Abstract
We have developed a one-step facile, flexible and readily scalable purification method for a recombinant protein, TM 1–99 (113 amino acid residues; 12,837 Da) based on reversed-phase high-performance liquid chromatography (RP-HPLC) from an E. coli cell lysate. Following cell lysis, the cell contents were extracted with 0.1% aqueous trifluoroacetic acid (TFA), applied directly under conditions of high sample load to a narrow bore RP-HPLC C8 column (150 mm × 2.1 mm I.D.) and eluted by a shallow gradient of acetonitrile (0.1%/min). Loads of 23 and 48 mg of lyophilized crude cell extract produced 2.4 and 4.2 mg of purified product (>94% pure), respectively, at >94% recovery. Our results show the excellent potential of one-step RP-HPLC for purification of recombinant proteins from cell lysates, where high yields of purified product and greater purity are achieved compared to affinity chromatography. Such an approach was also successful in purifying just trace levels (<0.1% of total contents of crude sample) of TM 1–99 from a cell lysate.
Keywords: Reversed-phase high-performance liquid chromatography, Recombinant proteins, Preparative purification, One-step purification
1. Introduction
Fusion-based affinity purification systems have frequently been used to carry out purification of a wide variety of different protein products [1,2]. Indeed, both affinity and immunoaffinity chromatography techniques are often critical in developing purification procedures for recombinant proteins, often as the first step in a multicolumn purification approach [3-6]. Such an approach may be required for the frequently arduous and time-consuming purification of recombinant proteins to homogeneity, particularly during sample scale-up. Although multicolumn protocols (generally employing combinations of affinity, ion-exchange and size-exclusion chromatography) are effective [7-13], complications remain with such issues as sample handling, concentration steps, dialysis, etc., leading to an increase in purification time as well as a concomitant loss of purified sample yield. Further, even if a one-step affinity approach to purification of a protein on a small scale was favoured, scale-up to larger sample amounts may become prohibitively expensive.
The excellent resolving power of reversed-phase high-performance liquid chromatography (RP-HPLC) has made it the predominant technique for peptide separations and many protein separations both for analytical purposes and for scale-up for preparative purification [13,14]. Despite the widespread use of this technique for such applications, there has often been reluctance on the part of researchers concerned with protein denaturation and, thus, activity loss, to take advantage of RP-HPLC due to its denaturing environment. Thus, the hydrophobicity of the stationary phases characteristic of RP-HPLC as well as typical mobile phases (organic solvents such as acetonitrile for solute elution; low pH values) are indeed denaturing of tertiary and quaternary structure [14-17]. However, researchers frequently employ denaturing agents (heat, acidic or basic conditions, miscible organic solvents, urea and guanidine hydrochloride, detergents) for protein purification purposes [18] which are at least as denaturing as RP-HPLC conditions. Organic solvents, urea and detergents (and, indeed, hydrophobic stationary phases of RP-HPLC) act primarily by disrupting hydrophobic interactions stabilizing protein tertiary and quaternary structure. The use of detergents is widespread for the solubilization of membrane proteins during isolation [19-21]. Further, low pH, which disrupts salt bridges, is commonly used for elution of proteins from affinity columns [22,23]. However, once the denaturants are removed (whether this involves, e.g., detergent, organic solvent, or hydrophobic stationary phase removal or a return to physiological pH), except where researchers are unaware of non-covalently-linked cofactors or metal ions or that chaperones are required for protein folding, proteins are generally able to refold and acquire their native conformation spontaneously together with recovery of activity when the correct refolding procedures are utilized [18,24-29].
Our laboratory has demonstrated the value of slow acetonitrile gradients (0.1–0.2% acetonitrile/min) to utilize more efficiently the hydrophobic stationary phase of RP-HPLC for peptide separations compared to the more traditionally employed conditions of 0.5–1% acetonitrile/min) [30-32]. Under such conditions, we have demonstrated efficient preparative purification of up to 30 mg of a 26-residue synthetic peptide crude on a 4.6 mm I.D. column and up to 200 mg of this same peptide on a 9.4 mm I.D. column on instrumentation designed primarily for analytical work [32]. Our approach, compared to standard scale-up approaches, avoids the necessity for the use of increasingly larger columns in order to maintain satisfactory levels of product purity and yield with concomitant higher operating costs in terms of packing, equipment and solvents. In an analogous manner, we now set out to develop a general one-step facile, flexible and readily scalable purification method for proteins, specifically recombinant proteins, based on RP-HPLC.
