Abstract
MicroRNAs are small regulatory RNAs that post-transcriptionally regulate gene expression and can be encoded by viral as well as cellular genomes. The functions of most viral miRNAs are unknown and few have been studied in an in vivo context. Here we show that the murine polyomavirus (PyV) encodes a precursor microRNA that is processed into two mature microRNAs, both of which are active at directing the cleavage of the early PyV mRNAs. Furthermore, we identify a deletion mutant of polyomavirus that is defective in encoding the microRNAs. This mutant replicates normally and transforms cultured cells with efficiencies comparable to wildtype PyV. The miRNA mutant is competent to establish a transient infection of mice following parenteral inoculation, and is cleared post infection at approximately the same rate as the wildtype virus. In addition, under these laboratory conditions, we observe no differences in anti-viral CD8 T cell responses. These results indicate that PyV miRNA expression is not essential for infection of cultured cells or experimentally inoculated mice, and raise the possibility that its role in natural infection might involve aspects of acquisition or spread that are not recapitulated by experimental inoculation.
Keywords: Polyomavirus, Simian Virus 40, SV40, virus, microRNA, RNA interference
MicroRNAs (miRNAs) are small (approximately 22 nucleotide) RNA molecules that regulate gene expression (reviewed in (Bartel, 2004; Carthew, 2006; He and Hannon, 2004; Kim, 2005)). MiRNAs are processed from a longer primary transcript containing a region of secondary structure that folds into an ~100–120 nucleotide hairpin structure. This long hairpin region is recognized and excised by the nuclear multi-protein microprocessor complex into a 50–70 nucleotide hairpin precursor miRNA (pre-miRNA) (Denli et al., 2004; Gregory et al., 2004; Seitz and Zamore, 2006). The pre-miRNA is exported to the cytoplasm and is further processed by the Dicer endonuclease (Bohnsack, Czaplinski, and Gorlich, 2004; Hutvagner et al., 2001; Ketting et al., 2001; Lund et al., 2004; Yi et al., 2003), eventually becoming the mature single-stranded, 22 nucleotide effector molecule bound by the multiprotein RNA Induced Silencing Complex (RISC) (Kim, 2005). Typically, only one arm of the pre-miRNA hairpin is favored to be active within RISC (Matranga et al., 2005). For the most part, RISC-bound miRNAs bind with imperfect complementarity to the 3′UTR of a target mRNA. This results in translational repression, probably through multiple mechanisms (Filipowicz, Bhattacharyya, and Sonenberg, 2008). In addition, if a miRNA has perfect complementarity to an mRNA target, it can guide RISC to specifically cleave the mRNA target in UTR as well as non-UTR regions of the transcript (Doench, Petersen, and Sharp, 2003; Hutvagner and Zamore, 2002; Zeng, Yi, and Cullen, 2003). This mechanism is analogous to cleavage mediated by small-interfering RNAs (siRNAs). However, miRNAs that mediate cleavage in an siRNA-like fashion are rare in animal cells. In fact, there is only a single mammalian miRNA known to function in this manner (Mansfield et al., 2004; Yekta, Shih, and Bartel, 2004). MiRNAs have generated enormous interest, in part, because they have been implicated as playing regulatory roles in numerous and diverse developmental and cellular processes. Recently, miRNAs encoded by viruses have been discovered, and increasing evidence suggests that both host and viral-encoded miRNAs play an important role in viral infection.
Viral-encoded microRNAs have been discovered in several virus families (reviewed in (Cullen, 2006; Grey, Hook, and Nelson, 2008; Nair and Zavolan, 2006; Pfeffer and Voinnet, 2006; Samols and Renne, 2006; Sarnow et al., 2006; Sullivan, 2008; Sullivan and Ganem, 2005b)). Most viral miRNAs have been identified in members of the herpesviral family. The macaque and human polyomaviruses Simian Virus 40 (SV40), JCV, and BKV have also been shown to encode miRNAs with shared, partial sequence identity (Seo et al., 2008; Sullivan et al., 2006; Sullivan et al., 2005). The function of the vast majority of viral miRNAs remains unknown, but recent progress has been made in identifying molecular targets of several viral miRNAs (Barth et al., 2008; Gottwein et al., 2007; Grey et al., 2007; Lo et al., 2007; Pfeffer et al., 2004; Samols et al., 2007; Skalsky et al., 2007; Stern-Ginossar et al., 2007). Viral miRNAs have been described that target host or viral transcripts, via either translational repression or through siRNA-like mediated cleavage. However, an in depth understanding of the in vivo functions of viral miRNAs is lacking.
Murine polyomavirus (PyV) was the first polyomavirus discovered (reviewed in (Benjamin, 2001)). When some strains of newborn mice are infected in a laboratory setting, PyV induces numerous tumors in multiple tissue types (Benjamin, 2001; Cole, 1996). As such, PyV has become a valuable model for understanding the mechanisms of viral-mediated tumorigenesis. However, despite its oncogenic potential, PyV establishes an asymptomatic persistent infection under natural conditions (Carroll et al., 2007). More recently, PyV has been used as a tool to better understand the host immune response to viral infection (Benjamin, 2001; Moser and Lukacher, 2001). Studies show that epitopes from the early proteins, middle T antigen (MT) and large T antigen (LT) are immunodominant, with most CTLs cloned from infected animals being responsive to MT or LT (Kemball et al., 2005; Lukacher et al., 1999). Thus, laboratory infection of mice with PyV has become a valuable model system for virological, immunological, and tumorigenesis studies.
PyV has a small, approximately 5 KB double-stranded circular DNA genome. Near the origin of replication (ori) are the early and late promoters that direct transcription in opposite directions. Through alternative splicing, three early proteins are encoded: small T antigen (sT), middle T Antigen (MT) and large T antigen (LT). These multifunctional proteins are involved in signaling, altering the cell cycle control mechanisms and promoting viral DNA replication (Cole, 1996). The late transcripts encode three structural protein components of the capsid: VP1, VP2, and VP3. Despite being separated by ~100 million of years of evolution, SV40 and PyV share extensive similarities in genomic architecture, transcript patterns, and amino acid identity of some proteins. Interestingly, PyV RNA structures compatible with a pre-miRNA were posited by Treisman in the late 1970s (for review see (Sullivan et al., 2006)). These studies (Treisman, 1981; Treisman and Kamen, 1981) combined with the need to develop an in vivo model to study the functions of viral miRNAs, led us to examine whether PyV encodes miRNAs.
We have previously shown that SV40 encodes a pre-miRNA (SVpre-miRNA) that is processed into two miRNAs, which function to downregulate early protein levels (Sullivan et al., 2005). Furthermore, a mutant SV40 virus that fails to make the pre-miRNA is more sensitive to cytotoxic T cell (CTL) lysis in in vitro assays. These data led to the suggestion that the SV miRNAs may augment evasion of the immune response during infection in vivo (Sullivan et al., 2006; Sullivan et al., 2005). Here we show that PyV encodes a pre-miRNA late during infection that is processed into two stable miRNAs, and explore the functions of these miRNAs both in culture and experimental inoculation of mice.
