Abstract
PtdIns4P is the major precursor for the synthesis of the multifunctional plasma membrane lipid, PtdIns(4,5)P2. Yet PtdIns4P also functions as a regulatory lipid in its own right, particularly at the Golgi apparatus. In the present study we define specific conditions that enable preservation of several organellar membranes for the immunocytochemical detection of PtdIns4P. We report distinct pools of this lipid in both Golgi and plasma membranes, which are synthesized by different PI4K (phosphatidylinositol 4-kinase) activities, and also the presence of PtdIns4P in cytoplasmic vesicles, which are not readily identifiable as PI4K containing trafficking intermediates. In addition, we present evidence that the majority of PtdIns4P resides in the plasma membrane, where it is metabolically distinct from the steady-state plasma membrane pool of PtdIns(4,5)P2.
Keywords: Golgi; immunofluorescence; phosphoinositides; plasma membrane; PtdIns(4,5)P2; PtdIns4P
Abbreviations: Arf1, ADP-ribosylation factor 1; DAPI, 4′,6-diamidino-2-phenylindole; DiD, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanineperchlorate; DiOC6, 3,3′-dihexyloxacarbocyanine iodide; DMEM, Dulbecco's modified Eagle's medium; DTT, dithiothreitol; EGF, epidermal growth factor; ER, endoplasmic reticulum; ERGIC, ER-Golgi intermediate compartment; FA, formaldehyde; FAPP1, phosphatidylinositol-4-phosphate adaptor protein 1; FYVE-Hrs, hepatocyte growth factor-regulated tyrosine kinase substrate; GA, glutaraldehyde; GFP, green fluorescent protein; GM130, cis-Golgi matrix protein of 130 kDa; GST, glutathione transferase; M6PR, mannose-6-phosphate receptor; NBD-C6-ceramide, N-(ϵ-7-nitrobenz-2-oxa-1,3-diazol-4-yl-aminocaproyl)-D-erythro-sphingosine; NGS, normal goat serum; OSBP, oxysterol-binding protein; PAO, phenylarsine oxide; PH, pleckstrin homology; PI3K, phosphoinositide 3-kinase; PI4K, phosphoinositide 4-kinase; PLC, phospholipase C; POPC, 1-palmitoyl 2-oleoyl phosphatidylcholine; TBS, tris-buffered saline; WGA, wheat germ agglutinin
INTRODUCTION
PtdIns4P is the most functionally diverse of the inositol lipids. Firstly, synthesis of the versatile signalling lipid PtdIns(4,5)P2 proceeds primarily via 5-phosphorylation of PtdIns4P [1]. PtdIns(4,5)P2 is in turn the precursor for generation of the signalling lipid PtdIns(3,4,5)P3 and PtdIns(3,4)P2 [2]. Therefore PtdIns4P synthesis can be considered to be a crucial step in the regulation of the numerous cellular processes under the control of these lipids [2–4]. Secondly, accumulating evidence implicates PtdIns4P as a regulatory molecule in its own right, via its interaction with a unique repertoire of binding proteins (reviewed in [5]). Synthesis of a Golgi pool of PtdIns4P is required for protein secretion in budding yeast [6,7] and PtdIns4P's interaction with a variety of adaptor proteins is required for vesicular traffic from the mammalian Golgi apparatus [8–10], where it is also required for non-vesicular transport and synthesis of sphingolipids [11,12]. Beyond the Golgi apparatus, PtdIns4P has been implicated in the control of vesicular traffic in the endocytic pathway [13–15] and to function in regulated secretory organelles [16–18].
PtdIns4P synthesis is catalysed by activity of PI4K (phosphatidylinositol 4-kinase). There are two evolutionarily diverse PI4Ks, the wortmannin-insensitive type II, and the wortmannin-sensitive type III. α and β isoforms of each are found in mammals (reviewed in [1]). Although much is known about the subcellular distribution of these enzymes, this does not necessarily inform as to the distribution of the active population, or localization of the lipid product. For example PI4K IIIα is found predominantly at the ER (endoplasmic reticulum) [19], yet is responsible for synthesis of a plasma membrane pool of PtdIns(4,5)P2 hydrolysed during PLC (phospholipase C) signalling [20]. Therefore, obtaining information on the subcellular distribution of PtdIns4P will be particularly useful for understanding the diverse functions of this molecule.
Imaging of living cells expressing fluorescent proteins fused to inositol lipid-binding domains has revolutionized the study of these lipids [21]. However, studies using PtdIns4P-binding fluorescent proteins have carried a number of caveats. Firstly, a family of PtdIns4P-binding PH (pleckstrin homology) domains, including those from FAPP1 (phosphatidylinositol-4-phosphate adaptor protein 1, also known as PLEKHA3) and OSBP (oxysterol-binding protein), were found to detect the lipid in Golgi membranes, but only when it was co-incident with the small GTPase Arf1 (ADP-ribosylation factor 1) [10,22]. Secondly, the Osh2p (a yeast homologue of OSBP) PH domain was found to bind PtdIns4P and PtdIns(4,5)P2 in vitro [23], but to bind plasma membrane PtdIns4P exclusively when expressed in cells [20,23].
To try and clarify the subcellular distribution of PtdIns4P, we have been developing techniques for the immunocytochemical detection of this lipid, using a commercially available antibody. In the present study, we describe conditions for the cytochemical preservation of a number of organellar membranes and the detection of PtdIns4P in a subset of these (the plasma membrane and Golgi). Furthermore, we have used these techniques to analyse the biosynthesis and biological features of different cellular pools of PtdIns4P and PtdIns(4,5)P2.
EXPERIMENTAL
Materials and antibodies
Monoclonal anti-PtdIns(4,5)P2 antibody (clone 2C11) and anti-PtdIns4P antibody (both IgM) are from Echelon Biosciences, and were used at 2.5 and 8 μg/ml respectively. We also used a monoclonal anti-γ-adaptin antibody [clone 100/3; Sigma–Aldrich, an IgG2b (a gift from M. Robinson, Cambridge Institute for Medical Research, Cambridge, U.K.)] at 1:500, anti-GM130 (cis-Golgi matrix protein of 130 kDa) antibody and anti-p115 antibody (BD Transduction Laboratories, both IgG1) at 1:400 and anti-δ-adaptin (a gift from A. Peden, Cambridge Institute for Medical Research, Cambridge, U.K.; [24]) at 1:5000. Polyclonal rabbit anti-giantin antibody and anti-M6PR (mannose-6-phosphate receptor) antibody (both from Abcam) were used at a 1:500 and 1:125 dilutions from the supplied concentration respectively. Antibodies were stored at −20 °C in 50% glycerol. Anti-GST (glutathione transferase) antibody (Chemicon) was stored at 4 °C and used at 5 μg/ml. Highly cross-absorbed Alexa Fluor® 488, Alexa Fluor® 555 and Alexa Fluor® 647 goat anti-mouse antibody or anti-rabbit IgGs (H+L) antibodies, as well as goat anti-mouse IgM, IgG1 and IgG2b antibodies were from Invitrogen. These were stored at −20 °C in 50% glycerol and used at a concentration of 5 μg/ml. Biotinylated EGF (epidermal growth factor) pre-conjugated with streptavidin Alexa Fluor® 488 (Invitrogen) was dissolved in PBS with 1% (w/v) BSA to 200 μg/ml, and stored in single-use aliquots at −20 °C.
16% (w/v) methanol-free FA (formaldehyde) and 25% (v/v) electron microscopy grade GA (glutaraldehyde) stock solutions were purchased from TAAB Laboratories. FA was diluted to 8% (v/v) in PBS and stored at −20 °C. Alcohol soluble, high-purity digitonin (Calbiochem) was prepared as a 20 mM stock solution in DMSO and stored in single-use aliquots at −20 °C. Saponin (from Quillaja bark; Sigma–Aldrich) was dissolved to 5% (w/v) in buffer A (20 mM Pipes, pH 6.8, 137 mM NaCl, 2.7 mM KCl) and stored in single use aliquots at −20 °C. Wortmannin was from Calbiochem and all other reagents were from Sigma–Aldrich. POPC (1-palmitoyl 2-oleoyl phosphatidylcholine; Avanti Polar Lipids) liposomes containing 5% (mol/mol) of the indicated dipalmitoyl inositol lipids (Echelon Biosciences) were prepared from a dried film of lipid (2 h under high vacuum) by hydration and sonication in buffer A to a final concentration of 140 μM total [lipid]. Liposomes were stored at room temperature and used on the day of preparation.
Plasmids encoding GFP (green fluorescent protein)–2×FYVE-Hrs (hepatocyte growth factor-regulated tyrosine kinase substrate) for mammalian expression and GST–2xFYVE-Hrs for bacterial expression [25], were a gift from H. Stenmark (The Norwegian Radium Hospital, Oslo, Norway). Bacterial expression vectors encoding GST–tagged PH-FAPP1 (human) and PH-PLCδ1 (rat) were gifts from O. Gozani (Stanford University, Stanford, Ca, U.S.A.) and M. Katan (The Institute of Cancer Research, London, U.K.) respectively. PH-FAPP1–GFP [26] was a gift from T. Balla (National Institute of Child Health and Human Development, Bethesda, MD, U.S.A.). GFP–Pharbin was used as described previously in [27], and the putative catalytic residue aspartate-559 was mutated to alanine by site-directed mutagenesis. All constructs were checked by dideoxy sequencing.