2. Experimental
2.1. Materials
HPLC-grade water was prepared by an E-pure water purification system from Barnstead International (Dubuque, IA, USA). Trifluoroacetic acid (TFA) was obtained from Halocarbon products (River Edge, NJ, USA). Acetonitrile was obtained from EM Science (Gibbstown, NJ, USA). Centricon centrifugal filter units were obtained from Millipore (Billerica, MA, USA).
2.2. Column and instrumentation
All preparative and analytical runs were carried out on a Zorbax 300SB-C8 column (150 mm × 2.1 mm I.D.; 3.5 μm particle size, 300 Å pore size) from Agilent Technologies (Little Falls Site, DE, USA). Preparative runs were carried out on a Beckman Coulter System Gold Liquid Chromatograph. Fraction analysis following preparative chromatography was carried out on an Agilent 1100 liquid chromatography system.
2.3. Construction of truncated α-tropomyosin recombinant protein
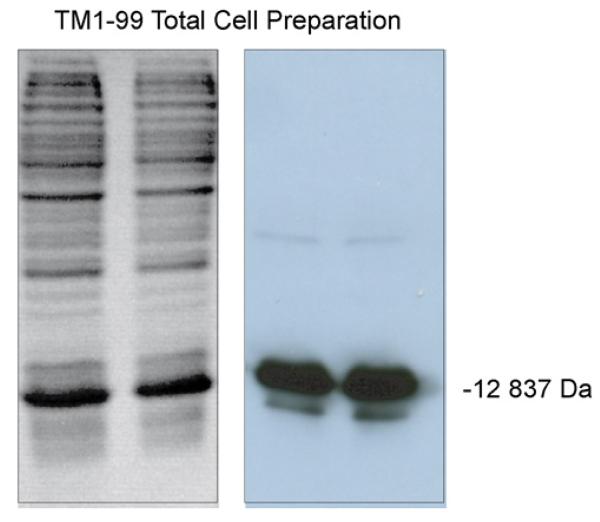
Polymerase chain reaction (PCR) primers were designed to utilize the native N-terminus of chicken skeletal α-tropomyosin but to truncate the downstream length to include residues 1–99. Thus, the downstream frame adds a stop codon after residue 99. Each primer contained a Bam H1 site for subcloning into the pET3a expression vector (Novagen). The construct was verified by DNA sequencing. The recombinant protein (TM 1–99) contains the T7 Tag at the N-terminus for easy identification during the original affinity purification strategy. The T7 Tag adds 14 amino acids (sequence: MASMTGGQQMGRGS) to the length of the protein for a total of 113 amino acids. The fusion protein was over-expressed in E. coli using induction with 0.4 mM IPTG (isopropylthio-β-galactoside) in 300 ml L-Broth culture containing ampicillin. TM 1–99 is then isolated from the soluble cytoplasmic fraction and extracted with 0.1% TFA for HPLC purification or with 20 mM Tris–HCl, pH 7.5, for antibody resin purification. The resuspended cells were sonicated and centrifuged at 10,000 × g for 10 min for a cleared lysate. Analysis of the crude lysate on 15% SDS–polyacrylamide gel electrophoresis (PAGE) gels stained with coomassie brilliant blue revealed a dominant protein in the soluble fraction with a molecular mass of 12,837 Da. Subsequent Western Blot analysis with detection by antibody specific for the T7 Tag (Novagen Lumiblot Kit) confirmed the correct size of the tagged product (Fig. 1).
Fig. 1.
Analysis of crude cell lysate by SDS–PAGE (15% gel). Left: SDS–PAGE gel of lysate stained with coomassie brilliant blue. Right: Western Blot analysis of gel with detection by antibody specific for the T7 tag on TM 1–99 (molecular mass of 12,837 Da).