Results
Identification of PyV miRNAs
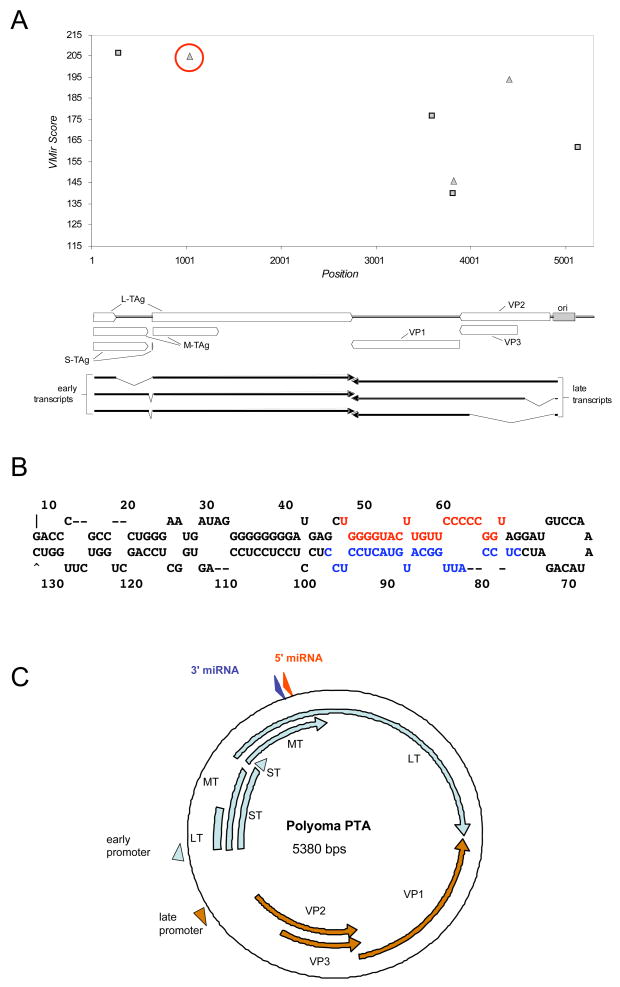
We set out to determine whether PyV, which is a well-characterized animal model of polyomavirus infection, encodes miRNAs. First we compared the sequence of the PyV genome to SV40 to look for sequence similarity to the SVpre-miRNA. On the whole, the genomic architecture of PyV and SV40 are similar, however, the region of SV40 that encodes the pre-miRNA is not conserved in PyV. Furthermore, no other region of the PyV genome contains significant nucleotide similarity with the SV40 pre-miRNA (data not shown). We next asked whether PyV might encode a pre-miRNA that is not homologous to the SVpre-miRNA. We applied Vmir (Grundhoff, Sullivan, and Ganem, 2006; Sullivan and Grundhoff, 2007; Sullivan et al., 2005), a pre-miRNA prediction algorithm, to the PyV genome. Two top scoring candidates (MF1 & MR1 for microRNA forward & reverse orientation) scored above an arbitrary cut off score of 200 (Fig. 1A) and were further pursued. Candidate MR1 has a predicted long hairpin stem and stable secondary structure (Fig. 1B) and maps in the late orientation approximately 1700 nucleotides downstream from the late polyadenylation site, complementary to the early RNAs (Fig. 1C). Neither MF1 nor MR1 has any sequence identity in common with the SV40 pre-miRNA, and both map to a different genomic location. However, because of its location and orientation, MR1 still could potentially generate miRNAs that downregulate early transcript levels.
Figure 1. Prediction of a PyV pre-miRNA.
(A) Vmir was used to predict candidate pre-miRNAs in the PyV genome (shown is a Vmir prediction plot with a cutoff score of 115). Top, The vertical axis indicates Vmir score, the horizontal axis indicates nucleotide position. The squares show individual candidates in the early (forward) orientation and triangles indicate candidates in the late (reverse) orientation. The top-scoring late orientation candidate, MR1 (for microRNA reverse 1) is circled in red. Bottom, is a linearized map of the PyV genome, corresponding to the nucleotide positions depicted in the Vmir plot. (B) Secondary structure prediction of candidate MR1 as called by Vmir. The red nucleotides correspond to the estimated position of the 5p miRNA, the blue nucleotides correspond to the estimated position of the 3p miRNA. (C) Genomic organization of PyV. The early (light blue) and late (orange) transcripts are indicated. Promoters are indicated with arrowheads. Locations of the 3p and 5p miRNAs (derived from the MR1 candidate pre-miRNA are shown, blue and red arrowheads, respectively).
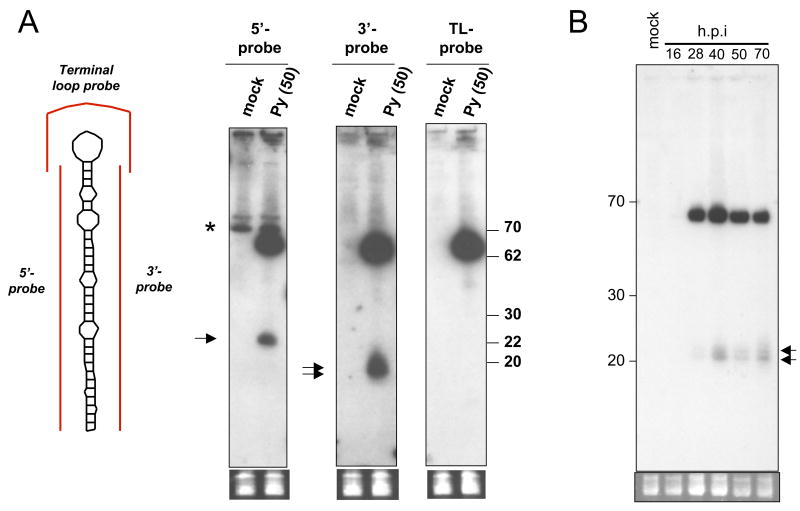
To test the validity of our computational predictions, we designed probes to both the 5 prime (5p) and three prime (3p) arms of each candidate and conducted Northern blot analysis. We did not detect any specific signal with probes directed against MF1 (data not shown), however, distinct bands migrating around 22 nucleotides (the typical size of a miRNA) and ~65 nucleotides (the typical size of the pre-miRNA) were detected with probes directed against either the 5p or 3p arms of the MR1 candidate (Fig. 2). A control probe designed to hybridize to the loop portion of the pre-miRNA (Terminal loop probe) did not recognize any signal migrating at 22 nucleotides but readily detected the ~65 nucleotide pre-miRNA. This demonstrates that there is not general degradation of transcripts in this region of the genome that could account for the ~22 nucleotide bands we detect with the 5p and 3p probes. Next we determined the kinetics of pre-miRNA/miRNA expression. We observed bands corresponding in size to both the pre-miRNA and miRNA that are readily detectable starting around 28 h.p.i. (Fig. 2B). Expression of these bands continued throughout the duration of the experiment. Combined, these results strongly suggest that we have identified a novel, late-expressed PyV pre-miRNA that is processed into two different miRNAs. Notably, the 5p and 3p miRNAs are both readily detectable and the pre-miRNA is present at higher amounts than the processed miRNAs. These are atypical but not unprecedented features of some pre-miRNA/miRNA derivatives.