Proteins
Bacterial expression vectors encoding GST-tagged proteins were transformed into BL21 (DE3) E. coli cells (Promega) by heat shock, and single colonies were grown in 500 ml of 2× YT media (1.6% (w/v) tryptone, 1.0% (w/v) yeast extract, 0.5% NaCl, 50 μg/ml ampicillin) until an attenuance at 600 nm (D600) of 0.6–1.5 was reached, at which time protein expression was induced by addition of IPTG (isopropyl β-D-thiogalactoside; Sigma–Aldrich) to a final concentration of 1 mM. After 3 h at 37 °C with vigorous shaking, cells were harvested by centrifugation at 3000 g, washed twice in ice-cold TBS (Tris-buffered saline; 10 mM Tris/HCl, pH 8.0, 150 mM NaCl) and re-suspended in 20 ml TBS with 1 mM EDTA, 0.5 mM PMSF, 4 μg/ml pepstatin A, 1 mM benzamidine HCl and 0.1% 2-mercaptoethanol. The suspension was lysed by incubation with 2 mg lysozyme for 30 min at 4 °C with gentle agitation, followed by sonication and further agitation for 30 min at 4 °C with Triton X-100 added to 1% (w/v). Insoluble material was removed by centrifugation at 18000 g for 20 min at 4 °C, and the cleared lysate added to 2.5 ml packed GSH–Sepharose beads; GST-tagged proteins were left to bind overnight at 4 °C with gentle agitation. After two washes with cold TBS, proteins were eluted twice by addition of 50 mM GSH in TBS (pH re-adjusted to 8.0). The eluates were pooled and concentrated by dialysis with a 3.5 kDa molecular mass cut-off dialysis membrane against 8 kDa poly(ethylene glycol) in buffer A, followed by further extensive dialysis in buffer A. Proteins were quantified by the Bradford reagent and by Coomassie Brilliant Blue staining of polyacrylamide gels with densitometric analysis of BSA standards; comparison of the two results estimated the proteins as 95% pure. Proteins were flash frozen in liquid nitrogen in single use aliquots and stored at −80 °C. For use as probes for lipids, aliquots were stored in 50% glycerol at −20 °C.
Cells
Cells (except DT-40) were grown in growth medium [DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) FBS (fetal bovine serum), 100 units/ml penicillin, 100 μg/ml streptomycin] at 37 °C, in a humidified 10% CO2 atmosphere, and were passaged twice per week by gentle dissociation with TrypLE (all from Invitrogen). DT-40 cells were cultured as described previously in [28].
Cells (1000–2000) were plated the day before transfection or staining in a final volume of 25 μl on Teflon-coated glass slides containing eight 6 mm wells (Scientific Laboratory Supplies) pre-coated with poly-L-lysine. For DT-40 cells, slides were also coated with Cell-Tak according to the manufacturer's instructions (BD Biosciences). For transfection, 5 μl of Opti-MEM (Invitrogen) containing 50 ng DNA and 150 ng Lipofectamine™ 2000 (Invitrogen), pre-complexed for 20 min, was added to the adherent cells in 25 μl growth medium. This was replaced with 25 μl of fresh growth medium after 4 h.
Where treated with inhibitors prior to fixation, the indicated agents were applied in DMEM (without Phenol Red or sodium pyruvate) supplemented with 25 mM Hepes, pH 7.4 (Invitrogen) and 10% (v/v) FBS.
Plasma membrane staining
This is modification from a previously published protocol described in [29]. Cells in 25 μl growth medium were rapidly fixed by the addition of 25 μl aldehyde in PBS to achieve a final concentration of 4% FA and 0.2% GA (glutaraldehyde); fixation was allowed to proceed for 15 min at room temperature (20–24 °C) before rinsing three times with PBS containing 50 mM NH4Cl. Slides were then placed on a metal plate in a deep ice bath and chilled for at least 2 min. All subsequent steps were performed on ice, with all solutions pre-chilled. Cells were blocked and permeabilized for 45 min with a solution of buffer A containing 5% (v/v) NGS (normal goat serum), 50 mM NH4Cl and 0.5% saponin. 100 nM GST–PH-PLCδ1 was included at this stage when the protein was used as a probe for PtdIns(4,5)P2, and GST-tagged protein was then removed by two rinses with buffer A. Primary antibodies were applied in buffer A with 5% NGS and 0.1% saponin for 1 h. After two washes in buffer A, a 45 min incubation with secondary antibody in buffer A with 5% NGS and 0.1% saponin was performed. Slides were then rinsed four times with buffer A, and cells were post-fixed in 2% FA in PBS for 10 min on ice, before warming to room temperature for an additional 5 min. FA was removed by three rinses in PBS containing 50 mM NH4Cl, followed by one rinse in distilled water. Wells were then dried, covered with 3 μl ProLong Gold (Invitrogen) supplemented with 1 μg/ml DAPI (4′,6-diamidino-2-phenylindole) and covered with 22 mm×22 mm glass cover slips (No. 1 thickness, Scientific Laboratory Supplies), and sealed with nail varnish.
Golgi staining
All steps were performed at room temperature. Cells in 25 μl medium were fixed by the addition of 25 μl pre-warmed FA in PBS to a final concentration of 2%, and incubated for 15 min. After removing FA with three rinses in PBS containing 50 mM NH4Cl, cells were permeabilized for 5 min by the addition of 20 μM digitonin in buffer A for 5 min. Digitonin was removed by three rinses in buffer A, and cells were blocked for 45 min with buffer A supplemented with 5% (v/v) NGS and 50 mM NH4Cl. When staining for PtdIns3P, 0.5 μg/ml GST–2×FYVE-Hrs was included at this stage. After removing GST-tagged proteins by two washes in buffer A, primary and secondary antibodies were applied in buffer A with 5% NGS, before post-fixation for 5 min in 2% FA and mounting, as described above for the plasma membrane protocol.
Lipid dyes
When staining for surface glycoproteins, 50 μg/ml Alexa Fluor® 555-conjugated WGA (wheat-germ agglutinin; Invitrogen) was applied to the cells on ice for 5 min in DMEM (without Phenol Red or sodium pyruvate) supplemented with 25 mM Hepes (Invitrogen) and 10% FBS, rinsed twice in the same solution and then fixed as above. Staining for plasma membrane PtdIns(4,5)P2 or endosomal PtdIns3P were performed as described in the preceding two sections. For staining of the endoplasmic reticulum, after post-fixation and rinsing in PBS/NH4Cl, cells were incubated for 1 min with DiOC6 (3,3′-dihexyloxacarbocyanine iodide; Invitrogen) in PBS, rinsed once with distilled water and mounted in ProLong Gold with 1 μg/ml DAPI as above. A similar protocol was used to stain cells with 500 μM DiD (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate; Invitrogen). For Golgi staining, the slides were chilled on ice for 2 min following the post-fixation and rinsing. They were incubated with a 5 μM solution of NBD-C6-ceramide [N-(ε-7-nitrobenz-2-oxa-1,3-diazol-4-yl-aminocaproyl)-D-erythro-sphingosine] complexed to BSA (Invitrogen) in PBS for 30 min on ice. Unbound ceramide was removed with two rinses in ice-cold PBS, and extra-Golgi ceramide was extracted by four washes with 3% (w/v) defatted-BSA (Sigma–Aldrich) in PBS over 1 h at room temperature. Finally, slides were stained for 5 min in 1 μg/ml DAPI in PBS, rinsed once in distilled water and mounted in Fluoromount-G (Southern Biotech). Note that we found retention of this latter dye was poor in glycerol-based mountants, hence the use of aqueous Fluoromount-G. Furthermore, slides stained with NBD-C6-ceramide were imaged promptly after staining, since we noticed progressive loss of the stain from the Golgi over a time course of hours.
Microscopy
Images were acquired on a Leica SP5 confocal microscope attached to a Leica DMI6000 inverted microscope stand. Excitation and emission settings for the different fluorophores were: for Alexa Fluor® 488 and DiOC6, excitation with the 488 nm line of an Argon-ion laser and emission detected at 500–550 nm; the same settings were used for NBD-C6-ceramide, except the 458 nm line of the laser was used; for Alexa Fluor® 555, excitation was with a 543 nm HeNe laser, and emission detected at 555–620 nm; for Alexa Fluor® 647 and DiD, excitation was with a 633 nm HeNe laser and emission detected at 650–700 nm; for DAPI, excitation was with a 405 nm diode laser and emission detected at 415–485 nm. Each colour fluorophore was scanned sequentially, to avoid bleed-through, or else scanned in pairs that were spectrally well separated (DAPI with Alexa Fluor® 555, Alexa Fluor® 488 with Alexa Fluor® 647 or DiD) to avoid cross-talk. Lack of cross-talk was confirmed by imaging samples where individual fluorophore-labelled probes had been omitted.
