Abstract
Spinal muscular atrophy (SMA) is attributed to mutations in the SMN1 gene, leading to loss of spinal cord motor neurons. The neurotropic Sindbis virus vector system was used to investigate a role for the survival motor neuron (SMN) protein in regulating neuronal apoptosis. Here we show that SMN protects primary neurons and differentiated neuron-like stem cells, but not cultured cell lines from virus-induced apoptotic death. SMN also protects neurons in vivo and increases survival of virus-infected mice. SMN mutants (SMNΔ7 and SMN-Y272C) found in patients with SMA not only lack antiapoptotic activity but also are potently proapoptotic, causing increased neuronal apoptosis and animal mortality. Full-length SMN is proteolytically processed in brains undergoing apoptosis or after ischemic injury. Mutation of an Asp-252 of SMN abolished cleavage of SMN and increased the antiapoptotic function of full-length SMN in neurons. Taken together, deletions or mutations of the C terminus of SMN that result from proteolysis, splicing (SMNΔ7), or germ-line mutations (e.g., Y272C), produce a proapoptotic form of SMN that may contribute to neuronal death in SMA and perhaps other neurodegenerative disorders.
Spinal muscular atrophy (SMA) is a common autosomal recessive disorder characterized by the loss of spinal cord motor neurons and weakness. Mutation or deletion of the telomeric copy of SMN (SMN1) occurs in over 96% of SMA patients (1, 2). A second copy of SMN, SMN2, produces a full-length SMN (survival motor neuron) protein that is identical to the SMN1 gene product but also produces splice variants of SMN, the most abundant of which lacks the last coding exon, exon 7 (SMNΔ7). Mutations found in SMA patients occur only in SMN1 and often result in the loss of exon 7 (2). Therefore, a deficiency in full-length SMN protein correlates with disease (3, 4). The converse implies that increased amounts of SMNΔ7 correlates with disease. However, it is currently not known if SMNΔ7 is partially protective, inactive, or detrimental in SMA.
SMN is a ubiquitously expressed 38-kDa protein that is highly conserved across widely diverse species. SMN has been shown to interact with several proteins involved in spliceosomal assembly including SMN-interacting protein-1 (SIP-1/Gemin2), SmB, and Gemin3/dp103 (5–8). Therefore, it is likely that one role of SMN is to facilitate the formation of spliceosomal complexes in the cytoplasm (5, 6, 9), to function in recycling small nuclear ribonucleoprotein in the nucleus, and potentially play a role in pre-mRNA splicing (10). A distinct function for SMN was suggested by the observation that SMN enhances the antiapoptotic function of Bcl-2 in microinjected HeLa cells (11, 12). Similarly, a human homologue of SMN, termed SMN-related protein is capable of modulating apoptosis of transfected cells (13).
We have developed a Sindbis virus (SV) vector system to deliver genes to neurons in culture and in vivo (14–16). This system has allowed us to evaluate the function of a variety of apoptotic regulators in the vertebrate central nervous system (14, 15, 17–21). In this model, the virus serves as both a cell death stimulus and a vector for expressing heterologous proteins in neurons of the central nervous system. SV specifically targets neurons including spinal cord motor neurons, triggering neuronal apoptosis as detected by terminal deoxynucleotidyltransferase-mediated UTP end labeling assay and DNA laddering (22, 23). We have used this system to show that full-length SMN protects neurons from apoptosis, whereas mutant SMN derivatives from patients with SMA accelerate neuronal death. These findings raise the possibility that the proapoptotic activity of SMN mutants may underlie the pathogenesis of SMA.
Materials and Methods
Viruses and Cells.
Virus was generated and plaque titrations performed on baby hamster kidney (BHK) cells (American Type Culture Collection) as described (14, 16). Primary cortical neurons were cultured in basal medium Eagle without glutamin, 2 mM glutaMAX, 10% FBS, 5% horse serum, 200 μM cysteine, 10 μM butylated hydroxyanisole, 1× N2 supplement, penicillin (100 units/ml), and streptomycin (100 μg/ml) (24). CSM 14.1.4 temperature-sensitive immortalized rat nigral neural cells (provided by D. Bredesen, Buck Center for Research in Aging, Novato, CA) were differentiated into neuron-like cells as reported (25). Cultured cells were infected with recombinant SV at a multiplicity of infection of five. Cells were stained with 10 μg/μl Hoescht/4.5 μg/μl propidium iodide in PBS with 25 mM glucose for 15 min at 37°C and examined by fluorescent microscopy. Random fields of cells were photographed, counted, and scored for viability/apoptotic morphology. Stable CSM 14.1.4 transfectants were selected and maintained with 10% FBS/DMEM containing 0.5 mg/ml G418. Individual clones expressing SMN were selected at random for further analyses.
In Vitro Translation and Cleavage Assays.
In vitro translation mix (5 μl) was incubated with 30 μl of brain lysate for 4 h at 37°C before analysis by SDS/PAGE (21). Brain lysates were prepared by harvesting whole brains 24 h postinfection and douncing in ICE buffer [100 mM Hepes, pH 7.5/10% sucrose/0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate/10 mM DTT].
Immunoblot Analysis.