Figure 2. Northern blot confirmation of MR1 candidate.
(A) Northern blot analysis was conducted using the probes diagramed in red (left side of Fig.) on RNA harvested from mock or PyV-infected BMK cells at 50 h.p.i.. Arrows indicate bands corresponding to miRNAs. Note predominant band detected by all three probes migrating at ~65 nucleotides corresponds to the pre-miRNA. Bottom panel shows pre-transferred, low molecular weight RNAs stained with ethidium bromide as a loading control (B) Time course of pre-miRNA/miRNA accumulation (hybridized with 5′ probe).
PyV miRNAs direct cleavage of the early RNAs
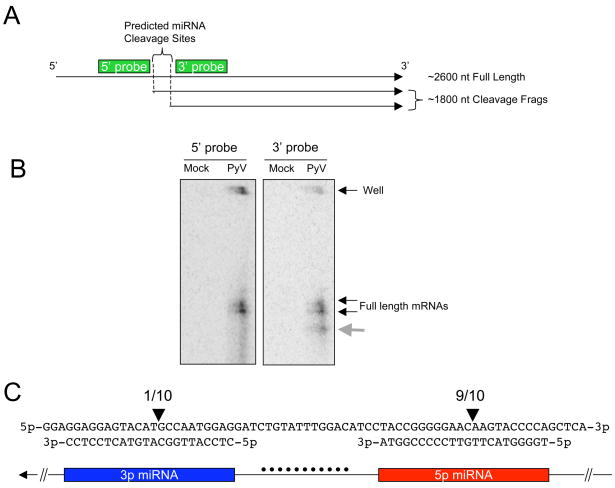
Both miRNAs we identified have perfect complementarity to the early RNA transcripts. Therefore, we tested the hypothesis that these miRNAs might direct cleavage of the early transcripts in a manner analogous to siRNA-mediated cleavage. In 1984, Fenton and Basillico identified an unexplained early transcript (Fenton and Basilico, 1982). Through Northern blot analysis, they identified an abundant early transcript migrating at approximately ~1800 nucleotides in length. It is notable that the transcript they identified is robustly present only at late times of infection, despite being of early orientation. Furthermore, they showed that this transcript is not capped. This is exactly the size range and 5′ end structure we would predict should exist if the PyV miRNAs we have identified are directing cleavage of the early transcripts. To map the early transcripts with more precision, we performed Northern blot analysis on polyadenylated-enriched RNAs using probes that recognize the early RNAs just proximal to the predicted miRNA cleavage sites on either the 5′ and 3′ ends (Fig. 3A & B). When probing with the 5′ probe, that should recognize only the full length mRNAs, we detect a doublet consistent with the multiple isoforms of the early mRNAs present during infection (Fenton and Basilico, 1982) (Fig. 3B, left panel). However, when probing with the 3′ probe, that should recognize both the full length mRNAs and the predicted cleavage fragments, we observe a faster migrating band consistent with the uncapped transcript first reported by Fenton and Basillico (Fenton and Basilico, 1982) (Fig. 3B, right panel). To map these cleavage fragment with single nucleotide precision, we performed 5′ RACE analysis using a modified protocol designed to enrich for sequencing of siRNA-like cleavage fragments. We sequenced 10 amplicons and all 10 mapped a 5′ cleavage fragment to the middle of a predicted miRNA region (Fig. 3A & C). 9 of the 10 amplicons mapped to a position within the predicted 5p miRNA, and 1 mapped to the 3p miRNA region. Combined with the results of Fenton and Basillico, our data suggest that miRNAs generated from each arm of the PyV pre-miRNA are active (albeit to varying degrees) at generating specific cleavage of the early RNAs.
Figure 3. 5′ RACE analysis identifies miRNA-mediate cleavage sites.
(A) Map of predicted cleavage fragments if the PyV 5p and 3p miRNAs are active at directing cleavage of the early mRNAs. Probes used in Northern blots analysis are shown as green rectangles. Note: the map is not drawn to scale. (B) Northern blot analysis for early mRNAs and cleavage fragments. Polyadenylated-enriched RNA was probed with oligonucleotide probes and visualized via phosphorimaging. The panel on the left depicts the blot probed with the 5′ probe. The panel on the left is the same blot that was stripped and re-probed with the 3′ probe. The grey arrow indicates the additional band detected only with the 3′ probe. (C) 5′ rapid amplification of cDNA ends (RACE) was employed using a protocol modified to enrich for cleavage fragments. Polyadenylated-enriched RNA was ligated to a linker, followed by reverse transcription and nested PCR to map the 5′ ends of early mRNA fragments in the vicinity of the pre-miRNA. Arrows indicate the position of mapped cleavage fragments. The numbers above the arrows indicate that 9 of 10 amplicons sequenced map antisense to the middle of the putative 5p miRNA and 1 of 10 amplicons sequenced map antisense to the middle of the putative 3p miRNA. The top line shows the top DNA strand, 5′ to 3′ in the early orientation. Below that is the predicted sequence of each miRNA (3′ to 5′). A cartoon showing how the nucleotides correspond to the stem (3p, blue box, 5p red box) or loop (black dots) portion of the pre-miRNA is shown at the bottom.
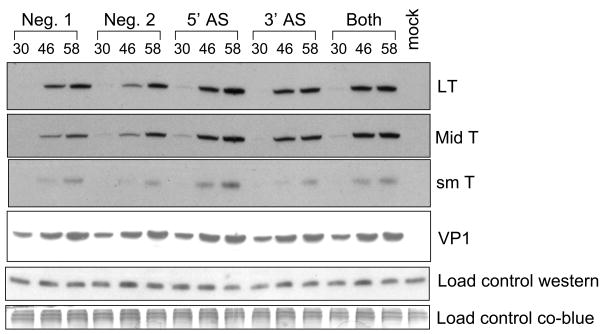
We next asked whether the PyV miRNA-mediated cleavage of the early RNAs results in a decrease in early protein levels. We designed 2′-O-methylated antisense inhibitors to either the 5p or 3p PyV miRNAs. We then transfected the inhibitors at two different times post- infection and assayed for early proteins levels by immunoblot analysis. Transfection of the 5′ miRNA inhibitor (5′ AS) had a modest but reproducible effect of increasing all three early protein levels (Fig. 4). Transfection of the 3p miRNA inhibitor (3′ AS) had significantly less of an effect (almost to negative control levels), and transfecting a pooled combination of ½ the concentration of both inhibitors (Both) had an intermediate effect. Importantly, transfection of an irrelevant negative control oligonucleotide or transfection reagent alone had no effect on early protein levels (Neg. 1 and Neg. 2). These results are consistent with the data of Fig 3B, which suggested that the 5p miRNA is the major species active in cleavage of PyV early RNA, the mRNA encoding the viral T antigens.
Figure 4. Immunoblot analysis shows inhibition of PyV miRNAs increases early protein levels.
Protein was harvested at various times post-infection and probed with relevant antibody. Shown are representative western blots for LT, MT, sT, VP1 as well as two different loading controls. The membrane was initially probed for LT (monoclonal antibody 762), then for MT and ST (monoclonal antibody F4), then it was stripped and re-probed with antibody that recognized PyV VP1. The top loading control corresponds to a non-specific, cross-reacting band with the F4 antibody, the bottom loading control corresponds Coomassie blue-stained high molecular weight proteins after transfer to the PVDF membrane.