For the high-resolution images shown in the Figures, a 63× 1.4 NA (numerical aperture) plan apochromatic oil-immersion objective was used, and the pinhole was set to a diameter corresponding to 1 Airy unit (optical section ∼240 nm) for true confocal images. When acquiring z-stacks, images were scanned at ∼120 nm intervals throughout the depth of the cells being imaged. For lower resolution images, used for quantification, a 20× 0.7 NA plan-apochromatic multi-immersion objective was used, and the pinhole was opened fully to give an optical section ∼3 μm. Single scans were acquired for Golgi-stained specimens, and z-stacks of three scans at 1.5 μm intervals were acquired for plasma membrane-stained specimens, such that the entire depth of the cells was captured. 8-bit images were acquired with gain and offsets adjusted such that the entire dynamic range was used, but with little or no pixel saturation.
Quantitative image analysis
All analysis was performed using the program ImageJ version 1.38 (http://rsb.info.nih.gov/ij/). Images were imported from the Leica image format (.lif) using the LOCI bioformats importer plugin (http://www.loci.wisc.edu/ome/formats.html). Automated custom macros were written to quantify inositol lipid staining in the images as described below.
Plasma membrane staining
Along with PtdIns4P and/or PtdIns(4,5)P2 staining, cells were also stained with DAPI for nuclei and DiD for whole cell fluorescence. Image stacks were reduced to a single, 8-bit image by use of the average intensity projection function. The nuclei and DiD images were subject to a 5×5 median-pixel intensity filter, the latter was background-subtracted, and both were converted to binary images. The watershed algorithm (http://rsb.info.nih.gov/ij/plugins/watershed.html) was run on the binary nuclei image, and the resulting watershed lines were subtracted from the binary DiD image via the image calculator command to produce segmentation of touching cells. This resulted in a segmented picture of cells, which was used as a mask for the inositol lipid staining. The in-built ‘analyze particles’ function was then used to measure the area and integrated pixel intensity of cells that were not touching the edges of the image field.
GFP-expressing cells
A similar procedure was used to that described above, except the GFP signal was used to produce the mask without need for DiD or DAPI staining (since GFP expression was sparse and rarely produced more than two touching cells). Essentially, the averaged stack was background subtracted and heavily median-filtered (8×8 pixels) to produce the binary mask, which was again subjected to watershed segmentation to separate touching cells.
Golgi staining
In addition to PtdIns4P, cells were also stained for giantin. The giantin staining was subject to a 5×5 median-pixel intensity filter and converted to a binary image by autothresholding; this was dilated by a single pixel using the ‘dilate’ command. The resulting binary image was used as a mask for the PtdIns4P staining image, which was analysed using the ‘analyze particles’ command as described above.
Thus integrated pixel intensity measurements for each cell (or Golgi complex) in a field was obtained. Typically, for each experiment three randomly selected fields were imaged per condition. In order for results to be pooled across independent experiments, integrated pixel intensity, I, was normalized as follows:
 |
where Ictrl is the intensity for the control sample and Ibg is the intensity of the background specimen in which anti-PtdIns4P/PtdIns(4,5)P2 probes were omitted. Pooled, normalized data were subjected to a Kruskal–Wallis test with Dunn's multiple comparison post-test using Prism4 Software (Graphpad). Results are presented as means±S.E.M. Statistically significant deviations from control are denoted as *P< 0.05, **P<0.01, ***P< 0.001.
Co-localization analysis
This was performed on magnified images of Golgi stacks (such as those shown in Figure 3a) using the Mander's coefficients plugin (http://www.macbiophotonics.ca/imagej/index.htm). This calculates the Pearson's correlation coefficient (r) between pixel intensities in the two channels.
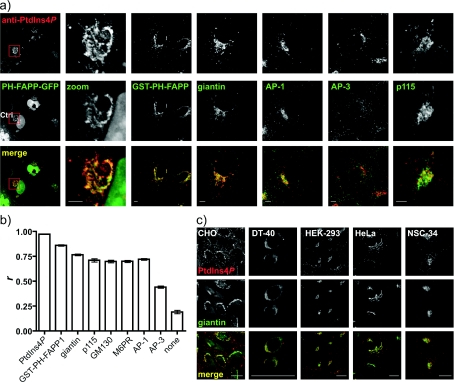
Figure 3. Anti-PtdIns4P antibody labels the Golgi apparatus at room temperature after 2% FA fixation and digitonin permeabilization.
(a) COS-7 cells were stained with anti-PtdIns4P antibody. Where indicated cells were expressing a GFP-tagged PH domain from FAPP1 (expressed for 24 h) and were co-stained using either an anti-GST antibody (against the purified GST-tagged FAPP1 PH domain) or antibodies against the indicated markers. Scale bars=5 μm. (b) Pearson's correlation coefficients for co-localized intensity were calculated between labelling for PtdIns4P and the indicated marker, as described in the Experimental section. Images not shown in (a) are in Supplementary Figure S2 at http://www.BiochemJ.org/bj/422/bj4220023add.htm. Results are presented as means±S.E.M. and a statistical comparison is shown in Table 1. (c) PtdIns4P and giantin labelling in a variety of cell types. Scale bars=20 μm. Images are single confocal optical sections.
RESULTS AND DISCUSSION
Preservation of cellular membranes for immunocytochemistry
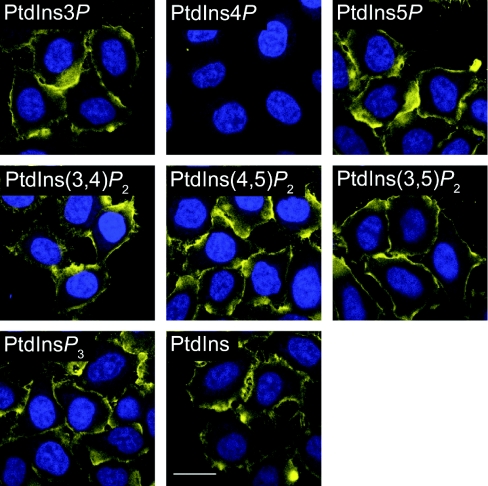
We have previously established conditions for staining of plasma membrane PtdIns(4,5)P2 using a specific monoclonal antibody [29]. Applying these conditions with a monoclonal anti-PtdIns4P antibody yielded plasma membrane staining in HeLa cells. The staining appeared specific, since it was abolished by pre-incubation of the antibody with liposomes containing 5 % (mol/mol) di-palmitoyl PtdIns4P, but not with any other inositol lipid (Figure 1). However, it was surprising to see such an exclusive plasma membrane staining, given the extensive evidence for a Golgi pool of PtdIns4P (described in the Introduction section).
Figure 1. Specificity of the anti-PtdIns4P antibody.
HeLa cells were fixed with 4% FA plus 0.2% GA and stained on ice using saponin permeabilization, as described in the Experimental section. Anti-PtdIns4P antibody was pre-absorbed with POPC liposomes containing 5% (mol/mol) of the indicated inositol lipid for 1 h at room temperature prior to applying to the cells on ice. Anti-PtdIns4P antibody staining is yellow, and DAPI-stained nuclei are in blue. Scale bar=20 μm. Images are single confocal optical sections.
Preservation of plasma membrane PtdIns(4,5)P2 requires a number of specific conditions, notably: (i) the inclusion of GA, either alone or in conjunction with FA; (ii) the use of saponin as the permeabilizing detergent; and (iii) maintaining temperatures ≤4 °C during fixation and staining [29]. However, staining of Golgi membranes with the fluorescent lipid NBD-C6-ceramide was abolished by low concentrations of saponin [30]. Therefore a comprehensive study of conditions that would preserve various cellular membranes was undertaken. Although there are many commercially available markers for cellular organelles, these tend to be antibodies raised against endogenous proteins, which will be cross-linked by aldehydes during fixation and hence become resistant to extraction by detergents. This is in marked contrast with the lipids, which generally do not contain primary amines and are therefore not cross-linked during fixation. For these reasons, we chose probes that either detect endogenous lipids, or are fluorescent lipids themselves and can intercalate into the bilayer. Such probes should provide an indication of the integrity of the endogenous organellar membranes. COS-7 fibroblasts were used for this purpose, as these cells have a large, flat cytoplasm in which visualization of various organelles is convenient.