Brains were collected and homogenized in RIPA buffer (50 mM Tris⋅HCl, pH 7.5/150 mM NaCl/1% NP-40/0.5% Na deoxycholate/0.1% SDS) containing 0.28 mg/ml aprotinin/2 mg/ml PMSF/2 mg/ml leupeptin. Lysate was centrifuged at 10,000 × g for 5 min after boiling. Brain extracts (150 μg) or CSM cell lysates (200 μg) were analyzed by SDS/PAGE and immunoblot analysis with SMN mAb, MANSMA2 clone 8F7 (26), mouse IgG 2B1 (27), exon-7 (G. Dreyfuss, University of Pennsylvania, Philadelphia), antihemagglutinin (anti-HA) 12CA5, or anti-HA.11.
Animals.
Three- or five-day-old CD1 mice were injected intracranially with 5 × 103 plaque-forming units of recombinant SV in 30 μl or buffer diluent alone. Transient ischemia (2 h of right middle cerebral artery occlusion) and reperfusion was performed as described (28, 29). For histologic analysis, animals were killed 3 days postinfection and perfused intracardially with 4% paraformaldehyde. The brains were postfixed in 4% paraformaldehyde or 4% paraformaldehyde/1% glutaraldehyde for electron microscopy.
Results
SMN Modulates Apoptosis in Cultured Neurons.
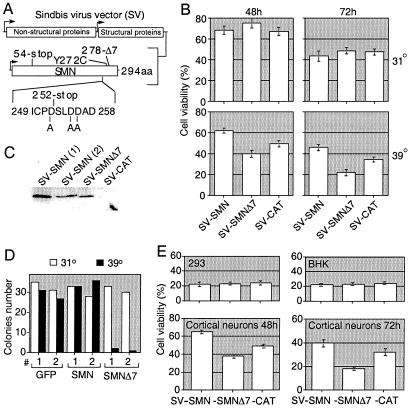
CSM 14.1.4 cells are immortalized with a temperature-sensitive SV40 T antigen and are capable of growing as undifferentiated cells (31°C, 10% FCS) but will mature into neuron-like cells in differentiation culture conditions (39°C, 1% FCS, bFGF). In differentiation medium, greater than 50% of the cells exhibited neuronal markers MAP2 and NF-H immunoreactivity (data not shown). Recombinant SV encoding HA-tagged human SMN, HA-tagged SMNΔ7 (an SMN mutant found in SMA patients lacking the C-terminal 16 amino acids), or an irrelevant control chloramphenicol acetyltransferase (CAT) were used to infect CSM 14.1.4 cells (Fig. 1A). Under nonneural conditions (31°C), infection with SV-SMN and SV-SMNΔ7 resulted in progressively declining cell viabilities that were indistinguishable from the SV-CAT control (Fig. 1B). However, under conditions favoring neuronal differentiation, SV-SMN resulted in significantly enhanced cell viability at 48 and 72 h (62 and 46%, respectively, compared with SV-CAT at 48 and 37%, respectively; Fig. 1B). In contrast, SV-SMNΔ7 resulted in diminished viability of cells relative to SV-SMN and SV-CAT at both 48 and 72 h (40 and 22%, respectively; Fig. 1B). The HA-tagged SMN constructs were functionally indistinguishable from untagged versions (data not shown). The 38-kDa SMN and 36-kDa SMNΔ7 were expressed at similar levels via the SV vector (Fig. 1C and data not shown).
Figure 1.
SMN modulates apoptosis of differentiated neural CSM 14.1.4 cells. (A) Map of the SV vector. (B) Cell viability of undifferentiated (31°C) and differentiated (39°C) CSM 14.1.4 cells was determined by propidium iodide/ Hoechst dual-staining after infection with the indicated recombinant SV. Data shown are for duplicate determinations in three independent experiments counting >400 cells per sample. Data from independently derived constructs were pooled. (C) Immunoblot analysis of undifferentiated CSM 14.1.4 cell lysates harvested at 24 h postinfection with the indicated recombinant viruses by using MANSMA2 anti-SMN antibody. Numbers in parenthesis distinguish two independently derived clones. (D) Individual clones of CSM 14.1.4 cells stably transfected with SMN, SMNΔ7, or green fluorescent protein were left undifferentiated (31°C) or were differentiated (39°C) and the number of colonies was determined 7 days after plating 100 cells. Results are the average of two similar independent experiments each using two independent clones per construct. (E) Cell viability was determined at 48 h postinfection of 293 and BHK cells, or at the indicated times after infection of primary rat cortical neurons with recombinant viruses. Data shown are the means ± SEM for duplicate determinations in three independent experiments counting >400 cells per sample.
To determine if SMNΔ7 can trigger cell death in the absence of a virus infection or other death stimulus, stably transfected CSM 14.1.4 cells expressing SMN were analyzed for survival after a shift to the differentiation conditions. Two independent cell lines expressing SMNΔ7 failed to produce colonies after a shift to differentiation conditions. Instead, these cells typically reached the 4–8 cell stage and then died. However, SMNΔ7 had no effect on colony formation when cells were maintained in the undifferentiated state (Fig. 1D). In contrast, cells expressing full-length SMN or control green fluorescent protein yielded similar numbers of colonies with and without differentiation.
