of wild-type MOF was required to generate mof1 and mof2 mutant male larvae for ChIP. We crossed w cv mof1; P[mof t6.8]18H1/CyO, twi>GAL4, UAS-2xEGFP females to y w/Y males for mof1, and y w mof2; P[mof t6.8]18H1/CyO, twi>GAL4, UAS-2xEGFP females to y w/Y males for mof2. We collected mof mutant (GFP+) and wild-type control (carrying the mof t6.8 genomic transgene, GFP-) male larvae.
Cell culture methods and RNAi
We maintained SL2 and Kc cells at 25°C in Schneider’s medium (Gibco) supplemented with 10% (v/v) heat-inactivated FBS (JRH) and antibiotic-antimycotic (Gibco). For ChIP-chip, we prepared chromatin from SL2 cells previously used to generate MSL3-TAP stable cell lines5 for direct comparison with MSL3-TAP data sets. For RNAi, we utilized SL2 cells that were obtained from Norbert Perrimon (Harvard Medical School), as these yielded optimal RNAi in our assays.
dsRNA to target the msl2 and mof genes was generated as described at www.flyrnai.org. In brief, we synthesized (T7 Megascript kit, Ambion) and purified (RNeasy kit, Qiagen) dsRNAs. The dsRNA targeting msl2 (DRSC00829) has two predicted off-targets at a 19 nt cutoff, and the dsRNA targeting mof (DRSC24125) has no predicted off-targets at 19 nt (www.flyrnai.org). We used GFP dsRNA as a negative control, as previously described36.
We performed RNAi treatments in T225 flasks by scaling up the protocol described at www.flyrnai.org. Briefly, we harvested cells by centrifugation and resuspended them in serum-free Schneider’s medium at 2×106 cells ml−1. We added 10 ml of cells to each flask with 225 μg of dsRNA in 5 mL of water. Following 45 min incubation at room temperature, we added 30 ml of Schneider’s medium containing serum to each flask. We incubated flasks for 6–7 days at 25°C, and harvested cells for Western analysis and chromatin preparation. To achieve maximal depletion of mof, we performed sequential RNAi for all samples in H4K16ac ChIP experiments, adapting the protocol described above for T75 flasks (75 μg of dsRNA and 7×106 cells). We repeated the treatment after four days of RNAi and harvested cells on the eighth day.
ChIP-chip from Tissue Culture Cells and Larvae
For ChIP-chip experiments in SL2 and Kc cells, we prepared chromatin as described previously5. We treated the Kc cells with GFP negative control RNAi as described above for other purposes. For ChIP-chip experiments in larvae, we collected 1000 sex-sorted y w third instar larvae and prepared chromatin as described previously11. We performed five IPs for each of the two biological replicates. For SL2 cells and male larvae, we performed one ChIP-chip experiment using anti-H4K16ac from Serotec (AHP417) (1–2 μl/IP). However, this antibody was discontinued, and we performed an additional experiment for each using anti-H4K16ac from Millipore (07–329) (3 μl/IP). The specificity of this antibody has been assessed by Western blotting with peptide competition (www.millipore.com). We performed both replicates for female larvae and Kc cells using anti-H4K16ac from Millipore. For detailed IP conditions, see Supplementary Methods. Following DNA isolation, we amplified the samples by ligation-mediated PCR, as described previously5. Replicate experiments were highly reproducible (Supplementary Fig. 10).
ChIP from Tissue Culture Cells and Larvae
For small-scale ChIP experiments, we modified protocols for sonication using the Bioruptor™ (Diagenode) (see Supplementary Methods for details). We performed SL2 cell RNAi ChIP experiments in triplicate and larval ChIP experiments in duplicate. We normalized chromatin samples for DNA content and incubated IPs overnight at 4 °C with 2 μl anti-MSL2 (ref. 8), 3 μl anti-MOF44, 3 μl anti-H3 (Abcam, ab1791), 3 μl anti-H4K16ac (Millipore, 07–329) or 10 μl anti-H4K16ac (Santa Cruz Biotechnology sc-8662-R). The specificity of sc-8662-R has been assessed by Western blotting with peptide competition37. The MOF antibody was raised against MOF (amino acids 365–766) fused to GST. We incubated IPs with 60 μl of 50% slurry Protein G beads preincubated with salmon sperm DNA (Millipore) for 1.5 h at 4°C to recover immune complexes. We performed wash, elution and DNA recovery steps as for ChIP-chip experiments (see Supplementary Methods).
Quantitation of ChIP DNA by qPCR
We performed qPCR analysis of ChIP DNA as described previously5. Briefly, we used primers of equivalent efficiency in duplicate qPCR reactions using Platinum SYBR Green qPCR SuperMix-UDG w/ROX (Invitrogen) on an ABI 7000 PRISM Sequence Detection System (Applied Biosystems). We performed relative quantification of each probe by the comparative CT method (User Bulletin #2, Applied Biosystems). Additional details about data normalization are provided in the Supplementary Methods Primer sequences are listed in Supplementary Table 2.