Characterization of a dl1013, a PyV miRNA mutant virus
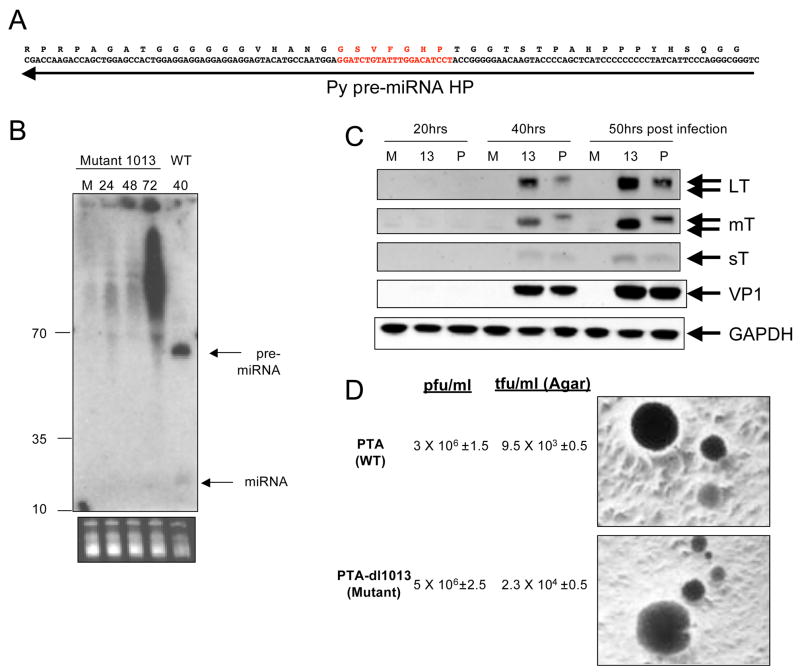
Next, we surveyed the literature to identify candidate PyV mutants that fail to express the pre-miRNA/miRNAs. A deletion mutant dl1013 (Magnusson and Berg, 1979) was the top candidate to emerge because it met the following criteria: (1) it is a small deletion (21 nucleotides) mapping to the middle of the pre-miRNA (Fig. 5A), (2) the deletion results in in-frame deletions in MT and LT (3) it is viable in cultured cells and (4) it transforms cells in culture with normal efficiency. These findings suggest that this mutation is not compromising the functionality of MT and LT. Thus, dl1013 was a prime candidate for a mutation that would disrupt pre-miRNA/miRNA processing while having no discernable effects on early protein function.
Figure 5. Characterization of mutant PTA-dl1013.
(A) Transcripts and amino acids of LT corresponding to the region of the early transcripts that is complimentary to the pre-miRNA (depicted with an arrow). The region in red identifies the nucleotides and LT amino acids deleted in PTA-dl1013. (B) Northern blot, probed for the 5p miRNA was conducted as shown in Fig. 2. Arrows indicate bands corresponding to the pre-miRNA and miRNA that not detectable in wildtype but not mutant PTA-dl1013-infected cells. RNA was harvested at 24, 48, or 72 hoi from PTA-dl1013-infected BMK cells or 40 h.p.i. from wildtype infected cells. M, mock infected. (C) Immunoblot analysis shows increased levels of LT and MT in PTA-dl1013-infected cells. BMK cells were infected with the wildtype strain PTA (P) or the miRNA mutant virus (PTA-dl1013, (13)) at an M.O.I. of 1 and immunoblot analysis was performed for large T antigen (LT), middles T antigen (MT), small T antigen (sT), VP1 or the load control protein GAPDH. (D) PTA-dl1013 transforms cultured cells as well as wildtype virus. Viral titers were determined by infecting UC1B cells at various dilutions, and the plaques arisen in soft-agar medium were counted. The ability of PTA-dl1013 to transform cultured cells was measured. by infecting rat fibroblast F111 with either PTA or PTA-dl1013 at M.O.I. of 0.5, 2.5 and 10. After 16 hrs post infection, the infected cells were mixed with soft-agar medium and immediately plated in 60mm plates. The transformed cells were counted after 2 weeks.
As a first step in characterizing the effects of this deletion, we created an isogenic mutant and wildtype pair by introducing the 21 bp deletion of dl1013 into the standard wild type laboratory strain PTA. We then compared these strains for pre-miRNA/miRNA production. Cells were infected with PTA-dl1013 or PTA and Northern blot analysis performed on RNA harvested at various times post-infection (Fig. 5B). This revealed that in PTA-dl1013 no specific bands were detected migrating in the pre-miRNA or miRNA size range, even at the latest times post-infection (when the pre-miRNA and miRNA are readily detectable in wildtype-infected cells). We note that by 72hpi, there is substantial accumulation of heterogeneous and seemingly aberrant RNA species in PTA-dl1013-infected cells; we speculate that these may result from abortive processing of transcripts arising from the deletion mutation. However, these transcripts lack any features of pre-miRNAs or miRNAs and it is doubtful that they can play analogous functional roles.
We then performed immunoblot analysis to examine the effects of the mutation on early protein production. As expected, we observed a reproducible increase in early protein LT and MT protein levels at late times post-infection in the mutant compared to wildtype PyV (Fig. 5c). Small T accumulation was also augmented by 50 hpi (Fig 5c), but the effects of the mutation on ST levels were not as large as those on LT and MT. The reason for the differential effects on the three early proteins is unclear and may reflect the non-linear limitations of western blot analysis, or the differences in abundance or timing of expression of the early transcripts. However, taken together with our antisense miRNA inhibitor studies (Fig. 4), these results demonstrate that the PyV miRNAs play some role in regulating the early protein levels.
No replication defect was noted in the original dl1013 isolate (Magnusson et al., 1981). Similarly, PTA-dl1013 in our hands grew to comparable titer and showed no difference in the size or time of appearance of plaques compared to PTA. (Fig. 5d). The original dl1013 mutant transformed cells, albeit with some dependence on the temperature (Magnusson et al., 1981). We observed normal transforming ability of PTA-dl1013 using F-111 established rat fibroblasts assayed by growth in soft agar (Fig 5d). These results indicate that the PyV miRNAs are not essential for virus replication or cell transformation in vitro.