Similar to the undifferentiated CSM 14.1.4 cell line, neither 293 nor BHK cell lines exhibited differential viability to any of the SV-SMN constructs relative to SV-CAT (Fig. 1E). In contrast, primary cortical neurons infected with SV-SMN exhibited enhanced survival compared with SV-CAT (65 versus 47% at 48 h), whereas neurons infected with SV-SMNΔ7 exhibited reduced survival compared with SV-CAT (19 versus 32% at 72 h; Fig. 1E). Thus, SMN modulates neuron-specific apoptosis.
SMN Modulates Neuronal Apoptosis in Vivo.
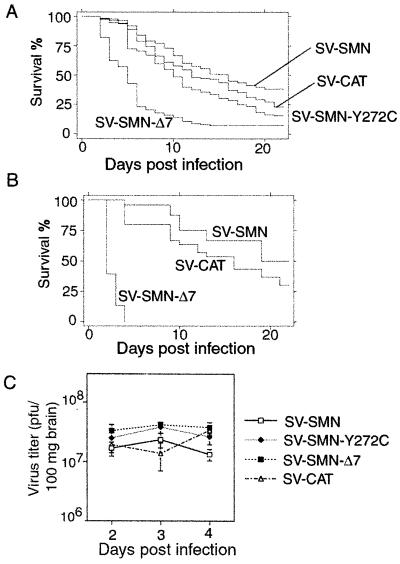
We have previously shown that SV causes widespread neuronal apoptosis in newborn mice and that animal mortality correlates with the extent of neuronal apoptosis (15, 20, 22). To investigate whether SMN alters the neuronal apoptotic response to SV infection in animals, newborn mice were infected intracerebrally with SV-SMN, SV-SMNΔ7, or SV-SMN-Y272C (Y272C, a missense mutation found in SMA patients). Kaplan Meier survival analysis revealed that only 23% of mice inoculated with SV-CAT control survived the infection (Fig. 2A). However, SV-SMN resulted in significantly increased survival relative to the control (39% at day 21; P < 0.001), whereas SV-SMN-Y272C and SV-SMNΔ7 constructs resulted in reduced survival compared with controls (15 and 7%, respectively; P < 0.001 for each construct relative to control). Additional control constructs including SV-SMN-Stop (stop codon inserted after codon 54) and SV-SMN-Rev (reverse orientation behind the HA-tag) produced survival curves that were superimposed on the SV-CAT survival curve (data not shown).
Figure 2.
SMN modulates survival of mice. (A) Survival of 3-day-old mice after infection with recombinant SV was determined by Kaplan Meier survival analysis. Data presented are from four independent experiments with more than 100 animals per group. (B) The same experiment shown in A was performed with 5-day-old mice. Data presented are from three independent experiments with 50 mice per group. (C) Titers of infectious virus were determined by standard plaque assay of whole brains. No statistical differences were observed. Each time point contains three animals from three independent experiments.
Because animal age at the time of infection is an important variable in the outcome of a SV virus infection with progressively less apoptosis and increased survival with older age (30), SMN function was also evaluated in 5-day-old mice (Fig. 2B). SV-SMN and SV-SMNΔ7 constructs were selected for analyses because of their disparate outcomes on infection of 3-day-old mice. Although survival of 5-day-old mice was increased for both SV-SMN and -CAT control as expected, SV-SMN was still significantly protective relative to SV-CAT (50 versus 27%; P < 0.001). Surprisingly, SV-SMNΔ7 was even more lethal in the older animals, with 100% mortality by 4 days postinfection (P < 0.001), suggesting that SMNΔ7 can overcome the age-dependent resistance to apoptosis. All viruses replicate within mouse brains to similar levels, indicating that differential virus replication does not explain differences in survival (Fig. 2C).
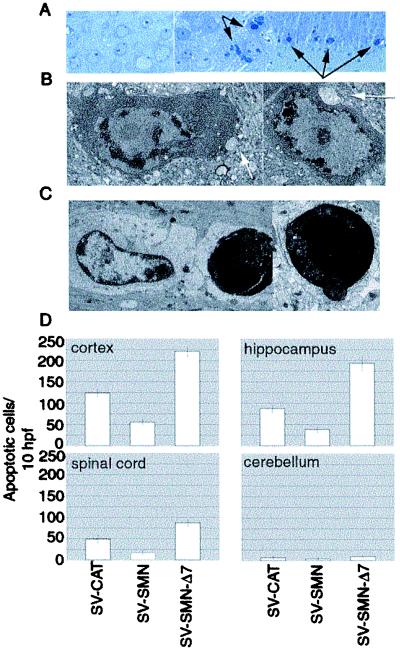
Histologic and ultrastructural analyses of infected mouse brains showed that neuronal death after infection with the SV-SMN-derived constructs occurred by an apoptotic mechanism, and that mouse survival correlated with the degree of neuronal apoptosis (Fig. 3 A–D). Cortical neurons showed normal morphology after injection with buffer only (Fig. 3A Left) whereas extensive nuclear fragmentation and cytoplasmic condensation characteristic of apoptotic death was apparent in cortical neurons two days after infection with SV-SMNΔ7 (Fig. 3A Center and Right). Electron microscopic analyses confirmed the classic features of neuronal apoptosis after infection with SV-SMNΔ7 (Fig. 3 B–C). Neurons exhibited chromatin condensation around the periphery of the nucleus and increased cytoplasmic density with preserved cytoplasmic organelles (Fig. 3B). Engulfment of an end-stage apoptotic neuron by a neighboring cell (Fig. 3C Left) and extrusion of an apoptotic body (Fig. 3C Right) provided additional evidence of apoptosis. The identification of neuronal synapses on the cell body of cells undergoing apoptosis confirmed their identity as neurons (Fig. 3B arrows). Quantitation of neuronal apoptosis by using morphologic criteria showed significantly fewer apoptotic neurons with SV-SMN but more neurons undergoing apoptosis in SV-SMNΔ7-infected hippocampus, cortex, and spinal cord compared with control SV-CAT (Fig. 3D). Spinal cord values were obtained by counting the large (motor) neurons in the ventral gray matter. Lower numbers of infected cells in the spinal cord is likely caused by the delay in trafficking of the virus from the site of infection (cerebrum). However, the relative killing function of SV-SMNΔ7 was similar in these regions. Cerebellar neurons showed no significant degree of apoptosis presumably because of inefficient targeting of SV to this brain region. These data support the hypothesis that SMN protects neurons from SV virus-induced apoptosis in vivo, whereas mutant SMN proteins found in SMA patients increase neuronal death in response to a SV infection.