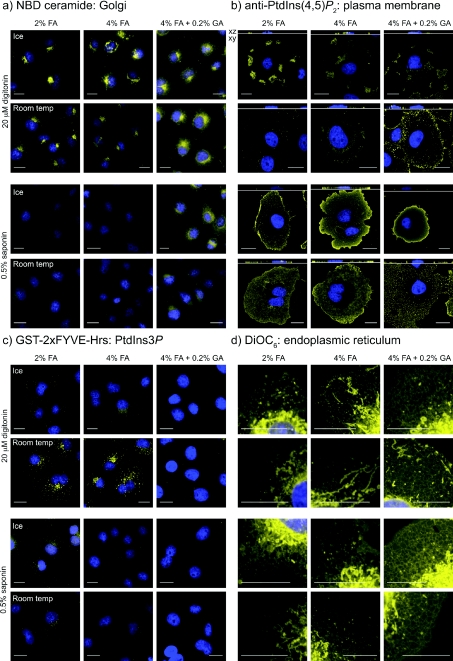
Figure 2 shows the results from such a study, where strength of fixation was varied (by increasing FA concentration and adding GA), staining at room temperature was compared to staining on ice, and the permeabilizing detergent digitonin was compared with saponin. Using NBD-C6-ceramide as a post-fixation marker for Golgi membranes [30], it is clear that staining is abolished by saponin, whereas it persists when digitonin is employed (Figure 2a). Fixation and temperature conditions did not seem to drastically affect the staining in the presence of digitonin, although distinct Golgi staining was clearer when GA was omitted (probably because there is reduced autofluorescence in the cells).
Figure 2. Effect of fixation, detergent and temperature on the retention of various organelle-specific lipid stains.
Cells were fixed with the indicated concentration of aldehydes, then stained at the designated temperature after permeabilization with digitonin or saponin. See the Experimental section for details. Agents were used that specifically label the (a) Golgi (NBD-ceramide), (b) plasma membrane (anti-PtdIns(4,5)P2 antibody), (c) endosomes (anti-GST against GST–2×FYVE-Hrs bound to PtdIns3P) or (d) endoplasmic reticulum (DiOC6). DAPI-stained nuclei are shown in blue. Scale bars=20 μm.
To follow the integrity of the plasma membrane under these conditions, we used anti-PtdIns(4,5)P2 antibodies, because this lipid is known to be enriched at the plasma membrane [31–34]. PtdIns(4,5)P2 staining was patchy, at best, when digitonin was used to permeabilize cells, whereas saponin gave a much more uniform pattern (Figure 2b). Staining was more even and consistent when performed on ice, and also after fixation with increasing concentrations of FA, with the best results obtained after fixation with a FA/GA mix (Figure 2b). Hence the conditions that preserve plasma membrane staining are diametrically opposed to those best for Golgi staining.
Given the different conditions required for preservation of Golgi and plasma membranes, further membranes were then tested under these conditions. PtdIns3P is enriched on endosomal membranes and can be detected using a tandem FYVE domain probe from Hrs, which labels scattered cytosolic puncta [25]. We therefore used a GST-tagged version as a probe for endosomal membrane integrity, using anti-GST antibodies to detect the bound protein. Punctate staining was abolished by use of GA in the fixative, or when staining on was performed on ice (Figure 2c). Whereas limited punctate staining was observed at room temperature in FA-fixed cells permeabilized with saponin, by far the clearest labelling was seen with digitonin permeabilization. Under these conditions, the labelling appeared specific, since it co-localized with an expressed GFP–2×FYVE-Hrs probe, and staining was inhibited by pre-treatment of cells with the PI3K (phosphoinositide 3-kinase) inhibitor wortmannin (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/422/bj4220023add.htm).
Finally, we used the fluorescent lipid DiOC6, which labels a continuous thread-like lattice corresponding to the ER, as opposed to the brightly stained snake- or sausage-like mitochondria [35]. No great disparity was seen in cells permeabilized with saponin or digitonin (Figure 2d), although ice-cold temperatures during permeabilization and staining produced the best preservation of the ER network, with optimal results observed after GA/FA fixation.
Taking all these results into account, we decided to proceed with two distinct staining protocols. The first, called the ‘Golgi staining protocol’, relies on fixation in 2% FA and a short permeabilization step with digitonin, with all steps performed at room temperature. This protocol is expected to produce good preservation of Golgi membranes, as well as other endomembranes such as those containing PtdIns3P (Figures 2a–2c), but at the expense of plasma membrane integrity (Figure 2b). Note that the best results, in terms of Golgi labelling, were obtained when a short 5 min permeabilization step with digitonin was performed, as longer incubations were detrimental. Similarly, we tried a number of concentrations of digitonin, and found 20 μM to be in the optimal range (results not shown).
The second procedure, called the ‘plasma membrane staining protocol’, was selected for its ability to preserve the plasma membrane, and involved fixation with a GA/FA mix, permeabilizing with saponin and performing all post-fixation steps on ice (Figure 2b). Under these conditions, good preservation of ER membranes is expected (Figure 2d), although Golgi and endosomal membrane staining will be abolished (Figures 2a and 2c). This procedure is a modification of the method previously published in [29]. First, fixation in the new protocol was performed at room temperature for 15 min, as temperature during fixation was not found to be critical for preservation of plasma membrane staining (G.R.V. Hammond, unpublished observations). Secondly, GA concentrations of 0.05% and greater were sufficient to produce optimal plasma membrane labelling, although higher concentrations caused elevated levels of autofluorescence. Thirdly, considerably shorter incubation periods (see the Experimental section) were found to be sufficient compared to the previous report, and even to produce better results. Finally, although the concentration of saponin (0.1–0.5%) is relatively high, we found staining of the plasma membrane was less even and intense at lower concentrations. Similarly, inclusion of saponin throughout the staining protocol was found to be optimal and exposure of cells to shorter periods of saponin permeabilization produced less labelling (results not shown). We have also tried using Tween 20 as the permeabilizing agent, as this was shown to produce staining of the plasma membrane and nuclear envelope with anti-PtdIns(3,4)P2 antibodies [36]. However, this did not produce such a consistent membrane labelling with anti-PtdIns(4,5)P2 when compared to saponin (results not shown).
The use of cooler temperatures during staining for PtdIns(4,5)P2 has also been shown to produce better labelling by other groups for various probes [31,32]. Furthermore, recent studies using various probes for PtdIns(4,5)P2 and PtdIns(3,4,5)P3 have described fixation with GA/FA, coupled with saponin permeabilization, resulting in good retention of labelling at the plasma membrane [37–40]. It has also been reported that calcium should be sequestered during fixation, with EGTA, to prevent PtdIns(4,5)P2 breakdown [32], although when comparing fixation using a GA/FA mix, we have seen no difference between cells fixed in the presence or absence of EGTA (G. R. V. Hammond, unpublished observations). Together, these studies demonstrate the general application of the above conditions for staining of inositol lipids.
Golgi staining protocol
Staining of COS-7 cells with anti-PtdIns4P using the ‘Golgi staining protocol’ described above produced labelling of peri-nuclear structures resembling the Golgi apparatus (see Figures 3–6). To formally identify this structure as the Golgi, we performed co-localization experiments with the PH domain from FAPP1, as this protein exhibits PtdIns4P-dependent localization to Golgi membranes [10,22,26,41]. COS-7 cells were either transfected with a GFP-tagged FAPP1 PH domain or were co-labelled with a GST-conjugated recombinant version. In either case, good overlap was seen between these probes and anti-PtdIns4P (Figure 3a). Green fluorescence of PH-FAPP1 was generally coincident with the red staining of the antibody, but there were regions of red fluorescence devoid of PH domain labelling (Figure 3a). This is perhaps explained by the fact that PH-FAPP1 detects PtdIns4P only where it is co-incident with the small GTPase Arf1 [10,22].
We also performed co-localization experiments with other markers of the Golgi apparatus, namely the cis/medial-resident giantin and GM130 proteins, the trans-Golgi recruited clathrin adaptor AP-1 (adaptor protein 1) and the Golgi/late endosome cycling M6PR. All showed staining tightly juxtaposed with the PtdIns4P antibody (Figures 3a and see Supplementary Figure S2 at http://www.BiochemJ.org/bj/422/bj4220023add.htm). Furthermore, PtdIns4P synthesis is important for ER-to-Golgi trafficking from ER exit sites [42], so we co-labelled the cells with p115, a marker of the ERGIC (ER-Golgi intermediate compartment) and cis-Golgi membranes ([43,44]; Figure 3a), which again showed a distribution tightly juxtaposed with PtdIns4P staining. We quantified the overlap using Pearson's correlation coefficient (r), a simple metric that measures the degree of correlation between pixel intensities in the two channels, and hence degree of overlap. This analysis was calibrated by establishing the maximum degree of co-localization (theoretically, r=1, actually r=0.97) by detecting the anti-PtdIns4P antibody with two different colours of secondary antibodies (Figure 3b). The minimum overlap (theoretically, r=0) was measured by correlating the anti-PtdIns4P antibody staining without another marker present (r=0.19, Figure 3b). All of the Golgi markers showed a very similar degree of overlap with PtdIns4P, regardless of their localization at cis-, medial- and trans-Golgi compartments, or at the ERGIC (Figure 3b and Table 1). In addition, none of the markers were as tightly distributed with PtdIns4P-labelling as with the PtdIns4P-reporter PH-FAPP1 (Figure 3b and Table 1). Together, these results confirm staining of the Golgi apparatus with the anti-PtdIns4P antibody, although it is not possible to ascertain the sub-Golgi localization of the signal at the resolution of the light microscope. We observed similar staining tightly juxtaposed with giantin in a number of different cell lines from diverse lineages, including epithelial cells [CHO (Chinese-hamster ovary), HEK (human embryonic kidney cells)-293, HeLa], a chicken B-cell lymphoma (DT40) and a neuroblastoma/motor neuron fusion cell line (NSC-34), as shown in Figure 3c.