In vivo infection with PTA-dl1013
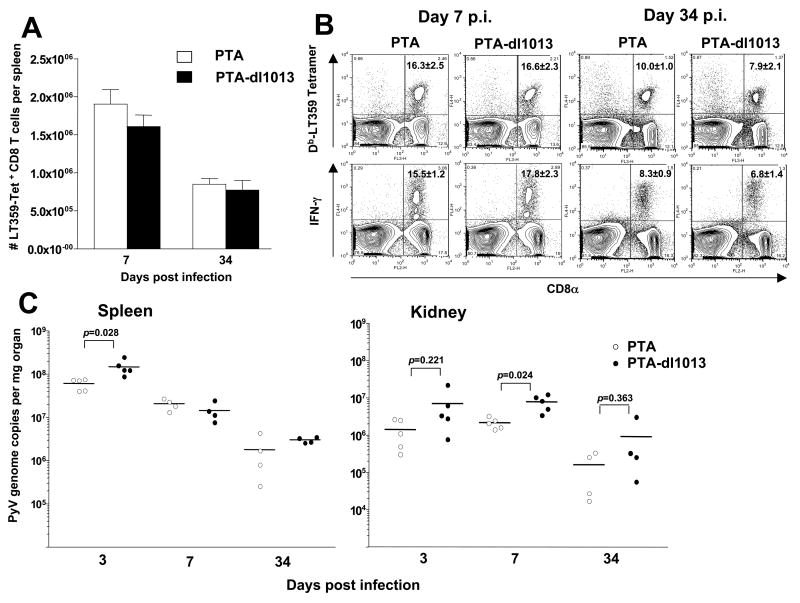
PyV infection of mice is a well-characterized animal model of viral infection. We took advantage of this established model and infected mice to look for a possible role for the PyV miRNAs. Adult C57BL/6 mice were subcutaneously (s.c.) inoculated with 1 × 105 PFU of either wildtype or dl1013 virus, and the spleens and kidneys were harvested at 3, 7, or 34 days post-infection.
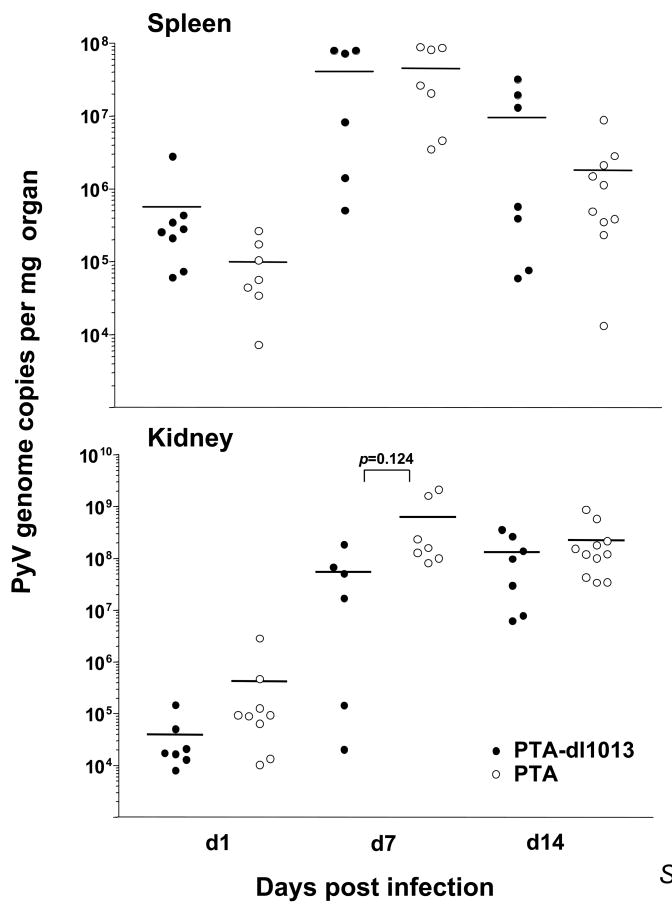
We have previously hypothesized that autoregulation of the early protein levels by the SV40 miRNAs may play a role in evading the CTL response in vivo (Sullivan et al., 2005). Because there are notable similarities between the PyV and SV40 pre-miRNAs (e.g. both are made late during infection, and both make two different miRNAs that direct cleavage of the early RNAs), we tested whether dl1013 has an altered PyV-specific CD8 T cell response in vivo. In order to visualize the frequency of PyV-specific CD8 T cells, we stained cells with a tetramer consisting of Db molecules bound to the dominant MHC Class-I restricted epitope from PyV (LT359-368) (Kemball et al., 2005). The magnitude of the Db-LT359 response was comparable between wildtype and dl1013-infected animals during both the acute and persistent phases of PyV infection (Fig. 6A). Furthermore, no differences were observed in secretion of IFNγ, a cytokine used to assess the functionality of T cells stimulated with the LT359 peptide at either 7 or 34 days post infection (Fig. 6B). Within the spleen and kidney, no major differences in viral clearance were observed (Fig. 6C). To see if a reduced viral infection would reveal differences in viral clearance or anti-viral CD8 T cell responses, we administered a smaller viral inoculum (1 × 103 PFU) of PTA or PTA-dl1013 to adult mice; however, Db LT359 CD8 T cell responses at day 10 post infection in the spleen and lung remained similar between the two groups, as was splenic viral DNA load (data not shown). To more closely approximate natural PyV exposure, we investigated whether neonates might exhibit differences in viral clearance rates. Neonatal mice were infected with 1 × 103 PFU of each virus within 24 hours after birth, and spleens and kidneys were removed at 1, 7, and 14 days post infection. As shown in Figure 7, no significant differences were noted in either the spleen or kidney at any of these timepoints.
Figure 6. Deletion of microRNA sequences does not significantly alter the magnitude of the acute or memory LT359-specific CD8 T cell response or clearance of virus in adult mice.
C57BL/6 mice (n=4) were infected s.c. with 105 PFU of either PTA PyV or PTA-dl1013 virus. The spleens and kidneys were removed and processed on d7 or d34. Absolute numbers of Db LT359-specific CD8 T cells in the spleen are depicted in (A), while the acute and memory responses are shown in (B). Spleen and kidney tissue samples were processed at the indicated timepoints and viral genome copy numbers were quantified using Taqman real-time PCR (C).
Figure 7. Infection of neonates with either PTA or PTA-dl1013 does not yield significant differences in viral clearance in neonatal mice.
Neonatal mice were infected subcutaneously (s.c.) with 103 PFU of virus and sacrificed at the indicated timepoints. Spleen and kidney tissue samples were processed and viral genome copy numbers were quantified using Taqman real-time PCR.
These results suggest that under the experimental conditions we employed, the PyV miRNAs have little effect on the induction of the CD8 T cell response to LT in vivo. Although the overall similarity of viral titers in the mice argues against a large and sustained effect in this regard, the possibility of a more modest effect at lower target-effector ratios cannot be excluded.
Discussion
Viral-encoded miRNAs have garnered much interest, in part, as possible targets for therapeutic intervention. Viral miRNAs have been described for several members of the Herpesvirus and Polyomavirus families, and possibly HIV (a lentivirus) (Cullen, 2006; Grey, Hook, and Nelson, 2008; Nair and Zavolan, 2006; Omoto and Fujii, 2005; Omoto et al., 2004; Pfeffer and Voinnet, 2006; Samols and Renne, 2006; Sarnow et al., 2006; Sullivan and Ganem, 2005b). In addition, a small percentage of the adenovirus VA RNAs made during infection is processed into RISC without Drosha processing (Andersson et al., 2005; Aparicio et al., 2006; Lu and Cullen, 2004; Xu et al., 2007). Thus, VA RNAs can be considered a pre-miRNA that gives rise to miRNAs through an atypical route of processing. Target mRNAs for some viral miRNAs have been described, but detailed biological functions have been ascribed to only a minority of such miRNAs, and even fewer studies have addressed the roles viral miRNA functions in the intact animal host.