Figure 3.
SMN modulates neuronal apoptosis in vivo. (A) Light microscopy of toluidine blue-stained sections of mouse brain cortex harvested at 48 h after injection of buffer only (Left) or SV-SMN-Δ7 (Center and Right). (B and C) Electron microscopy of cortical neurons after infection of mice with SV-SMN-Δ7. Arrows mark neuronal synapses. See text for description. (D) Apoptosis was quantified by counting the number of apoptotic neurons per 10 randomly selected high-power fields (hpf) of brain and spinal cord sections. Tissues were harvested at 48 h after infection with the indicated recombinant viruses. Data are presented as the means ± SEM.
Processing of SMN Protein During Cell Death.
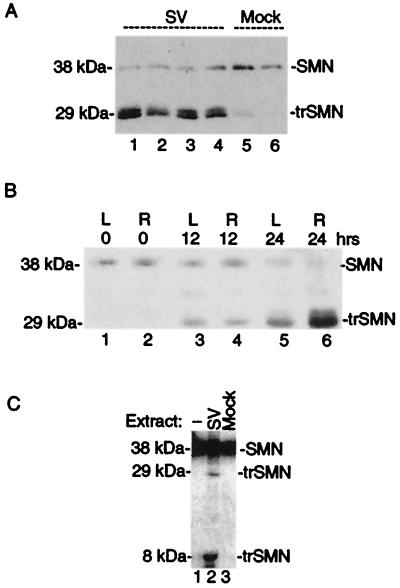
Immunoblot analyses of mock-infected brain extracts detected endogenous full-length SMN (38 kDa) as well as trace amounts of a 29-kDa immunoreactive species (Fig. 4A, lanes 5 and 6). In contrast, the amount of full-length SMN was reduced and the 29-kDa SMN immunoreactive species was prominent in the brains of animals infected with a control SV vector (lanes 1–4). The same 38- and 29-kDa species were detected with a second SMN mAb 2B1 (Fig. 5), suggesting that both bands are related to SMN and that the smaller 29-kDa form is more abundant in brains undergoing neuronal apoptosis. Furthermore, the smaller 29-kDa form appeared to be more stable than the full-length SMN in SV-infected brains, and this accumulation within cells could be important for the prodeath activity of truncated SMN.
Figure 4.
SMN is cleaved in brain. (A) Immunoblot analysis of brain extracts prepared from SV-infected (72 h) or control animals by using SMN mAb MANSMA2. (B) Brain extracts prepared at the indicated times after ischemia/reperfusion injury were analyzed by immunoblotting with SMN mAb MANSMA2. L (left, contralateral), and R (right, ipsilateral). (C) [35S]Met, in vitro-translated SMN was left untreated or treated with brain extracts prepared from uninfected or Sindbis virus (NSV)-infected mice. Samples were analyzed by SDS/PAGE and autoradiography.
Figure 5.
Asp-252 is required for cleavage of SMN. (A) Immunoblot analysis of BHK cell lysates harvested at 48 h postinfection with two independently derived recombinant viruses for each SMN construct by using anti-HA (12CA5) or anti-SMN (2B1). (B) Immunoblot analysis of mouse brain lysates prepared at 24 and 48 h after infection with the indicated viruses by using SMN monoclonal 2B1. (C) Immunoblot analysis of mouse brains after infection with two independently derived recombinant SV expressing SMN. The same lysates were analyzed with HA (12CA5), SMN (2B1), and exon-7 antibodies.
To determine if production of the apparent 29-kDa truncated SMN protein is observed in other models of neuronal injury, we examined the expression pattern of endogenous SMN in brains after transient focal ischemia of the right middle cerebral artery. At 24 h after the ischemic event, little full-length SMN was detected in either hemisphere and a 29-kDa truncated SMN-immunoreactive species was found to accumulate primarily ipsilateral to the stroke (Fig. 4B, lanes 5 and 6). Although no pathology was observed on the contralateral side, appearance of some truncated SMN is consistent with molecular signs of damage induced by reperfusion injury on the contralateral side (R.J.T., unpublished data). Taken together, the generation of a truncated 29-kDa SMN is not unique to virus-induced apoptosis, and instead may be a feature of other mechanisms of neuronal injury.