2.4. HPLC conditions
2.4.1. Analytical RP-HPLC
Analytical runs and fraction analyses were carried out by a linear AB gradient (1% B/min for analytical profiles and 2% B/min for fraction analysis) at a flow-rate of 0.3 ml/min where Eluent A is 0.05% aq. TFA and Eluent B is 0.05% TFA in acetonitrile; temperature, 25 °C. TFA concentrations of 0.05–0.1% are characteristic of most separations of peptides and proteins by RP-HPLC (for both analytical and scale-up purposes [14,15]. In addition, a gradient rate of 1% B/min is a good compromise between run time and sample resolution prior to scale-up to significantly higher sample loads.
2.4.2. Preparative RP-HPLC
Preparative purification was carried out by a linear AB gradient (Eluent A is 0.05% aq. TFA, pH 2.0, and Eluent B is 0.05% TFA in acetonitrile) of 2% B/min up to 24% B, followed by a slow (0.1% B/min) gradient up to 40% B. A rapid rise (4% B/min) up to 60% B was then followed by an isocratic wash with 60% B. An analytical run of a crude sample mixture of TM 1–99 demonstrated that the desired product and impurities were completely eluted from the column by the time this acetonitrile concentration had been reached (Fig. 2). Flow-rate, 0.3 ml/min; temperature, 25 °C; 2 min fractions were collected.
Fig. 2.
Analytical RP-HPLC of crude sample mixture containing recombinant protein TM 1–99. Conditions: linear AB gradient (1% B/min) at a flow-rate of 0.3 ml/min, where Eluent A is 0.05% aq. TFA and Eluent B is 0.05% TFA in acetonitrile; temperature, 25 °C. P denotes desired product.
Following fraction analysis, three fraction pools containing either hydrophilic impurities, purified TM 1–99 or hydrophobic impurities were freeze-dried and subsequently dissolved in an equal volume of 0.05% aq. TFA prior to analysis.
2.5. Affinity purification of TM 1–99
Affinity purification of the fusion protein TM 1–99 was performed using the T7 Tag Affinity Purification Kit (Novagen). A 1 ml bed volume of the T7 Tag Antibody Agarose was packed by gravity flow in a 90 mm × 8 mm I.D. column and washed with 10 vol. of 1× bind/wash buffer (4.29 mM Na2HPO4, 1.46 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, 0.1% Tween 20, 0.002% sodium, pH 7.3). The crude lysate was then loaded onto the column with concomitant collection of flow-through eluent. The bound fusion protein was subsequently washed with 10 volumes of 1× bind/wash buffer to elute unbound contaminants and eluted with 5× 1-ml aliquots of 100 mM citric acid, pH 2.2, into 0.15 ml of neutralization buffer (2 M Tris base, pH 10.4). The column was then washed with 10 volumes of 1× bind/wash buffer for regeneration prior to subsequent purifications. The purified protein was then concentrated in a Centricon centrifugal filter unit.
3. Results and discussion
Fig. 2 shows the analytical RP-HPLC profile of the crude cell lysate where the cell contents were extracted with 0.1% aq. TFA.
3.1. Affinity purification of TM 1–99
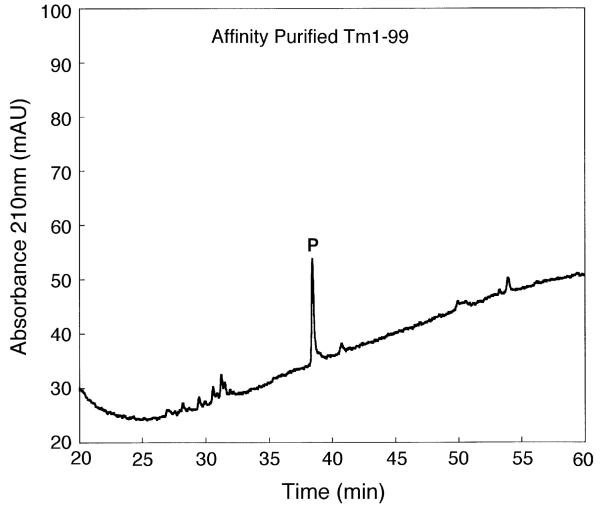
Fig. 3 shows a one-step affinity purification of TM 1–99 from the crude cell lysate (see Section 2.5). This purification resulted in partially purified TM 1–99 (64% purity as measured by peak integration). Subsequent purification of this partially purified product by RP-HPLC (i.e., a two-step affinity/RP-HPLC protocol) produced a product with an excellent purity of >99% (separation not shown). However, this two-step protocol resulted in a considerable reduction in protein yield compared to the one-step affinity purification (Table 1). Such sample losses are not uncommon during the frequent sample handling characteristic of multi-purification protocols and, hence, our desire to simplify such a process.