Microarray analysis
We analyzed microarrays according to the methods described in ref. 60 for direct comparison to MSL3-TAP data sets analyzed in ref. 11. We performed two replicates for each experiment, with two-channel Nimblegen arrays (ChIP vs. input). These arrays contained 388,000 50mer probes, covering the X and 2L chromosomes at 100 bp resolution. For each array, we corrected dye-specific bias based on the average intensity vs. log-ratio relationship60, and then shifted the log-ratio distribution so that the mode of the distribution for the control probes (2L) is 0. To define a bound region, we first computed the noise level (σ*) of each array (see ref. 11 for details) and considered probe values above 3σ* as statistically significant. After rescaling based on their noise levels (3σ* defined as log-ratio of 1), we took the mean of the two replicates and applied running median smoothing with window size of 7 probes (700 bp) along their genomic locations. Since the average DNA fragment size from sonication was about 800 bp, clusters consisting of at least 8 probes above the enrichment threshold were considered significant. The curves in Figure 2b are smoothed through a binned kernel density estimation function with a Gaussian kernel, using the bkde() function in the R package KernSmooth. The default bandwidth was used. For the distributions in Figures 1 and 4, we generated histograms with bin size 0.1 and then connected the values at the midpoints. Given the small bin size, application of bdke() does not make noticeable difference. We performed all computations using the statistical language R.
Accession codes
Data are deposited at the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE14884.
Supplementary Material
Acknowledgments
We thank O. Lee (Brigham & Women’s Hospital) for excellent technical assistance, A. Alekseyenko (Brigham & Women’s Hospital) for male larval chromatin, A. Alekseyenko and P. Kharchenko (Children’s Hospital) for helpful advice and discussions, and members of the Kuroda lab (Brigham & Women’s Hospital and Harvard Medical School) for critical reading of the manuscript. We are grateful to Y. Zhang and B. Oliver (National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health) for sharing information prior to publication. This work was supported by the National Institutes of Health (GM45744 to M.I.K. and GM67825 to P.J.P.). M.E.G. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-1913-06) and E.L. is supported by the Medical Foundation Charles A. King Trust.
Footnotes
Author contributions
M.E.G. and E.L. performed ChIP and ChIP-chip experiments and genetic analysis of mof mutants. S.P. performed all bioinformatics analyses. P.J.P. and M.I.K. supervised the analyses, and M.E.G. and M.I.K. prepared the manuscript in consultation with all co-authors.
References
- 1.Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–92. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- 2.Gelbart ME, Kuroda MI. Drosophila dosage compensation: a complex voyage to the X chromosome. Development. 2009;136:1399–410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alekseyenko AA, et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 2008;4:e1000302. doi: 10.1371/journal.pgen.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–57. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilfillan GD, et al. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev. 2006;20:858–70. doi: 10.1101/gad.1399406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer MJ, Richman R, Richter L, Kuroda MI. Sex-specific regulation of the male-specific lethal-1 dosage compensation gene in Drosophila. Genes Dev. 1994;8:698–706. doi: 10.1101/gad.8.6.698. [DOI] [PubMed] [Google Scholar]
- 8.Lyman LM, Copps K, Rastelli L, Kelley RL, Kuroda MI. Drosophila male-specific lethal-2 protein: structure/function analysis and dependence on MSL-1 for chromosome association. Genetics. 1997;147:1743–53. doi: 10.1093/genetics/147.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu W, Szauter P, Lucchesi JC. Targeting of MOF, a putative histone acetyl transferase, to the X chromosome of Drosophila melanogaster. Dev Genet. 1998;22:56–64. doi: 10.1002/(SICI)1520-6408(1998)22:1<56::AID-DVG6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 10.Sass GL, Pannuti A, Lucchesi JC. Male-specific lethal complex of Drosophila targets activated regions of the X chromosome for chromatin remodeling. Proc Natl Acad Sci U S A. 2003;100:8287–91. doi: 10.1073/pnas.1332749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larschan E, et al. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–33. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Bell O, et al. Transcription-coupled methylation of histone H3 at lysine 36 regulates dosage compensation by enhancing recruitment of the MSL complex in Drosophila melanogaster. Mol Cell Biol. 2008;28:3401–9. doi: 10.1128/MCB.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sural TH, et al. The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat Struct Mol Biol. 2008;15:1318–1325. doi: 10.1038/nsmb.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legube G, McWeeney SK, Lercher MJ, Akhtar A. X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes Dev. 2006;20:871–83. doi: 10.1101/gad.377506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta V, et al. Global analysis of X-chromosome dosage compensation. J Biol. 2006;5:3. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith ER, Allis CD, Lucchesi JC. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J Biol Chem. 2001;276:31483–6. doi: 10.1074/jbc.C100351200. [DOI] [PubMed] [Google Scholar]
- 17.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. Embo J. 1997;16:2054–60. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith ER, et al. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol Cell Biol. 2000;20:312–8. doi: 10.1128/mcb.20.1.312-318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5:367–75. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 20.Thomas T, Voss AK. The diverse biological roles of MYST histone acetyltransferase family proteins. Cell Cycle. 2007;6:696–704. doi: 10.4161/cc.6.6.4013. [DOI] [PubMed] [Google Scholar]