To explore the possibility that SMN is cleaved to a 29-kDa form during apoptosis, 35S-labeled, in vitro-translated SMN was treated with brain extracts. Apoptotic extracts prepared from virus-infected brains but not mock-infected brain extracts cleaved SMN to produce a 29- and 6-kDa species (Fig. 4C). This finding suggests that the 29-kDa SMN observed in injured brains may be a proteolytic cleavage product of full-length SMN rather than the product of a splice variant.
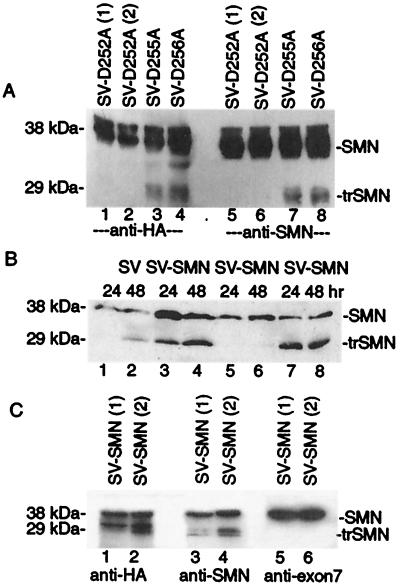
Caspases are a family of cysteine proteases that are activated during apoptosis and cleave after specific Asp residues in key substrates to facilitate apoptotic cell death (31). A C-terminal region of SMN, amino acids 249–258, contains several potential caspase cleavage sites (ICPD252, DSLD255, and SLDD256; Fig. 1A). To determine if cleavage of SMN during apoptosis is dependent on specific amino acids in this region, single point mutants D252A, D255A, and D256A were generated. These SMN mutants (with N-terminal HA tags) were expressed via the SV vector in BHK cells. Immunoblot analysis of infected cell lysates revealed that Asp-252 was required to generate the 29-kDa fragment whereas mutation of Asp-255 and Asp-256 had no effect on SMN cleavage (Fig. 5A). Detection of the cleavage fragment of SMN with both anti-HA and anti-SMN (2B1) antibodies is consistent with a proteolytic cleavage site near the C-terminal end.
To determine if SV-SMN-D252A is resistant to cleavage in brain, immunoblot analyses (with 2B1 antibody) were performed on brain lysates after intracerebral inoculation of 5-day-old mice (Fig. 5B). Lysates from control SV-, SV-SMN-, and SV-SMN-D256A-infected animals revealed the appearance of the truncated 29-kDa SMN species at 24 and 48 h postinfection (lanes 1–4, 7, and 8) which was significantly reduced with SV-SMN-D252A (lanes 5 and 6). A low level of the 29-kDa species after infection with SV-SMN-D252A is likely because of endogenous SMN protein. Thus, the D252A mutant is resistant to proteolytic processing.
To verify that the 29-kDa truncated SMN is lacking the C terminus, SV-SMN-infected brain lysates were analyzed with three different antibodies. As expected, the HA antibody, which detects the N-terminal tag, and the 2B1 SMN antibody recognized both the full-length and truncated protein (Fig. 5C). In contrast, an antibody-specific for SMN exon 7 failed to recognize truncated SMN (lanes 5 and 6), confirming that the 29-kDa SMN species lacks amino acids contained within this exon. Thus, several lines of evidence suggest that SMN is cleaved at amino acid 252 during apoptosis.
Cleavage-Resistant SMN Mutant Has Enhanced Antiapoptotic Activity.
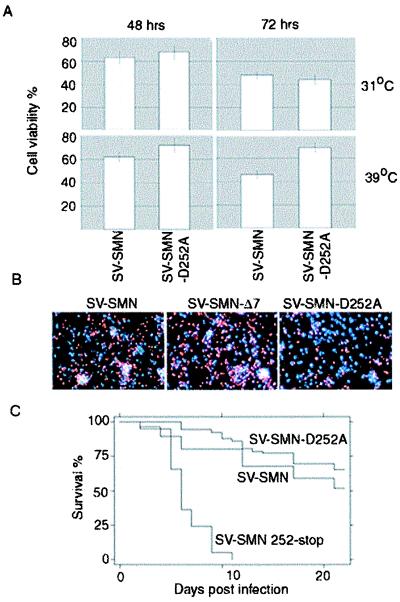
If SMN is cleaved during apoptosis, then mutation of the cleavage site would be expected to enhance the antiapoptotic function of SMN. To test this possibility, undifferentiated or neuronally differentiated CSM 14.1.4 cells were infected with SV-SMN and SV-SMN-D252A (Fig. 6 A and B). In undifferentiated cells, SMN-D252A did not alter the viability, but in neuronally differentiated cells, infection with SV-SMN-D252A resulted in significantly increased survival relative to SV-SMN. Similar results were obtained with the mouse model (Fig. 6C). Compared with SV-SMN, animals infected with SV-SMN-D252A had improved survival (P < 0.01) with 70% long-term survivors after infection with SV-SMN-D252A compared with 50% with SV-SMN. In contrast, infection of animals with a virus encoding the predicted cleavage product, SV-SMN (), resulted in high mortality that was indistinguishable from that caused by SV-SMNΔ7. We propose that cleavage of SMN during apoptosis abrogates its antiapoptotic function and creates a proapoptotic fragment.