Fig. 3.
One-step affinity purification of TM 1–99. Column and conditions: see Section 2. P denotes desired product.
Table 1.
Comparison of one-step RP-HPLC and affinity chromatography of TM 1–99
| Chromatography | Purity | Yield | Figure |
|---|---|---|---|
| One-step affinity | >64% | 1× | 3 |
| Two-step affinity + RP-HPLC | >99% | 0.1× | – |
| One-step RP-HPLC | >94% | 10× | 4 |
It should be noted that maximum capacity of the affinity resin is reported by the manufacturer as being 300 μg T7 tagged protein/ml of resin. In the present study, the yield of protein from one pass of lysate through the T7 Tag Antibody resin was approximately only 20 μg, with an abundance of TM 1–99 in the flow-through. Subsequent multiple passes of the flow-through yielded the same amount of protein/pass. Interestingly, use of the T7 Tag Antibody resin with a different fusion protein (T7 Tag plus full length calmodulin) gave an excellent yield per protein pass through the resin, much closer to the reported potential maximum, i.e., the amount of protein bound is dependent on the specific protein under investigation. Such variability is possibly due to placement and accessibility of the tag within the folded protein affecting binding efficiency. Thus, the maximum load on the affinity column must be determined empirically for each protein of interest.
3.2. One-step preparative RP-HPLC of TM 1–99
3.2.1. Purification protocol
The protocol developed as a flexible purification approach was as follows: a rapid (2% acetonitrile/min) rise to an acetonitrile concentration 15% below that of the eluting concentration during an analytical gradient elution run. This is followed by a slow gradient (e.g., 0.1–0.2% acetonitrile/min, depending on the complexity of the crude sample), a rapid rise to a concentration of acetonitrile known from an analytical run to be enough to ensure elution of all crude sample components from the column and, finally, by an isocratic wash at this high concentration of acetonitrile. A key advantage of such a slow gradient approach is the high sample loading concentrating the desired material on the column, thus aiding its separation from closely adjacent impurities. In addition, the slow gradient spreads the desired product over a large number of fractions, the bulk of which would contain purified product only, with just the first and last product fractions containing hydrophilic and hydrophobic impurities, respectively. The above “rule of thumb” approach whereby the shallow gradient is started 15% below the acetonitrile concentration required to elute the protein of interest in an analytical run can be applied as a general rule for polypeptides of 10-residue length and longer. This is due to the mechanism of multisite binding of peptides and proteins to the hydrophobic matrix (as opposed to the partitioning mechanism of small organic molecules) preventing their elution until the acetonitrile concentration approaches that required to elute them during an analytical run, i.e., peptides and proteins exhibit only narrow partitioning windows [33]. Indeed, such windows become narrower with increasing polypeptide chain length [33]. Another key advantage of starting the shallow gradient at this acetonitrile concentration is that a large range of sample loads may be separated using this same approach. Finally, in our experience, starting the shallow gradient at 15% acetonitrile concentration below that required to elute the product of interest is a good compromise between the time taken for efficient product purification and obtaining maximum sample load, i.e., a lower or greater starting acetonitrile concentration would extend the run time or decrease maximum sample load, respectively.