In this report we show that the well-characterized model virus, PyV encodes a pre-miRNA. We have previously shown that SV40, a monkey polyomavirus encodes a pre-miRNA with several distinct features: (1) the SV40 pre-miRNA is made at robust levels late in the infectious cycle, (2) it is processed into two active miRNAs, one from each arm of the hairpin precursor, and (3) both SV40 miRNAs are active at directing the cleavage of the same substrates - the early mRNAs. The PyV pre-miRNA shares all of these characteristics with its cognate pre-miRNA in SV40 (Figs. 2 & 3). However, there are notable differences as well: no sequence similarity is detected between the SV40 and PyV pre-miRNAs and they are encoded in different regions of the genome (although both are complementary to the early mRNAs). Additionally, we note that although the viral RNA targets of these miRNAs are well-documented, this in no way excludes the possibility that the polyomaviral miRNAs might also target host transcripts.
Interestingly, the PyV miRNAs appear to be dispensable for PyV infection after experimental inoculation of either cultured cells or intact mice. PTA-dl1013 fails to make the PyV pre-miRNA or miRNAs yet replicates and transforms well in culture and replicates and spreads efficiently in the mouse (Fig. 6 C). Furthermore, no differences are observed in the CD8 T cell response to LT in mice that are sacrificed at 7 or 34 days post infection dpi. How can one account for the fact that the PyV miRNAs are not required in experimental murine infection? While it is formally possible that the PyV pre-miRNA serves no important function in PyV biology, the conservation of abundantly expressed pre-miRNA structures with perfect homology to early mRNAs in multiple members of the Polyomaviridae makes this proposal highly unlikely. We note in this connection that in several other viruses (e.g. HSV and HCMV), a large fraction of the genome can be regarded as dispensable by similar laboratory criteria, yet many of these genes play critical roles in the biology of natural infection (Mocarski Jr., Shenk, and Pass, 2007; Roizman, Knipe, and Whitley, 2007).
Clearly, whatever function these miRNAs serve is not required for infection as it is commonly studied in experimental settings. However, laboratory PyV infection bears little resemblance to authentic PyV infection as it occurs in the wild – and it is the latter type of infection that has shaped the evolution of the PyV genome. Laboratory protocols for infection have been optimized for experimental convenience – especially for high efficiency of infection and potential for tumorigenesis. These considerations favor parenteral exposure with large amounts of virus. However, in nature PyV is transmitted by mucosal exposure of the upper respiratory tract to very low levels of virus – conditions which might make the virus substantially more vulnerable to attack by either innate immune factors or CTLs. (Moreover, typically such transmission events occur among neonates or juveniles, rather than the adult animals we employed for much of the study). Under natural conditions, even a slight decrement in susceptibility to such attack (as described in vitro for the SV40 miRNAs (Sullivan and Ganem, 2005b)) could confer a substantial selective advantage. Our short-term experimental inoculation protocol also does not examine another aspect of infection – the persistent or intermittent shedding of virus in the urine (which is the presumed source of the virus that is spread between individuals in the population). If the miRNAs promoted even a modest extension of such shedding, this too could have important epidemiologic consequences for the virus and be positively selected during viral evolution. Future studies of polyomavirus miRNA biology will need to focus on these and other subtler phenotypes affecting the innate immune response, viral persistence in individuals, and transmission in the population, rather than on effects on single-cycle infection in vitro or short-term replication in vivo.
Note added in proof: while this work was under review, we discovered that the recently identified human polyomavirus– Merkel Cell Polyomavirus, encodes a pre-miRNA in a similar region of the genome as the muPyV pre-miRNA described here (Seo, Chen, and Sullivan, 2009).
Materials and methods
Cells and virus strains
Baby mouse kidney (BMK), UC1B and NIH 3T3 cells were grown in DMEM with 10% newborn calf serum and used for viral infection or transfection. Mouse polyoma virus strains PTA and PTA-dl1013 were used in this study. PTA-dl1013, a PTA version of dl1013 (a derivative of the strain A2) was created by using polymerase chain reaction (PCR) and site-directed mutagenesis methods. Briefly, a 21bp corresponding to the dl1013-deleted region was targeted for deletion by PCR using the QuikChange site-directed mutagenesis kit (Stratagene), a primer set (5′-GGAGTACATGCCAATGGAACCGGAGGAACAAGTACCCC/5′-GGGGTACTTGTTCCTCCGGTTCCATTGGCATGTACTCC) and PTA-pBluescript plasmid DNA as a template. The amplified circular DNA was transformed into bacterial cells, and the structure of the plasmid DNA from each bacterial colony was determined by sequencing. The plasmid having the targeted deletion was transfected into NIH 3T3 cells, and the viruses were harvested at 9 days post transfection. Then the mutant virus PTA-dl1013 was plaque purified on UC1B cells, and its structure was confirmed again by sequencing.
Immunoblot analysis for Polyoma proteins
For the studies performed in Fig. 4, the methodology for immunoblots was performed as previously described (Sullivan and Ganem, 2005a). NIH 3T3 cell were infected with an MOI of 10 with the wildtype (PyV PTA) strain. 20 micrograms of total protein was run on a denaturing SDS polyacrylamide gel. Proteins were then electroblotted onto PVDF membranes (Millipore) and were blocked in a solution of TBST and 5% condensed milk (Carnation). All probings were carried out in TBST, 0.5% TBST. MT was probed with monoclonal antibody 762 (obtained from Steve Dilworth, Imperial College, London). For sT and LT, monoclonal antibody F4 (Pallas et al., 1986) that recognizes all three T antigens was used. Initially blots were probed with a 1: 25,000 dilution of Ab 762 over night at 4 degrees C, were washed in TBST and probed with HRP-conjugated goat anti-mouse secondary antibody (Sigma) at a 1: 50,000 dilution. Then blots were probed with hybridoma supernatant from cells expressing the monoclonal Ab F4, at a dilution 1:125 for 2 hours. Blots were stripped with commercial stripping reagent according to manufacturer’s suggestions (Pierce/Thermo Scientific) and re-probed with anti-VP-1 antibody (obtained from Bob Garcea, University of Colorado Health Science Center) at 1:12,000 dilution.
BMK cells were infected at an M.O.I. of 1, and lysates of the infected cells were prepared in PBS with 1% NP40 and protease inhibitor cocktail (Roche) at indicated times post infection. Collected samples were subjected to gel electrophoresis using 4–20% gradient Tris-HCl gels (Invitrogen), and was followed by transfer to a nitrocellulose membrane. This membrane was blocked overnight or for 2 hours in PBST buffer (PBS with 0.1% Tween 20) containing 5% non-fat dry milk, and then probed with antibodies against T antigens (F4), VP1 and GAPDH (Calbiochem; 1:90,000 dilution). After washing in 20 ml of PBST five times at room temperature, the membrane was incubated with Alexa fluor 680 anti-mouse IgG and 800 anti-rabbit IgG antibodies (1:15,000 dilution; LI-COR Biosciences, Lincoln, NE) in PBST containing 5% non-fat milk solids for 45 min at room temperature and washed in PBST five times. The membrane was scanned using Odyssey infrared imaging system (LI-COR Biosciences).