Figure 6.
SMN-D252A has enhanced antiapoptotic function in vitro and in vivo. (A) Cell viability of undifferentiated (31°C) or differentiated (39°C) CSM 14.1.4 cells was determined as described for Fig. 1B at 48 and 72 h after infection with the indicated recombinant viruses. The means and SEM were determined in three independent experiments counting >300 cells per sample. (B) Representative fluorescence microscopy of differentiated CSM 14.1.4 cells stained with propidium iodide (red) and Hoechst (blue) at 72 h postinfection with recombinant viruses expressing SMN or derivatives. (C) Kaplan Meier survival analysis of 5-day-old mice infected with recombinant viruses as described in the legend to Fig. 2.
Discussion
The SV vector system provides a unique model system to evaluate the apoptosis regulatory functions of SMN in neurons both in culture and in vivo. By using this system, we show that full-length SMN inhibits virus-induced neuronal apoptosis whereas the SMN mutants derived from SMA patients (Y272C and SMNΔ7) enhance neuronal apoptosis. This functional correlation with wild-type and mutant SMN proteins in the SV model lends credence to the hypothesis that the loss of antiapoptotic function of SMN is important in the death of ventral motor neurons in SMA patients.
Genetic studies in mice also support a role for programmed neuronal death in SMA disease. SMN-deficient mice die during embryogenesis with massive apoptosis (32–34). SMN heterozygous mice lose 40% of their motor neurons in the first 6 months of life (35). In addition, SMN-deficient mice carrying a copy of human SMN2, which expressed predominantly SMNΔ7 transcripts, have normal numbers of motor neurons at one day of age but begin to show neuronal loss by day 3 (36). By day 5 after birth, these mice exhibit a 35% reduction in spinal cord motor neurons and increased mortality. Similarly, we observed that 5-day-old mice were more susceptible to SMNΔ7 than were 3 day olds. Taken together, these findings imply an age-dependent death of neurons in SMA rather than a defect in neurogenesis.
Our finding that SMNΔ7 is proapoptotic in neurons is somewhat surprising in light of the observation that higher copy numbers of the human SMN2 transgene in SMN-deficient mice correlate with improved prognosis (33, 36). SMN2 produces predominantly SMNΔ7 transcripts (80%) and lower levels of full-length transcripts (20%), raising the possibility that SMNΔ7 is partially protective. However, the relative protein levels of full-length vs. SMNΔ7 in motor neurons is currently unresolved and are otherwise estimated on the basis of reverse transcription–PCR analysis of mRNA levels. The generation of a stable processed form of SMN may also play a role (Figs. 4 and 5). An alternative possibility is that full-length SMN produced by SMN2 is solely responsible for the observed protective effects. This possibility is further supported by the observation that mice homozygous for SMNΔ7 (SMNΔ7/Δ7) and therefore have no full-length SMN are embryonic lethal (34). Another possibility is that the apoptotic function of SMNΔ7 is only realized after a death stimulus such as SV, ischemic injury, or developmental stimuli. However, no additional death stimulus was required for SMNΔ7 to induce apoptosis of differentiated CSM 14.1.4 cells (Fig. 1D).
Our observation that the anti- and proapoptotic functions of SMN are detected only in neurons or neuronally differentiated stem cells is consistent with the neuronal phenotype of SMA, but does not explain why SMA primarily involves ventral motor neurons. However, we have shown that another apoptosis regulator, Bax, is anti- or proapoptotic depending on the neuron subtype (20). Similarly, the potency and function of SMN may vary between neuron subsets. However, whereas ventral motor neurons are the most susceptible neuronal population in SMA, other neuronal populations may also be affected by SMN mutations. Chromatolytic cells can be found in SMA in the ventrolateral thalamus, mesencephalic nucleus, pallidum, nucleus basalis of Meynert, brainstem motor nuclei, and even diffusely throughout the cerebral cortex (37–39). Protein expression levels may also contribute to the cell type specificity of SMN because SMN protein may be more abundantly expressed in spinal cord motor neurons than in other cell types (3, 4, 33).
A 29-kDa SMN immunoreactive species was detected in apoptotic cells and brains but was largely absent from healthy cells and tissues. Cleavage of in vitro-translated SMN with apoptotic but not control cell extracts to generate a 29-kDa fragment suggests that SMN is cleaved in apoptotic cells. This 29-kDa species appears to be truncated at the C terminus and mutation of Asp-252 abolished cleavage, implying a role for caspases, the only mammalian proteases other than granzyme B that cleaves specifically after Asp. However, the 29-kDa species migrates on gels as a smaller protein than either SMNΔ7 (amino acids 1–278) or the predicted cleavage product containing amino acids 1–252 (data not shown). Therefore, an additional proteolytic event may be required to generate the 29-kDa fragment. However, this second cleavage event apparently does not occur when Asp-252 is mutated to Ala because the D252A mutant was no longer susceptible to cleavage. A similar mechanism was proposed for the generation of the β-amyloid peptide in Alzheimer's disease where cleavage of the amyloid-beta precursor protein by caspase-3 was shown to enhance formation of β-amyloid through subsequent cleavage events (40). Taken together, our results suggest that SMN may be converted from an antiapoptotic factor into a proapoptotic factor by one of several mechanisms including splicing to generate SMNΔ7, by mutations (e.g., SMN Y272C), and possibly by proteolysis. If SMN is cleaved to release a proapoptotic fragment during neuronal injury or with an apoptotic stimulus, then SMN could potentially contribute to neuronal death in other disease states. This possibility is supported by our observation that the 29-kDa processed form of SMN was also found after ischemic brain injury.