3.2.2. Preparative RP-HPLC of TM 1–99
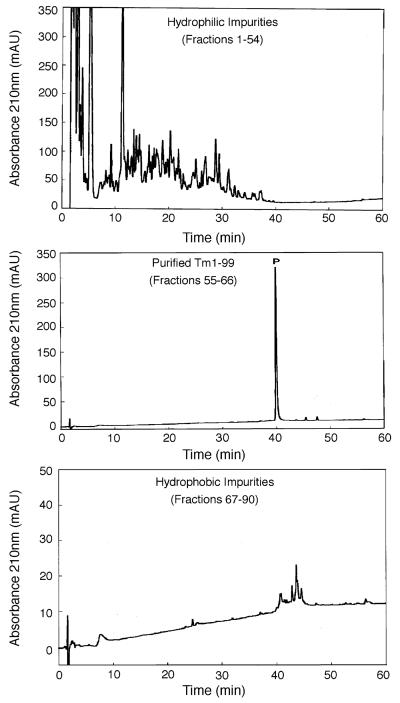
Fig. 4 shows the one-step RP-HPLC purification of TM 1–99 (analytical RP-HPLC profile of crude sample shown in Fig. 2), where the desired protein is clearly identifiable due to over-expression. This purification was achieved by the purification protocol described above (and under Section 2) using a gradient rate of 0.1% acetonitrile/min. The sample load (23 mg of lyophilized crude cell extract) represented 7× the sample load used in the one-step affinity purification shown in Fig. 3. From Fig. 4, an excellent purification of TM 1–99 was achieved, with negligible overlap between purified product and hydrophilic or hydrophobic impurities. The purified product (2.4 mg in total) was >94% pure (Table 1) with >90% recovery.
Fig. 4.
One-step preparative RP-HPLC purification of TM 1–99. Conditions: linear AB gradient (Eluent A is 0.05% aq. TFA, pH 2.0 and Eluent B is 0.05% TFA in acetonitrile) of 2% B/min up to 24% B, followed by a gradient of 0.1%/min up to 40% B. A gradient of 4% B/min up to 60% B was then followed by an isocratic wash with 60% B; flow-rate, 0.3 ml/min; temperature, 25 °C. 2 min fractions were collected and subjected to the same analytical conditions as Fig. 1 except for a gradient rate of 2%B/min. Sample volume: 4 ml, containing 23 mg of lyophilized crude cell extract. P denotes desired product.
A second purification was carried out with approximately double the sample load (48 mg) of the run shown in Fig. 4. Excellent purity (>94% pure) and yield of purified product (4.2 mg, representing about 90% recovery) was again achieved. However, a further increase in sample load to 100 mg of cell extract produced just 6.4 mg of purified product, representing about 65% recovery. TM 1–99 was observed in the flow-through following sample loading, together with some overlap of the desired product and hydrophilic impurities, indicating that the capacity of this small column had been exceeded. Nevertheless, even under such non-optimal load conditions, a significant amount of purified product (>95% pure) was still obtained, highlighting the “user-friendliness” of this preparative approach.
3.2.3. One-step preparative RP-HPLC of trace levels of TM 1–99
On occasion, problems may occur which reduce desired yields of expressed proteins, e.g., inadequate lysing of cells. A marked example of such a problem is illustrated in Fig. 5 (top), which shows the analytical elution profile of a cell lysate where expression of TM 1–99 was poor and represented <0.1% of total contents of the crude sample. Following a one-step preparative separation (0.1% acetonitrile/min), a very effective separation of TM 1–99 was achieved (Fig. 5, bottom panels) with a purity of >94%. These results highlight the efficacy of the slow gradient approach, with concomitant concentration of desired product, to purifying even trace levels of product from a crude lysate.
Fig. 5.
One-step preparative RP-HPLC purification of trace levels of TM 1–99. Conditions: same as Fig. 4. Sample volume: 5 ml (60 ml of lysate had been freeze-dried and redissolved in 5 ml of 0.1% aq. TFA). The top panel shows the analytical RP-HPLC profile of the crude sample mixture. P denotes desired product which represents <0.1% of total contents of crude sample.