Antisense miRNA inhibition studies
Cells were infected at an M.O.I. of 1, and were transfected twice, once at 5 h.p.i., and again and 24 h.p.i. with an irrelevant 2′-O-methylated oligonucleotide, an anti-5p miRNA 2′-O-methylated oligonucleotide, an anti-3p miRNA 2′-O-methylated oligonucleotide, or a combination of the anti-5p and anti-3p 2′-O-methylated oligonucleotide. Cells were transfected with 30 nM of total oligonucleotide using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions.
The sequences of the 2′-O-methylated oligonucleotides used in this study:
Negative control: UUAAUGCGGAGAGGAAUGGUGGCCUGGUUGACA
Anti-3P: GAGGAGGAGGAGUACATGCCAATGGAGGAUCU
Anti-5p: CAUCCUACCGGGGGAACAAGUACCCAGCTCA
Northern blot analysis for early cleavage fragments
Total RNA was harvested utilizing RNABee (Tel-Test Laboratories, Friendswood Texas). The samples were enriched for polyadenylated RNA using the Oligotex mRNA mini kit (Qiagen) according to the manufacturer’s protocol. Polyadenylated-enriched RNA, obtained from approximately 30 micrograms/sample of total RNA, was loaded into each well. Samples were electrophoresed through a 5% polyacrylamide (acrylamide:bis, 29:1, BioRad), 0.5X TBE, urea denaturing gel for approximately 9 hours at 300 volts. A large, wire-bound transfer apparatus (BioRad) was used to electroblot transfer the gels to Hybond N+ membrane (Amersham) in 0.5X TBE for 1.5 hours as follows: 30 volts for 1 hour, 40 volts for an additional 30 minutes. Blotted membranes were UV crosslinked and allowed to dry overnight at room temperature. Membranes were hybridized with 10 picomoles of radiolabeled oligonucleotide probes (end-labeled with T4 polynucleotide kinase) in ExpressHyb hybridization solution (Clontech) at the temperatures listed below. Blots were washed at room temperature ~4 times with 2X SSC, 0.05% SDS followed by 2 washes with 0.1X SSC, 0.1% SDS. Blots were exposed overnight to a Phosphorimager screen (Kodak).
Probes used:
PY_5′Probe: TAGGTCGGGTTGCTCAGAAGAC (53 degrees C hybridization temperature)
PY_3′Probe: GAGGCCTGGCTTATTTTCTGTT (48 degrees C hybridization temperature)
5′ RACE analysis of early cleavage fragments
A modified version of Llave et al. was used (Llave et al., 2002). The RLM RACE kit (Ambion) was used according to manufacturer’s directions, except the calf intestine alkaline phosphatase and tobacco acid pyrophosphatase steps were omitted so as to enrich for up-capped potential miRNA-mediated cleavage fragments. Amplicons were cloned into the TOPO TA cloning kit (Invitrogen) and plasmid DNA was harvested and sequenced from 10 different colonies previously screened to have an insert.
The primers used for this study:
PyV-specific outer primer: AGGCTTGGTGACTGCCTTACACAT
PyV-specific inner primer: GGCATTTGAATTCGGGCCTGAACT
Small RNA Northern Blots
miRNAs were detected as previously described (Grundhoff, Sullivan, and Ganem, 2006; Sullivan and Grundhoff, 2007). Briefly, BMK cells were infected at an M.O.I. of 10, and total RNA was extracted with RNABee (TEL-TEST, INC., Friendswood, TX) at various times post-infection. Fifteen micrograms of total RNA were run on a urea denaturing 15% polyacrylamide gels and electroblot transferred to Hybond N+ membrane (Amersham). Blots were probed at 38.5 degrees C in Expresshyb (Clontech) hybridization solution and washed 4 times with room temperature 2X SSC, 0.1% SDS. T4 PNK (USB)-labeled oligonucleotides were used as probes.
The sequences of the oligonucleotide probes used were:
PymiRNA5p: TCCTACCGGGGGAACAAGTACCCCAGCTCA
PymiRNA3p: GGAGGAGGAGTACATGCCAATGGAGGA
Control PymiRNALoop: AGGATCTGTATTTGGACATCC
In vitro transformation assays
F111 rat embryo fibroblasts were infected with WT PTA or PTA-dl1013 at M.O.I. of 0.5, 2.5 and 10. After 16 hrs post infection, the infected cells were trypsinized, collected by centrifugation, resuspended in soft-agar medium and immediately plated in 60mm plates as (Raptis, Lamfrom, and Benjamin, 1985). The transformed cells were counted after 14 days.
Mice
C57BL/6J mice used for neonatal infection were purchased from the Jackson Laboratory (Bar Harbor, Maine). They were kept in the SPF barrier facility at Harvard Medical School prior to use. C57BL/6NCr female mice were purchased from the Frederick Cancer Research and Development Center of the National Cancer Institute (Frederick, MD). Mice were housed and bred in accordance with the guidelines of the Institutional Animal Care and Use Committee, and the Department of Animal Resources at Emory University.
Virus inoculations
Each mouse was inoculated s.c. in the hind footpads with 1×103 or 1×105 PFU of virus. Mice were infected at 6–12 weeks of age. Neonatal mice were infected s.c. with 1×103 PFU within 24 h after birth.
Peptides and intracellular IFN-γ staining
LT359-368C7Abu peptide was synthesized and stored as previously described (Kemball et al, 2005). Cells were stimulated directly ex vivo with 1 mM peptide and stained for CD8a and IFN-γ as described (Kemball et al, 2005).
Flow cytometry
Cells were stained in PBS containing 2 FBS and 0.1% sodium azide (FACS buffer) for 40 min at room temperature (tetramer staining) or according to the manufacturer’s instructions for intracellular staining (BD Biosciences). Samples were washed twice in FACS buffer. Samples were immediately acquired on a FACSCalibur (BD Biosciences). Cells were stained with allophycocyanin-conjugated tetramer. Anti-CD8a and IFN-g antibodies were purchased from BD Pharmingen. Data were analyzed using FloJo software (Tree Star).
Quantification of PyV genomes
DNA isolation and Taqman PCR were performed as described (Kemball et al, 2005). The PyV DNA quantity is expressed in genome copies per milligram of tissue and is calculated based on a standard curve of known PyV genome copy number vs threshold cycle of detection. The detection limit with this assay is 10 copies of genomic viral DNA.