The concept that antiapoptotic proteins can be converted into proapoptotic factors by proteolysis has been observed previously for other apoptosis regulators including Bcl-2, Bcl-xL, and Bid, and has been proposed for a number of neurologic disease gene products (refs. 17, 41, and 42; R. J. Clem, T.-T. Sheu, B. W. M. Richter, W.-W. He, N. A. Thornberry, C. S. Duckett, and J. M. Hardwick, unpublished work). Bcl-2 is cleaved by caspase-3 to release a C-terminal fragment that can induce the release of cytochrome c from mitochondria, leading to amplification of the caspase cascade and cell death (43). The mechanism by which mutant/cleaved SMN stimulates cell death is not known. Mutant SMN may gain a new function like cleaved Bcl-2 and/or may function as a dominant-negative regulator of full-length SMN. Perhaps SMNΔ7 interferes with the role of full-length SMN in premRNA splicing. SMNΔ7 has lost its ability to bind SmB but retains its ability to complex with SIP-1, possibly sequestering SIP-1 in a nonfunctional state. In support of this hypothesis, infection of animals with a Sindbis-SMN construct encoding only the central region of SMN (amino acids 29–255) has neither proapoptotic nor antiapoptotic function (data not shown). The ability of SMN, Bcl-2, and other protective factors to be converted into death factors raises the possibility that they may be sensors of cellular damage and function as molecular switches. Delineation of the SMN killing pathway will provide new potential targets for therapy.
Acknowledgments
We thank G. Dreyfuss and G. Morris for antibodies; D. Irani, D. Bellows, and L. Martin for advice; and N. Crouse and S. Zou for technical assistance. This work was supported by grants from the Muscular Dystrophy Association (J.M.H. and D.A.K.), the Parkinson's Disease Foundation (D.A.K.), and Grants KO8 (D.A.K.) and NS34175 (J.M.H.) from the National Institutes of Health.
Abbreviations
- SMN
survival motor neuron
- SMA
spinal muscular atrophy
- BHK
baby hamster kidney
- CAT
chloramphenicol acetyltransferase
- HA
hemagglutinin
- SV
Sindbis virus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230364197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230364197
References
- 1.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 2.Wirth B. Hum Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Nat Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 4.Coovert D D, Thanh T L, McAndrew P E, Strasswinner J, Crawford T O, Mendell J R, Coulson S E, Androphy E J, Prior T W, Burghes A H M. Hum Mol Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 5.Liu Q, Fischer U, Wang F, Dreyfuss G. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 6.Fischer U, Liu Q, Dreyfuss G. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 7.Campbell L, Hunter K M D, Mohaghegh P, Tinsley J M, Brasch M A, Davies K E. Hum Mol Genet. 2000;9:1093–1100. doi: 10.1093/hmg/9.7.1093. [DOI] [PubMed] [Google Scholar]
- 8.Charroux B, Pellizzoni L, Perkinson R A, Shevchenko A, Mann M, Dreyfuss G. J Cell Biol. 1999;147:1181–1193. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattaj I W. Curr Biol. 1998;8:R93–R95. doi: 10.1016/s0960-9822(98)70055-7. [DOI] [PubMed] [Google Scholar]
- 10.Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 11.Iwahashi H, Eguchi Y, Yasuhara N, Hanafusa T, Matsuzawa Y, Tsujimoto Y. Nature (London) 1997;390:413–417. doi: 10.1038/37144. [DOI] [PubMed] [Google Scholar]
- 12.Sato K, Eguchi Y, Kodama T S, Tsujimoto Y. Cell Death Differ. 2000;7:374–383. doi: 10.1038/sj.cdd.4400660. [DOI] [PubMed] [Google Scholar]
- 13.Talbot K, Miguel-Aliaga I, Mohaghegh P, Ponting C P, Davies K E. Hum Mol Genet. 1998;7:2149–2156. doi: 10.1093/hmg/7.13.2149. [DOI] [PubMed] [Google Scholar]
- 14.Cheng E H Y, Levine B, Boise L H, Thompson C B, Hardwick J M. Nature (London) 1996;379:554–556. doi: 10.1038/379554a0. [DOI] [PubMed] [Google Scholar]
- 15.Levine B, Goldman J E, Jiang H H, Griffin D E, Hardwick J M. Proc Natl Acad Sci USA. 1996;93:4810–4815. doi: 10.1073/pnas.93.10.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardwick J M, Levine B. Methods Enzymol. 2000;322:492–508. doi: 10.1016/s0076-6879(00)22045-4. [DOI] [PubMed] [Google Scholar]