3.2.4. Comparison of one-step RP-HPLC and affinity chromatography of TM 1–99
From Table 1, the one-step RP-HPLC procedure produced a 10× improvement in yield of highly purified material over that of one-step affinity chromatography. Even with optimum binding of the TM 1–99 to the resin, a considerable number of affinity chromatography runs and/or expensive scale-up of the resin would be required to achieve yields of purified product approaching that of the one-step RP-HPLC procedure. In addition, the affinity-purified material would still require further purification to match that of the RP-HPLC protocol, with subsequent loss of product due to a multi-step procedure. The one-step RP-HPLC procedure produced a 100× improvement in yield of highly purified product compared to a two-step affinity/RP-HPLC approach, i.e., a considerable amount of product was lost when combining the affinity and RP-HPLC modes. Further, only a small improvement in product purity was achieved by the two-step approach (>99%) compared to our favoured one-step protocol (>94%).
Although other methods have been developed for the purification of peptides which make effective use of the hydrophobic stationary phase (e.g., sample displacement chromatography, whereby separation is achieved in the absence of organic modifier [34-37], and a two-step, step gradient isocratic approach [38]), such methods are not suitable for proteins. In addition, these methods may require more developmental time compared to the present slow gradient approach.
4. Conclusions
The present study demonstrated that a one-step RP-HPLC slow gradient approach shows excellent potential for purification of recombinant proteins from cell lysates, where high yields of purified product are achieved compared to affinity chromatography.
Acknowledgements
This work was supported by an NIH grant (RO1 GM61855 to R.S.H.) and the John Stewart Chair in Peptide Chemistry. We thank Dr. L.B. Smillie at the University of Alberta for providing the tropomyosin gene.
References
- 1.Terpe K. Appl. Microbiol. Biotechnol. 2003;60:523. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- 2.Waugh DS. Trends Biotechnol. 2005;23:316. doi: 10.1016/j.tibtech.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Goshe MB, Conrads TP, Panisko EA, Angell NH, Veenstra TD, Smith RD. Anal. Chem. 2001;73:2578. doi: 10.1021/ac010081x. [DOI] [PubMed] [Google Scholar]
- 4.Feng B, Patel AH, Keller PM, Slemmon JR. Rapid Commun. Mass Spectrom. 2001;15:821. doi: 10.1002/rcm.276. [DOI] [PubMed] [Google Scholar]
- 5.Wehr T. Pharmagenomics. 2003;3:36. [Google Scholar]
- 6.Sakanayan V. J. Chromatogr. B. 2005;815:77. doi: 10.1016/j.jchromb.2004.08.045. [DOI] [PubMed] [Google Scholar]
- 7.Wagner K, Racaityte K, Unger KK, Miliotis T, Edholm LE, Bischoff R, Marko-Varga G. J. Chromatogr. A. 2000;893:293. doi: 10.1016/s0021-9673(00)00736-6. [DOI] [PubMed] [Google Scholar]
- 8.Washburn MP, Wolters D, Yates JR., III Nature Biotech. 2001;19:242. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 9.Wagner K, Miliotis T, Marko-Varga G, Bischoff R, Unger KK. Anal. Chem. 2002;74:809. doi: 10.1021/ac010627f. [DOI] [PubMed] [Google Scholar]
- 10.Issaq HJ, Conrads TP, Janini GM, Veenstra TD. Electrophoresis. 2002;23:3048. doi: 10.1002/1522-2683(200209)23:17<3048::AID-ELPS3048>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Hanash S. J. Chromatogr. B. 2003;787:11. doi: 10.1016/s1570-0232(02)00335-5. [DOI] [PubMed] [Google Scholar]