- 21.Turner BM, Birley AJ, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–84. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 22.Bone JR, et al. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- 23.Dou Y, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–85. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 24.Kapoor-Vazirani P, Kagey JD, Powell DR, Vertino PM. Role of hMOF-dependent histone H4 lysine 16 acetylation in the maintenance of TMS1/ASC gene activity. Cancer Res. 2008;68:6810–21. doi: 10.1158/0008-5472.CAN-08-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci U S A. 2005;102:5501–6. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–7. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 27.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet. 2002;32:378–83. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 28.Clapier CR, Nightingale KP, Becker PB. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 2002;30:649–55. doi: 10.1093/nar/30.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corona DF, Clapier CR, Becker PB, Tamkun JW. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 2002;3:242–7. doi: 10.1093/embo-reports/kvf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 31.Robinson PJ, et al. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–25. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell O, et al. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. Embo J. 2007;26:4974–84. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kind J, et al. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell. 2008;133:813–28. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 34.Schwaiger M, et al. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev. 2009;23:589–601. doi: 10.1101/gad.511809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Straub T, et al. Stable chromosomal association of MSL2 defines a dosage-compensated nuclear compartment. Chromosoma. 2005;114:352–64. doi: 10.1007/s00412-005-0020-x. [DOI] [PubMed] [Google Scholar]
- 36.Hamada FN, Park PJ, Gordadze PR, Kuroda MI. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 2005;19:2289–94. doi: 10.1101/gad.1343705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu CL, et al. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendjan S, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–23. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Corona DF, et al. ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol. 2007;5:e232. doi: 10.1371/journal.pbio.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deuring R, et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell. 2000;5:355–65. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Zhang W, Jin Y, Johansen J, Johansen KM. The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell. 2001;105:433–43. doi: 10.1016/s0092-8674(01)00325-7. [DOI] [PubMed] [Google Scholar]
- 43.Spierer A, Seum C, Delattre M, Spierer P. Loss of the modifiers of variegation Su(var)3–7 or HP1 impacts male X polytene chromosome morphology and dosage compensation. J Cell Sci. 2005;118:5047–57. doi: 10.1242/jcs.02623. [DOI] [PubMed] [Google Scholar]
- 44.Oh H, Park Y, Kuroda MI. Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes Dev. 2003;17:1334–9. doi: 10.1101/gad.1082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Wit E, Greil F, van Steensel B. Genome-wide HP1 binding in Drosophila: developmental plasticity and genomic targeting signals. Genome Res. 2005;15:1265–73. doi: 10.1101/gr.3198905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–7. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 47.Bhadra U, Pal-Bhadra M, Birchler JA. Role of the male specific lethal (msl) genes in modifying the effects of sex chromosomal dosage in Drosophila. Genetics. 1999;152:249–68. doi: 10.1093/genetics/152.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta A, et al. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol Cell Biol. 2008;28:397–409. doi: 10.1128/MCB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas T, Dixon MP, Kueh AJ, Voss AK. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol Cell Biol. 2008;28:5093–105. doi: 10.1128/MCB.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith ER, et al. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–88. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taipale M, et al. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol. 2005;25:6798–810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gupta A, et al. Involvement of human MOF in ATM function. Mol Cell Biol. 2005;25:5292–305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sykes SM, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–51. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz YB, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–5. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 55.Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–54. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kahn TG, Schwartz YB, Dellino GI, Pirrotta V. Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J Biol Chem. 2006;281:29064–75. doi: 10.1074/jbc.M605430200. [DOI] [PubMed] [Google Scholar]
- 57.Beisel C, et al. Comparing active and repressed expression states of genes controlled by the Polycomb/Trithorax group proteins. Proc Natl Acad Sci U S A. 2007;104:16615–20. doi: 10.1073/pnas.0701538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parker DS, Ni YY, Chang JL, Li J, Cadigan KM. Wingless signaling induces widespread chromatin remodeling of target loci. Mol Cell Biol. 2008;28:1815–28. doi: 10.1128/MCB.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu W, Wei X, Pannuti A, Lucchesi JC. Targeting the chromatin-remodeling MSL complex of Drosophila to its sites of action on the X chromosome requires both acetyl transferase and ATPase activities. Embo J. 2000;19:5202–11. doi: 10.1093/emboj/19.19.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peng S, Alekseyenko AA, Larschan E, Kuroda MI, Park PJ. Normalization and experimental design for ChIP-chip data. BMC Bioinformatics. 2007;8:219. doi: 10.1186/1471-2105-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.