Table 1. Co-localization between anti-PtdIns4P antibody labelling and Golgi markers.
Results of a Kruskal–Wallis test with Dunn's multiple comparison test for differences between the Pearson's correlation coefficients (r) presented in Figure 3(b). Numbers in parentheses represent number of cells for each marker (n). Anti-PtdIns4P antibody was visualized using the Alexa Fluor® 555-conjugated secondary antibody, and Pearson's correlation coefficient (r) between this and the Alexa Fluor® 488-conjugated antibody staining of the second probe was quantified as described in the Experimental section. For anti-PtdIns4P antibody with itself, Alexa Fluor® 488 and Alexa Fluor® 555 goat anti-mouse secondary antibodies were used to produce a ‘perfect’ co-localization. For ‘None’, an Alexa Fluor® 488-conjugated goat anti-rabbit secondary was used without primary antibody. n.s., not significant; *, P<0.05; **, P<0.01; ***, P<0.001.
| PtdIns4P | GST–PH-FAPP1 | giantin | p115 | GM130 | M6PR | AP-1 | AP-3 | |
|---|---|---|---|---|---|---|---|---|
| PtdIns4P (34) | ||||||||
| GST–PH-FAPP1 (29) | n.s. | |||||||
| giantin (110) | *** | ** | ||||||
| p115 (36) | *** | *** | n.s. | |||||
| GM130 (32) | *** | *** | n.s | n.s. | ||||
| M6PR (58) | *** | *** | * | n.s. | n.s. | |||
| AP-1 (47) | *** | *** | n.s | n.s. | n.s. | n.s. | ||
| AP-3 (73) | *** | *** | *** | *** | *** | *** | *** | |
| None (33) | *** | *** | *** | *** | *** | *** | *** | n.s. |
Whereas Golgi staining is apparent using anti-PtdIns4P antibody, no such staining is observed with anti-PtdIns(4,5)P2 antibody under the same conditions (see Supplementary Figure S3 at http://www.BiochemJ.org/bj/422/bj4220023add.htm), consistent with the observation that PtdIns(4,5)P2 is not prevalent in the Golgi [31]. Replacing digitonin with Triton X-100, which would be expected to completely solubilize the Golgi membranes in fixed cells [30], abolishes Golgi staining with anti-PtdIns4P (see Supplemental Figure S3). On the other hand, the same treatment unmasks a speckled nuclear staining with the anti-PtdIns(4,5)P2 antibody, as has already been well-characterized with this antibody [45,46]. This observation underscores the importance of selecting appropriate conditions for detection of lipids in specific membranes (compare anti-PtdIns(4,5)P2 labelling in Figure 2(b) with that after Triton X-100 in Supplemental Figure S3).
In addition to the peri-nuclear Golgi staining, the anti-PtdIns4P antibody also labelled a few scattered cytosolic puncta (see PtdIns4P channel in Figures 3–6). As several studies have identified the presence of type II PI4Ks in endocytic compartments [13–15], we undertook co-labelling experiments to test whether anti-PtdIns4P stained these structures. Specifically, PtdIns4P has been reported to interact directly with the clathrin adaptor AP-1 at trans-Golgi membranes [8], and PI4K IIα has been shown to localize to lysosomal/endosomal structures containing the clathrin adaptor AP-3 [15]. However, the anti-PtdIns4P positive puncta were clearly distinct from the cytosolic structures labelled with probes against either adaptor (Figure 3a). Despite the peri-nuclear enrichment of AP-3 staining, labelling was poorly co-incident with the Golgi PtdIns4P signal (Figure 3a) as shown by the co-localization analysis (Figure 3b and Table 1).
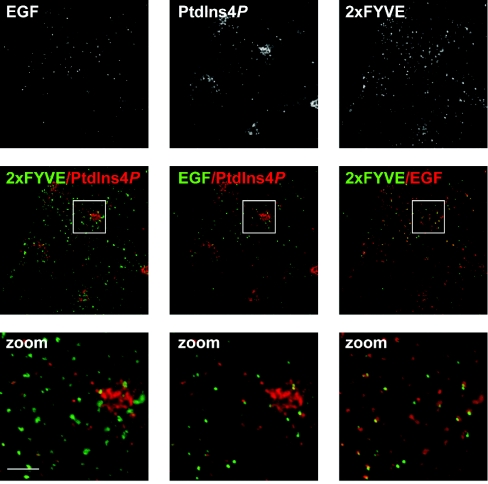
Type IIα PI4K has been reported at specific endosomal compartments containing internalized EGF receptors [14]. To further explore the distribution of such endosomes, COS-7 cells were loaded with fluorescent EGF for 30 min, before fixing and co-staining with the anti-PtdIns4P antibody. The cells were also stained using anti-GST antibodies to detect GST-2×FYVE-Hrs, a probe for PtdIns3P, which is known to be prevalent in endosomes [25]. Figure 4 shows that cytosolic PtdIns4P-labelled puncta were excluded from the EGF-containing structures, as well as from PtdIns3P-containing endosomes, despite the fact that the majority of EGF puncta appeared to be within PtdIns3P-labelled endosomes (Figure 4). Therefore, the punctate cytosolic structures labelled with anti-PtdIns4P do not correlate with membranes known to contain PtdIns4P (or enzymes that synthesize it); the specific identity and functional relevance of PtdIns4P in such compartments is a future avenue of research.
Figure 4. Co-localization of PtdIns4P and PtdIns3P with internalized EGF.
COS-7 cells were serum-starved overnight, then incubated for 30 min with 10 μg/ml Alexa Fluor® 488-conjugated EGF, prior to fixing and staining for PtdIns4P and PtdIns3P as described in Figure 3. Images labelled ‘zoom’ show the enlarged region outlined in the panels above. Scale bars=5 μm. Images are single confocal optical sections.
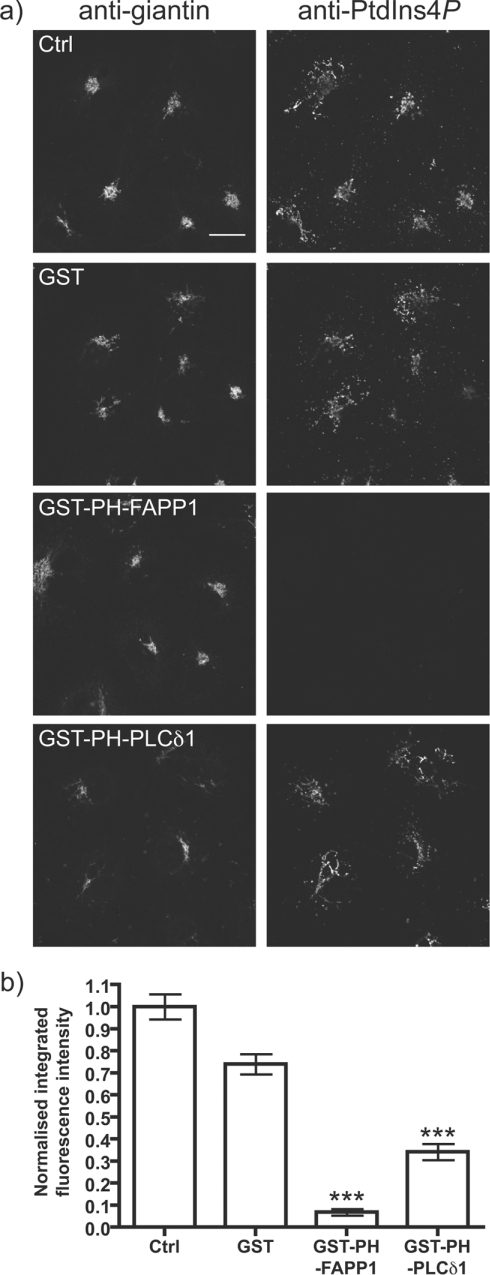
To further assess the specificity of Golgi staining with anti-PtdIns4P, we used GST-tagged PH-FAPP1 at high concentration (10 μM) to sequester the endogenous PtdIns4P. This protein indeed abolishes staining, whereas GST alone had no affect (Figures 5a and 5b). GST–PH-PLCδ1, which should be more specific for PtdIns(4,5)P2 compared with PtdIns4P [47,48] did not abolish Golgi staining (Figure 5a), but quantitative analysis revealed an approx. 50% decrease in staining intensity (Figure 5b). This might suggest that some of the observed signal with the anti-PtdIns4P antibody is due to the presence of PtdIns(4,5)P2, as PH-FAPP1 can bind both lipids in vitro [23]. This, however, seems unlikely given the specificity of the antibody for exogenous competing lipid (Figure 1), as well as the fact that anti-PtdIns(4,5)P2 does not label the Golgi (see Supplemental Figure S3), which is known to anyway contain relatively little PtdIns(4,5)P2 [31]. Rather, we suspect the partial competition is due to some weak PtdIns4P-binding by PH-PLCδ1, which may be exacerbated by the high concentration of protein used, and the presence of the GST tag, which effectively increases affinity [49].