Acknowledgments
We thank Steve Dilworth and Bob Garcea for kindly providing antibodies used in these studies. The initial impetus for this work was greatly aided by helpful suggestions from and the PhD thesis work of Richard Treisman (London Research Institute). In addition, the authors acknowledge helpful conversations regarding this work with Brian Schaffhausen, Janet Butel, and Jim Pipas. CSS is supported by start-up funds from the University of Texas at Austin and a fellowship from the Institute for Cellular and Molecular Biology at the University of Texas at Austin. The worked performed in TB’s lab is supported by grant RO1 CA 90992 from the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79(15):9556–65. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol. 2006;80(3):1376–84. doi: 10.1128/JVI.80.3.1376-1384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36(2):666–75. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin TL. Polyoma virus: old findings and new challenges. Virology. 2001;289(2):167–73. doi: 10.1006/viro.2001.1124. [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. Rna. 2004;10(2):185–91. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J, Dey D, Kreisman L, Velupillai P, Dahl J, Telford S, Bronson R, Benjamin T. Receptor-binding and oncogenic properties of polyoma viruses isolated from feral mice. PLoS Pathog. 2007;3(12):e179. doi: 10.1371/journal.ppat.0030179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16(2):203–8. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Cole CN. Polyomaviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. 3. Lippincott-Raven Publishers; Philadelphia: 1996. pp. 1997–2043. [Google Scholar]
- Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38(Suppl 30):S25–30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–5. doi: 10.1038/nature03049. Epub 2004 Nov 7. [DOI] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17(4):438–42. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton RG, Basilico C. Changes in the topography of early region transcription during polyoma virus lytic infection. Proc Natl Acad Sci U S A. 1982;79(23):7142–6. doi: 10.1073/pnas.79.23.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, Cullen BR. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450(7172):1096–9. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–40. doi: 10.1038/nature03120. Epub 2004 Nov 7. [DOI] [PubMed] [Google Scholar]
- Grey F, Hook L, Nelson J. The functions of herpesvirus-encoded microRNAs. Med Microbiol Immunol. 2008;197(2):261–7. doi: 10.1007/s00430-007-0070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3(11):e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. Rna. 2006;12(5):733–50. doi: 10.1261/rna.2326106. Epub 2006 Mar 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297(5589):2056–60. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Kemball CC, Lee ED, Vezys V, Pearson TC, Larsen CP, Lukacher AE. Late priming and variability of epitope-specific CD8+ T cell responses during a persistent virus infection. J Immunol. 2005;174(12):7950–60. doi: 10.4049/jimmunol.174.12.7950. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15(20):2654–9. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;15:15. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297(5589):2053–6. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A. 2007;104(41):16164–9. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78(23):12868–76. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacher AE, Moser JM, Hadley A, Altman JD. Visualization of polyoma virus-specific CD8+ T cells in vivo during infection and tumor rejection. J Immunol. 1999;163(6):3369–78. [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–8. doi: 10.1126/science.1090599. Epub 2003 Nov 20. [DOI] [PubMed] [Google Scholar]
- Magnusson G, Berg P. Construction and analysis of viable deletion mutants of polyoma virus. J Virol. 1979;32(2):523–9. doi: 10.1128/jvi.32.2.523-529.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson G, Nilsson MG, Dilworth SM, Smolar N. Characterization of polyoma mutants with altered middle and large T-antigens. J Virol. 1981;39(3):673–83. doi: 10.1128/jvi.39.3.673-683.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, Sharp PA, Tabin CJ, McManus MT. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36(10):1079–83. doi: 10.1038/ng1421. Epub 2004 Sep 12. [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123(4):607–20. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Mocarski ES, Jr, Shenk T, Pass RF. Cytomegaloviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott Williams & Wilkins; Philadelphia: 2007. [Google Scholar]
- Moser JM, Lukacher AE. Immunity to polyoma virus infection and tumorigenesis. Viral Immunol. 2001;14(3):199–216. doi: 10.1089/088282401753266738. [DOI] [PubMed] [Google Scholar]
- Nair V, Zavolan M. Virus-encoded microRNAs: novel regulators of gene expression. Trends Microbiol. 2006;14(4):169–75. doi: 10.1016/j.tim.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Omoto S, Fujii YR. Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J Gen Virol. 2005;86(Pt 3):751–5. doi: 10.1099/vir.0.80449-0. [DOI] [PubMed] [Google Scholar]
- Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, Saksena NK, Fujii YR. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1(1):44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas DC, Schley C, Mahoney M, Harlow E, Schaffhausen BS, Roberts TM. Polyomavirus small t antigen: overproduction in bacteria, purification, and utilization for monoclonal and polyclonal antibody production. J Virol. 1986;60(3):1075–84. doi: 10.1128/jvi.60.3.1075-1084.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Voinnet O. Viruses, microRNAs and cancer. Oncogene. 2006;25(46):6211–9. doi: 10.1038/sj.onc.1209915. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–6. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Raptis L, Lamfrom H, Benjamin TL. Regulation of cellular phenotype and expression of polyomavirus middle T antigen in rat fibroblasts. Mol Cell Biol. 1985;5(9):2476–86. doi: 10.1128/mcb.5.9.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman BR, Knipe DM, Whitley RJ. Herpes Simplex Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2502–2601. [Google Scholar]
- Samols MA, Renne R. Virus-encoded microRNAs: a new chapter in virus-host cell interactions. Fut Virol. 2006;1:233–42. [Google Scholar]
- Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3(5):e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P, Jopling CL, Norman KL, Schutz S, Wehner KA. MicroRNAs: expression, avoidance and subversion by vertebrate viruses. Nat Rev Microbiol. 2006;4(9):651–9. doi: 10.1038/nrmicro1473. [DOI] [PubMed] [Google Scholar]
- Seitz H, Zamore PD. Rethinking the microprocessor. Cell. 2006;125(5):827–9. doi: 10.1016/j.cell.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Seo GJ, Chen CJ, Sullivan CS. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology. 2009;383(2):183–7. doi: 10.1016/j.virol.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Seo GJ, Fink LH, O’Hara B, Atwood WJ, Sullivan CS. Evolutionarily conserved function of a viral microRNA. J Virol. 2008;82(20):9823–8. doi: 10.1128/JVI.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81(23):12836–45. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O. Host immune system gene targeting by a viral miRNA. Science. 2007;317(5836):376–81. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS. New roles for large and small viral RNAs in evading host defences. Nat Rev Genet. 2008;20:20. doi: 10.1038/nrg2349. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Ganem D. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J Virol. 2005a;79(12):7371–9. doi: 10.1128/JVI.79.12.7371-7379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Ganem D. MicroRNAs and viral infection. Mol Cell. 2005b;20(1):3–7. doi: 10.1016/j.molcel.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff A. Identification of Viral MicroRNAs. Methods Enzymol. 2007;427:1–23. doi: 10.1016/S0076-6879(07)27001-6. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff A, Tevethia S, Treisman R, Pipas JM, Ganem D. Expression and function of microRNAs in viruses great and small. Cold Spring Harb Symp Quant Biol. 2006;71:351–6. doi: 10.1101/sqb.2006.71.065. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435(7042):682–6. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- Treisman R. Ph. D. Thesis. University College; London: 1981. Structures of polyomavirus nuclear and cytoplasmic RNA molecules. [Google Scholar]
- Treisman R, Kamen R. Structure of polyoma virus late nuclear RNA. J Mol Biol. 1981;148(3):273–301. doi: 10.1016/0022-2836(81)90539-8. [DOI] [PubMed] [Google Scholar]
- Xu N, Segerman B, Zhou X, Akusjarvi G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J Virol. 2007;81(19):10540–9. doi: 10.1128/JVI.00885-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–6. doi: 10.1101/gad.1158803. Epub 2003 Dec 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA. 2003;100(17):9779–84. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]