- 17.Clem R J, Cheng E H Y, Karp C L, Kirsch D G, Ueno K, Takahashi A, Kastan M B, Griffin D E, Earnshaw W C, Hardwick J M. Proc Natl Acad Sci USA. 1998;95:554–559. doi: 10.1073/pnas.95.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duckett C S, Nava V E, Gedrich R W, Clem R J, Van Dongen J L, Gilfillan M C, Shiels H, Hardwick J M, Thompson C B. EMBO J. 1996;15:2685–2694. [PMC free article] [PubMed] [Google Scholar]
- 19.Nava V E, Rosen A, Veliuona M A, Clem R J, Levine B, Hardwick J M. J Virol. 1998;72:452–459. doi: 10.1128/jvi.72.1.452-459.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis J, Oyler G A, Ueno K, Fannjiang Y, Chau B N, Vornov J, Korsmeyer S J, Zou S, Hardwick J M. Nat Med. 1999;5:832–835. doi: 10.1038/10556. [DOI] [PubMed] [Google Scholar]
- 21.Bellows D S, Chau B N, Lee P, Lazebnik Y, Burns W H, Hardwick J M. J Virol. 2000;74:5024–5031. doi: 10.1128/jvi.74.11.5024-5031.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis J, Wesselingh S L, Griffin D E, Hardwick J M. J Virol. 1996;70:1828–1835. doi: 10.1128/jvi.70.3.1828-1835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine B, Huang Q, Isaacs J T, Reed J C, Griffin D E, Hardwick J M. Nature (London) 1993;361:739–742. doi: 10.1038/361739a0. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh A, Greenberg M E. Neuron. 1995;15:89–103. doi: 10.1016/0896-6273(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 25.Anton R, Kordower J H, Maidment N T, Manaster J S, Kane D J, Rabizadeh S, Schueller S B, Yang J, Rabizadeh S, Edwards R H, et al. Exp Neurol. 1994;127:207–218. doi: 10.1006/exnr.1994.1097. [DOI] [PubMed] [Google Scholar]
- 26.Young P J, Le T T, thi Man N, Burghes A H M, Morris G E. Exp Cell Res. 2000;256:365–374. doi: 10.1006/excr.2000.4858. [DOI] [PubMed] [Google Scholar]
- 27.Liu Q, Dreyfuss G. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 28.Britton P, Lu X C, Laskosky M S, Tortella F C. Life Sci. 1997;60:1729–1740. doi: 10.1016/s0024-3205(97)00132-x. [DOI] [PubMed] [Google Scholar]
- 29.Longa E Z, Weinstein P R, Carlson S, Cummins R. Stroke (Dallas) 1989;20:84–91. doi: 10.1161/01.str.20.1.84. [DOI] [PubMed] [Google Scholar]
- 30.Taylor R M, Hurlbut H S, Work T H, Kingston J R, Frothingham T E. Am J Trop Med Hyg. 1955;4:844–862. doi: 10.4269/ajtmh.1955.4.844. [DOI] [PubMed] [Google Scholar]
- 31.Thornberry N A, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 32.Schrank B, Gotz R, Gunnersen J M, Ure J M, Toyka K S, Smith A G, Sendtner M. Proc Natl Acad Sci USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh-Li H M, Chang J-G, Jong Y-J, Wu M-H, Wang N M, Tsai C H, Li H. Nat Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 34.Frugier T, Tiziano F D, Cifuentes-Diaz C, Miniou P, Roblot N, Dierich A, Le Meur M, Melki J. Hum Mol Genet. 2000;9:849–858. doi: 10.1093/hmg/9.5.849. [DOI] [PubMed] [Google Scholar]
- 35.Jablonka S, Schrank B, Kralewski M, Rossoll W, Sendtner M. Hum Mol Genet. 2000;9:341–346. doi: 10.1093/hmg/9.3.341. [DOI] [PubMed] [Google Scholar]
- 36.Monani U R, Sendtner M, Coovert D D, Parsons D W, Andreassi C, Thanh T T, Jablonka S, Schrank B, Rossol W, Prior T W, et al. Hum Mol Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 37.Towfighi J, Young R S, Ward R M. Acta Neuropathol. 1985;65:270–280. doi: 10.1007/BF00687008. [DOI] [PubMed] [Google Scholar]
- 38.Bingham P M, Shen N, Rennert H, Rorke L B, Black A W, Marin-Padilla M M, Nordgren R E. Neurology. 1997;49:848–851. doi: 10.1212/wnl.49.3.848. [DOI] [PubMed] [Google Scholar]
- 39.Crawford T O, Pardo C A. Neurobiol Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 40.Gervais F G, Xu D, Robertson G S, Vaillancourt J P, Zhu Y, Huang J, LeBlanc A, Smith D, Rigby M, Shearman M S, et al. Cell. 1999;97:395–406. doi: 10.1016/s0092-8674(00)80748-5. [DOI] [PubMed] [Google Scholar]
- 41.Cheng E H Y, Kirsch D G, Clem R J, Ravi R, Kastan M B, Bedi A, Ueno K, Hardwick J M. Science. 1997;278:1966–1968. doi: 10.1126/science.278.5345.1966. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Zhu H, Xu C, Yuan J. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 43.Kirsch D G, Doseff A, Chau B N, Lin D-S, de Souza-Pinto N C, Hansford R, Kastan M B, Lazebnik Y A, Hardwick J M. J Biol Chem. 1999;274:21155–21161. doi: 10.1074/jbc.274.30.21155. [DOI] [PubMed] [Google Scholar]