Figure 5. Competition for Golgi PtdIns4P staining by PH-FAPP1.
(a) The indicated GST-tagged fusion proteins (10 μM) were included with the primary anti-PtdIns4P antibody and anti-giantin antibodies applied to COS-7 cells. Staining conditions are as Figure 3. Single confocal optical sections are shown. Scale bar=20 μm. (b) Quantification of anti-PtdIns4P antibody labelling (n=207). See the Experimental section for details. ***P<0.001.
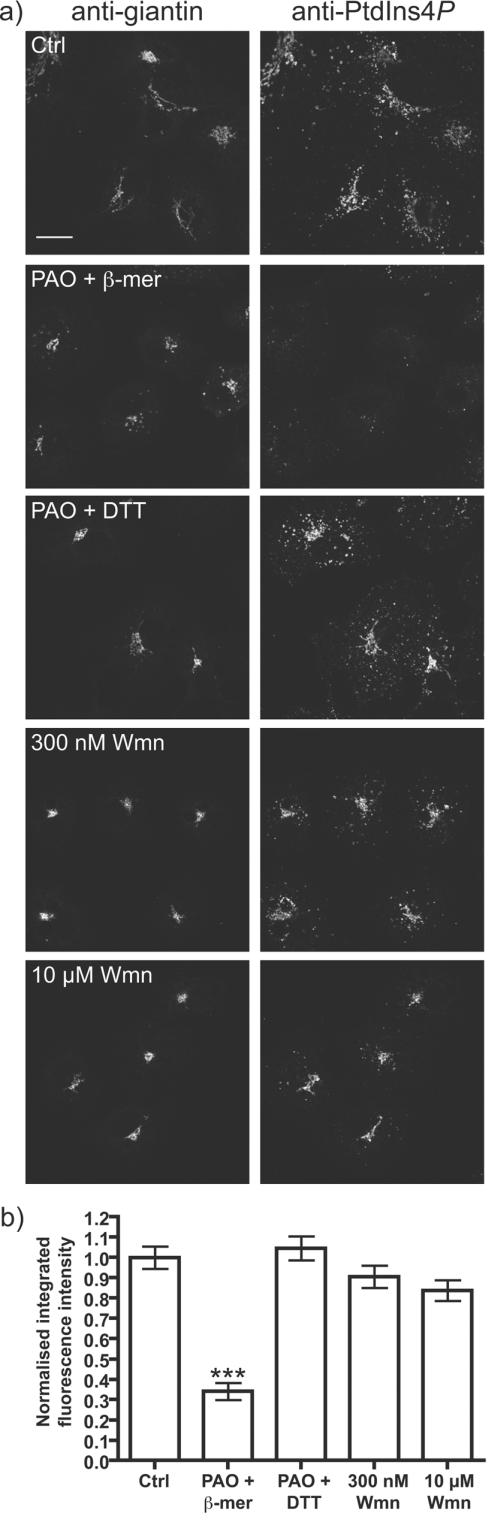
PtdIns4P is synthesized by at least two Golgi-resident PI4Ks, PI4K IIα and PI4K IIIβ [1,41], although the evidence suggests that PI4K IIβ may also play a role [50]. To address the relative contributions of these isoforms to Golgi PtdIns4P staining, we incubated cells for 30 min before fixation with a high concentration of wortmannin (10 μM), which should abolish type IIIβ (and α) activity [11]. Figure 6 shows that wortmannin addition produces no detectable change in the PtdIns4P staining intensity at the Golgi, despite the fact that it induces partial dissociation of PtdIns4P-bound proteins [26] and a redistribution of Golgi PtdIns4P [41] over this time period. This result suggests that the type III enzyme contributes a relatively small, or slowly turned-over fraction, of the steady-state PtdIns4P levels in the Golgi, despite the functional importance of this enzyme to Golgi function [11,51]. It can also be inferred that a type II activity generates the majority of the steady-state pool. However, unambiguous assignment of PI4K activities to distinct pools of PtdIns4P are hampered by a lack of pharmacological inhibitors of the type II enzymes, and RNA-interference based methods have been shown to produce only very minor effects on cellular PtdIns4P levels [20].
Figure 6. Effect of PI4K inhibitors on Golgi PtdIns4P labelling.
(a) COS-7 cells were pre-treated for 30 min at 37 °C with 0.1% DMSO (Ctrl), 10 μM PAO with 1 mM 2-mercaptoethanol or DTT (PAO+β-mer or PAO+DTT) and 300 nM or 10 μM wortmannin (Wmn) as indicated, prior to fixation and staining as described in Figure 3. Single confocal optical sections are shown. Scale bar=20 μm. (b) Quantification of anti-PtdIns4P labelling (n=237). See Experimental section for details. *** P<0.001.
We also tested the arsenical compound PAO (phenylarsine oxide) (Figure 6). Surprisingly, 10 μM PAO dramatically reduced Golgi PtdIns4P staining (Figure 6b), resulting in scattered cytosolic puncta (Figure 6a). PAO reacts with sulfhydryl groups, and its effects are reversed by a powerful reducing agent such as DTT (dithiothreitol), but not by the weaker 2-mercaptoethanol (Figure 6b), consistent with earlier reports [20]. PAO primarily inhibits PI4K IIIα at this concentration, which should also be blocked by 10 μM wortmannin [13]. Therefore, this result is not a direct effect of PAO on Golgi PI4Ks.
In parallel experiments, we measured the effects of these compounds on staining using the anti-GST antibodies to detect GST–2×FYVE-Hrs-bound endosomal PtdIns3P (see Supplemental Figure S1). Whereas PAO had no effect on staining, wortmannin caused a dramatic diminution of staining at 300 nM, a concentration selective for PI3K over PI4K.
Plasma membrane staining
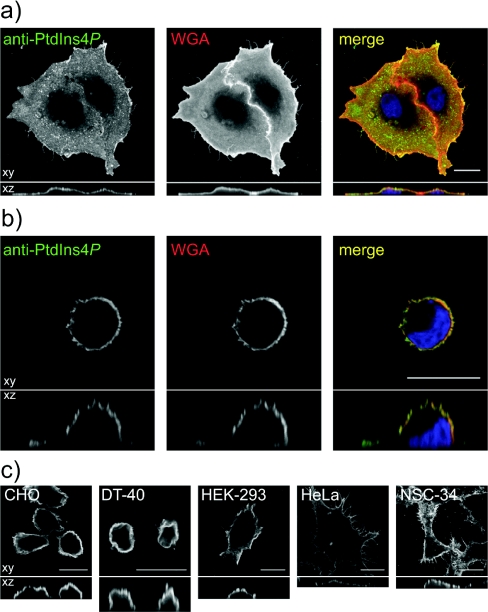
Consistent with the observations from Figure 1, staining of cells using the ‘plasma membrane staining’ protocol generates surface labelling of cells with anti-PtdIns4P antibody. This can be seen in COS-7 cells co-stained with Alexa® Fluor 555-conjugated WGA to label surface glycoproteins (Figures 7a and 7b). Owing to the flattened nature of adherent COS-7 cells, the plasma membrane does not appear as a ring of fluorescence in confocal optical sections, although sections through the z-axis reveal the surface nature of the labelling (Figure 7a). A clearer example of this surface labelling is seen in COS-7 cells adhered to glass for just 20 min prior to fixation, (i.e. before they have had time to spread). These cells clearly show a ring of surface labelling in both x-, y- and z-confocal sections (Figure 7b). As for Golgi staining, this surface labelling was also observed in a number of other cell types (Figure 7c). Note that in subsequent experiments, to clearly show changes in whole-cell plasma membrane labelling, maximum intensity projections from confocal sections spanning the entire depth of the cells are shown. Hence the staining appears rather uniform across the cells.
Figure 7. Anti-PtdIns4P antibody labels the plasma membrane of cells fixed with FA and GA, permeabilized with saponin and stained on ice.
(a) COS-7 cells grown overnight prior to fixation and staining or (b) fixed and stained 20 min after seeding on poly-L-lysine. In both cases, cells were incubated for 5 min on ice with 50 μg/ml Alexa Fluor® 555-conjugated WGA prior to fixing to label the plasma membrane. (c) Different cell types stained with anti-PtdIns4P antibody using the above conditions. All images show single confocal optical sections in the xy plane (upper panels), or sections through the z plane (lower panels). Scale bars=20 μm.