- 12.Millea KM, Kass IJ, Cohen SA, Krull IS, Gebler JC, Berger SJ. J. Chromatogr. A. 2005;1079:287. doi: 10.1016/j.chroma.2005.04.048. [DOI] [PubMed] [Google Scholar]
- 13.Neverova I, Van Eyk JE. J. Chromatogr. B. 2005;815:51. doi: 10.1016/j.jchromb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Mant CT, Hodges RS, editors. High-Performance Liquid Chromatography of Peptides and Proteins: Separation, Analysis and Conformation. CRC Press; Boca Raton, FL: 1991. [Google Scholar]
- 15.Mant CT, Hodges RS. In: HPLC of Biological Macromolecules. second ed. Gooding KM, Regnier FE, editors. Marcel Dekker; New York: 2002. p. 433. [Google Scholar]
- 16.Lau SYM, Taneja AK, Hodges RS. J. Biol. Chem. 1984;259:13253. [PubMed] [Google Scholar]
- 17.Mant CT, Zhou NE, Hodges RS. J. Chromatogr. 1989;476:363. doi: 10.1016/s0021-9673(01)93882-8. [DOI] [PubMed] [Google Scholar]
- 18.Marston FAO. Biochem. J. 1986;240:1. doi: 10.1042/bj2400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bordier C. J. Biol. Chem. 1981;256:1604. [PubMed] [Google Scholar]
- 20.Winkler G, Heinz FX, Guirakhoo F, Kunz C. J. Chromatogr. 1985;326:113. doi: 10.1016/s0021-9673(01)87436-7. [DOI] [PubMed] [Google Scholar]
- 21.Welling-Wester S, Feijbrief M, Koedijk DGAM, Welling GW. J. Chromatogr. A. 1998;816:29. doi: 10.1016/s0021-9673(98)00288-x. [DOI] [PubMed] [Google Scholar]
- 22.Janson J-C, Rydén L, editors. Protein Purification. VCH Publishers, Inc.; New York: 1989. [Google Scholar]
- 23.Burke D. In: Basic HPLC and CE of Biomolecules. Cunico RL, Gooding KM, Wehr T, editors. Bay Bioanalytical Laboratory; Richmond, CA: 1998. p. 223. [Google Scholar]
- 24.Anfinsen CB. Science. 1973;181:223. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 25.Wulfing C, Pluckthun A. Mol. Microbiol. 1994;12:685. doi: 10.1111/j.1365-2958.1994.tb01056.x. [DOI] [PubMed] [Google Scholar]
- 26.Krek W, Ewen ME, Shirodkar S, Arany Z, Kaelin WG, Jr., Livingston DM. Cell. 1994;78:161. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 27.Roberts SG, Green MR. Nature. 1994;371:717. doi: 10.1038/371717a0. [DOI] [PubMed] [Google Scholar]
- 28.Burgess RR. Methods Enzymol. 1996;273:145. doi: 10.1016/s0076-6879(96)73014-8. [DOI] [PubMed] [Google Scholar]
- 29.Joneson T, McDonough M, Bar-Sagi D, Van Aelst L. Science. 1996;274:1374. doi: 10.1126/science.274.5291.1374. [DOI] [PubMed] [Google Scholar]
- 30.Mant CT, Burke TWL, Hodges RS. J. Chromatographia. 1987;24:565. [Google Scholar]
- 31.Parker JMR, Mant CT, Hodges RS. Chromatographia. 1987;24:832. [Google Scholar]
- 32.Chen Y, Mant CT, Hodges RS. J. Chromatogr. A, submitted for publication. [Google Scholar]
- 33.Hodges RS, Mant CT. In: High-Performance Liquid Chromatography of Peptides and Proteins: Separation, Analysis and Conformation. Mant CT, Hodges RS, editors. CRC Press; Boca Raton, FL: 1991. p. 3. [Google Scholar]
- 34.Hodges RS, Burke TWL, Mant CT. J. Chromatogr. 1988;444:349. doi: 10.1016/s0021-9673(01)94036-1. [DOI] [PubMed] [Google Scholar]
- 35.Burke TWL, Mant CT, Hodges RS. J. Liq. Chromatogr. 1988;11:1229. [Google Scholar]
- 36.Hodges RS, Burke TWL, Mant CT. J. Chromatogr. 1991;548:267. doi: 10.1016/s0021-9673(01)88608-8. [DOI] [PubMed] [Google Scholar]
- 37.Husband DL, Mant CT, Hodges RS. J. Chromatogr. A. 2000;893:81. doi: 10.1016/s0021-9673(00)00751-2. [DOI] [PubMed] [Google Scholar]
- 38.Mehok AR, Mant CT, Gera L, Hodges RS. J. Chromatogr. A. 2002;972:87. doi: 10.1016/s0021-9673(02)01076-2. [DOI] [PubMed] [Google Scholar]