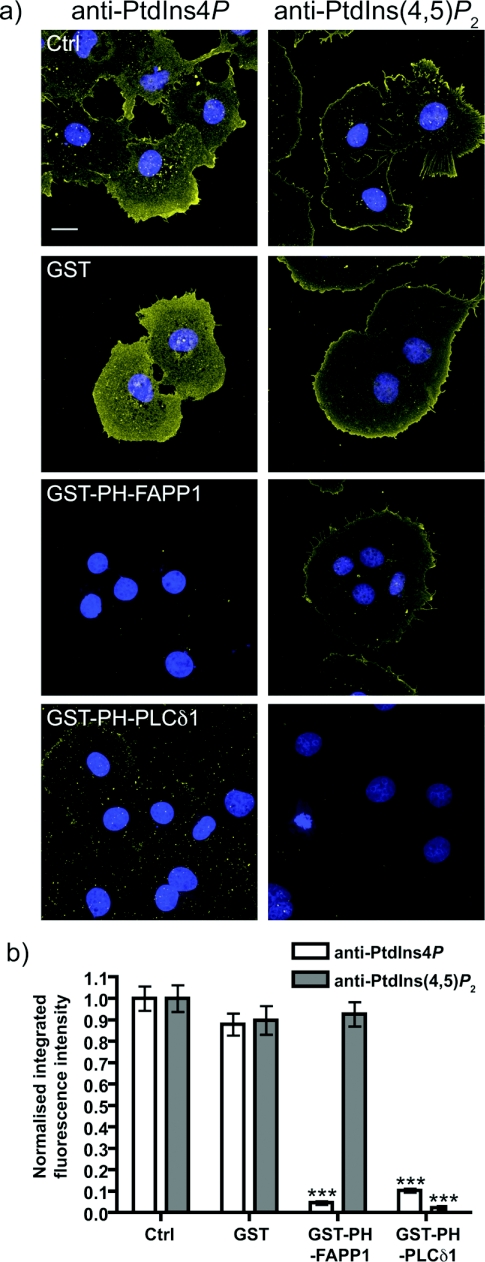
The specificity of the staining was again assessed by sequestration of endogenous PtdIns4P with GST–PH-FAPP1. As seen for Golgi staining, this completely abolished staining, whereas GST alone had no effect (Figure 8). However, the PtdIns(4,5)P2-sequestering PH domain from PLCδ1 produced a near ablation of staining (Figure 8). This is in marked contrast with parallel experiments using anti-PtdIns(4,5)P2 antibody, where only GST–PH-PLCδ1 was able to abolish staining, consistent with our earlier report [29]. Given the specific competition of PtdIns4P staining by exogenous PtdIns4P compared with PtdIns(4,5)P2 (Figure 1), it seems unlikely that this result reflects reactivity of anti-PtdIns4P antibody with plasma membrane PtdIns(4,5)P2. As argued above, it may simply reflect binding of high concentrations of GST–PH-PLCδ1 to plasma membrane PtdIns4P. However, given the enrichment of PtdIns(4,5)P2 at the plasma membrane, cross-reactivity was more of a concern than for Golgi staining, so we conceived further experiments to eliminate this possibility.
Figure 8. Competition for plasma membrane PtdIns4P staining by PH-FAPP1.
(a) The indicated GST-tagged fusion proteins were included at 10 μM with the primary anti-PtdIns4P or anti-PtdIns(4,5)P2 antibodies applied to COS-7 cells. Staining conditions are as in Figure 7. Images are maximum intensity projections of confocal sections spanning the entire depth of the cells. Scale bar=20 μm. DAPI stained nuclei are in blue indicating the presence of cells where lipid staining is absent. (b) Quantification of staining (n=126 cells for anti-PtdIns4P antibody and 123 cells for anti-PtdIns(4,5)P2 antibody). See the Experimental section for details. ***P<0.001.
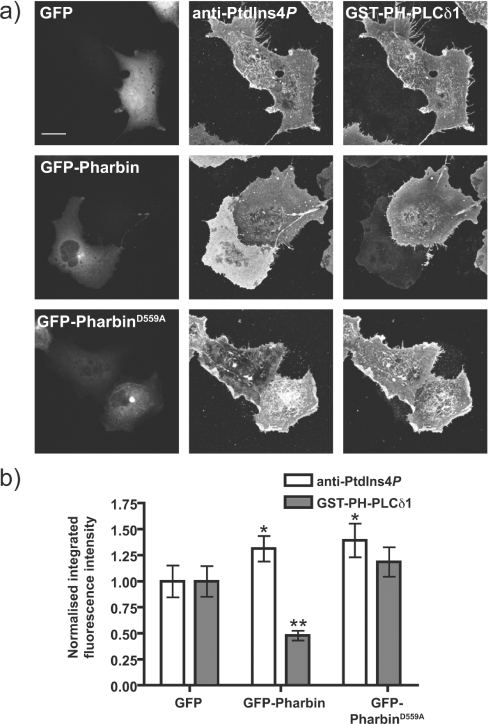
Expression of an inositol 5-phosphatase activity would be expected to decrease PtdIns(4,5)P2 levels in the plasma membrane without depletion of PtdIns4P. To this end, we transfected COS-7 cells with GFP-tagged pharbin, an enzyme with PtdIns(4,5)P2 5-phosphatase activity [52]. Cells were then fixed and co-stained with an anti-PtdIns4P antibody and anti-GST antibody to stain for GST–PH-PLCδ1 and follow PtdIns(4,5)P2 [31]. Over-expressed pharbin was localized throughout the cytosol and was particularly enriched on the Golgi, as reported previously in [52,53]. Despite its preference for PtdIns(3,4,5)P3 as a substrate [53], the over-expressed enzyme caused reduction in the intensity of staining against PtdIns(4,5)P2 compared with neighbouring cells without visible GFP expression. This effect was not seen in GFP-expressing cells or in cells expressing a mutant pharbin, with the putative catalytic aspartate-559 residue mutated to alanine (Figure 9a). Quantification of fluorescence images showed that pharbin caused an approx. 50% decrease in staining for PtdIns(4,5)P2 (Figure 9b). Importantly, no reduction in staining for PtdIns4P was detected (Figure 9b), and in fact the quantification revealed a slight increase in PtdIns4P staining. This is probably not due to catalytic production of PtdIns4P by pharbin as it is also seen with the inactive mutant (Figure 9b).
Figure 9. Effect of a 5-phosphatase on plasma membrane PtdIns4P/PtdIns(4,5)P2 labelling.
COS-7 cells were transfected with GFP, GFP-Pharbin or the putatively inactive mutant of the latter, D559A, for 18 h prior to fixation and staining as described in Figure 7. (a) Representative images of COS-7 cells transfected with each construct. Images are maximum intensity projections of confocal sections spanning the entire depth of the cells. Scale bar=20 μm. (b) Quantification of staining (n=93 cells). See the Experimental section for details. *P<0.05, **P<0.01.
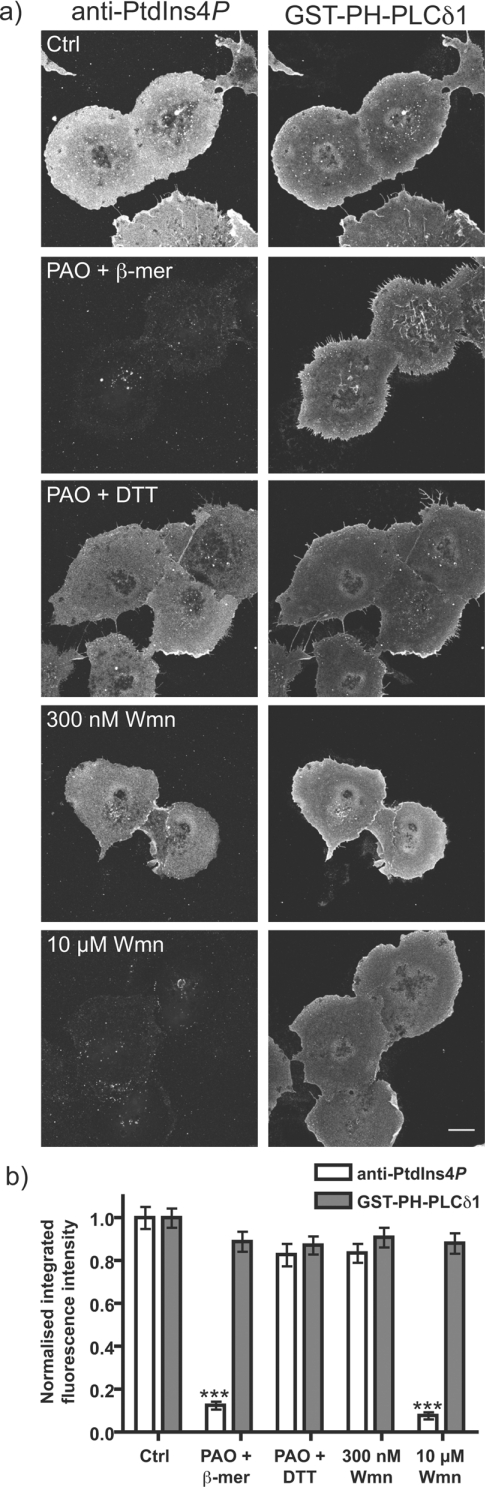
Together, these experiments provide evidence for the existence of a substantial pool of plasma membrane PtdIns4P, as has been reported in cells expressing GFP-fusions of the PtdIns4P-binding PH domain from Osh2p [20,23]. It has been shown that the plasma membrane pool of PtdIns(4,5)P2 required for PLC signalling is synthesized via PI4K IIIα [20,26,54]. PI4K IIIα is uniquely sensitive to wortmannin and 10 μM PAO [13], so we pre-incubated cells with these drugs for 30 min prior to fixation and staining for plasma membrane PtdIns4P and PtdIns(4,5)P2. Wortmannin abolishes plasma membrane PtdIns4P staining at the high (10 μM), but not lower (300 nM), concentrations specific for PI3K (Figure 10). PAO also abolishes plasma membrane PtdIns4P staining (Figure 10), and this is reversed by DTT and not by 2-mercaptoethanol, as reported previously [20]. Surprisingly, plasma membrane staining for PtdIns(4,5)P2 was completely unaffected by treatment with either drug (Figure 10). A similar observation has been made after metabolic labelling of inositol lipids with 2-[3H]-inositol or [32P]-phosphate [20,55], and suggests that the steady-state plasma membrane PtdIns(4,5)P2 is turning-over much more slowly than the PtdIns4P pool, or else is synthesized from PtdIns4P made by an entirely different PI4K activity. Either way, this observation indicates that despite being the precursor for PtdIns(4,5)P2 synthesis, plasma membrane PtdIns4P pools are metabolically distinct from the steady-state PtdIns(4,5)P2 pool.
Figure 10. Effect of PI4K inhibitors on plasma membrane PtdIns4P and PtdIns(4,5)P2 labelling.
(a) COS-7 cells were pre-treated for 30 min at 37 °C with 0.1% DMSO (Ctrl), 10 μM PAO with 1 mM 2-mercaptoethanol or DTT (PAO+β-mer or PAO+DTT), 300 nM or 10 μM wortmannin (Wmn) as indicated, prior to fixation and staining with anti-PtdIns4P antibody and GST–PH-PLCδ1 as described in Figure 7. Images are maximum intensity projections of confocal sections spanning the entire depth of the cells. Scale bar=20 μm. (b) Quantification of staining (n=136 cells). See the Experimental section for details. ***P<0.001.
Previous reports using anti-PtdIns4P antibodies have not generated evidence for plasma membrane PtdIns4P [8,56], although the conditions used for staining in those experiments were not particularly amenable for the preservation of the plasma membrane (Figure 2). In the present study, there is unambiguous evidence for distinct pools of PtdIns4P in Golgi and plasma membranes. What is the relative abundance of these two pools? Pre-treatment of cells with 10 μM wortmannin causes an approx. 80% drop in total cellular PtdIns4P levels, as measured by metabolic labelling with 32P [20,54,55], and complete ablation of plasma membrane PtdIns4P (Figure 10). Conversely, selective depletion of Golgi PtdIns4P with a Golgi-localized Sac1 mutant causes a reduction in radiolabelled PtdIns4P levels of approx. 20% [56]. Therefore, it appears that the major pool of PtdIns4P turning over in resting cells is present in the plasma membrane.
Online data
AUTHOR CONTRIBUTION
Gerald Hammond, Giampietro Schiavo and Robin Irvine designed experiments. Gerald Hammond performed experiments and analysed data. Gerald Hammond, Giampietro Schiavo and Robin Irvine wrote the paper.
ACKNOWLEDGEMENTS
We thank colleagues cited in the Experimental section for providing reagents, and members of the Irvine laboratory for helpful discussion. We are also indebted to our colleagues Jon Clarke, Tamas Balla and Shane Minogue for critical reading of the manuscript prior to publication.
FUNDING
The work was supported by a Wellcome Trust programme grant [grant number WT063581] to R. F. I.; and by Cancer Research UK funding to G. S.
References
- 1.Balla T. Phosphoinositide-derived messengers in endocrine signaling. J. Endocrinol. 2006;188:135–153. doi: 10.1677/joe.1.06595. [DOI] [PubMed] [Google Scholar]
- 2.Vanhaesebroeck B., Leevers S. J., Ahmadi K., Timms J., Katso R., Driscoll P. C., Woscholski R., Parker P. J., Waterfield M. D. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 3.Balla A., Balla T. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 2006;16:351–361. doi: 10.1016/j.tcb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Di Paolo G., De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 5.D'angelo G., Vicinanza M., Di Campli A., De Matteis M. The multiple roles of PtdIns(4)P – not just the precursor of PtdIns(4,5)P2. J. Cell Sci. 2008;121:1955–1963. doi: 10.1242/jcs.023630. [DOI] [PubMed] [Google Scholar]
- 6.Hama H., Schnieders E. A., Thorner J., Takemoto J. Y., DeWald D. B. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- 7.Walch-Solimena C., Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y. J., Wang J., Sun H. Q., Martinez M., Sun Y. X., Macia E., Kirchhausen T., Albanesi J. P., Roth M. G., Yin H. L. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- 9.Wang J., Sun H. Q., Macia E., Kirchhausen T., Watson H., Bonifacino J. S., Yin H. L. PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol. Biol. Cell. 2007;18:2646–2655. doi: 10.1091/mbc.E06-10-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godi A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D. R., Kular G. S., Daniele T., Marra P., Lucocq J. M., De Matteis M. A. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- 11.Tóth B., Balla A., Ma H., Knight Z. A., Shokat K. M., Balla T. Phosphatidylinositol 4-kinase IIIbeta regulates the transport of ceramide between the endoplasmic reticulum and Golgi. J. Biol. Chem. 2006;281:36369–36377. doi: 10.1074/jbc.M604935200. [DOI] [PubMed] [Google Scholar]
- 12.D'Angelo G., Polishchuk E., Di Tullio G., Santoro M., Di Campli A., Godi A., West G., Bielawski J., Chuang C. C., van der Spoel A. C., et al. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 2007;449:62–67. doi: 10.1038/nature06097. [DOI] [PubMed] [Google Scholar]
- 13.Balla A., Tuymetova G., Barshishat M., Geiszt M., Balla T. Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J. Biol. Chem. 2002;277:20041–20050. doi: 10.1074/jbc.M111807200. [DOI] [PubMed] [Google Scholar]
- 14.Minogue S., Waugh M. G., De Matteis M. A., Stephens D. J., Berditchevski F., Hsuan J. J. Phosphatidylinositol 4-kinase is required for endosomal trafficking and degradation of the EGF receptor. J. Cell Sci. 2006;119:571–581. doi: 10.1242/jcs.02752. [DOI] [PubMed] [Google Scholar]
- 15.Craige B., Salazar G., Faundez V. Phosphatidylinositol-4-kinase type II alpha contains an AP-3-sorting motif and a kinase domain that are both required for endosome traffic. Mol. Biol. Cell. 2008;19:1415–1426. doi: 10.1091/mbc.E07-12-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiedemann C., Schäfer T., Burger M. M. Chromaffin granule-associated phosphatidylinositol 4-kinase activity is required for stimulated secretion. EMBO J. 1996;15:2094–2101. [PMC free article] [PubMed] [Google Scholar]
- 17.Barylko B., Gerber S. H., Binns D. D., Grichine N., Khvotchev M., Südhof T. C., Albanesi J. P. A novel family of phosphatidylinositol 4-kinases conserved from yeast to humans. J. Biol. Chem. 2001;276:7705–7708. doi: 10.1074/jbc.C000861200. [DOI] [PubMed] [Google Scholar]
- 18.Wiedemann C., Schäfer T., Burger M. M., Sihra T. S. An essential role for a small synaptic vesicle-associated phosphatidylinositol 4-kinase in neurotransmitter release. J. Neurosci. 1998;18:5594–5602. doi: 10.1523/JNEUROSCI.18-15-05594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong K., Meyers D. D. R., Cantley L. C. Subcellular locations of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 1997;272:13236–13241. doi: 10.1074/jbc.272.20.13236. [DOI] [PubMed] [Google Scholar]
- 20.Balla A., Kim Y. J., Varnai P., Szentpetery Z., Knight Z., Shokat K. M., Balla T. Maintenance of hormone-sensitive phosphoinositide pools in the plasma membrane requires phosphatidylinositol 4-kinase IIIα. Mol. Biol. Cell. 2008;19:711–721. doi: 10.1091/mbc.E07-07-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Várnai P., Balla T. Live cell imaging of phosphoinositides with expressed inositide binding protein domains. Methods. 2008;46:167–176. doi: 10.1016/j.ymeth.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine T. P., Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- 23.Roy A., Levine T. P. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J. Biol. Chem. 2004;279:44683–44689. doi: 10.1074/jbc.M401583200. [DOI] [PubMed] [Google Scholar]
- 24.Peden A. A., Oorschot V., Hesser B. A., Austin C. D., Scheller R. H., Klumperman J. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J. Cell Biol. 2004;164:1065–1076. doi: 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G., Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balla A., Tuymetova G., Tsiomenko A., Várnai P., Balla T. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-IIIα: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol. Biol. Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts H. F., Clarke J. H., Letcher A. J., Irvine R. F., Hinchliffe K. A. Effects of lipid kinase expression and cellular stimuli on phosphatidylinositol 5-phosphate levels in mammalian cell lines. FEBS Lett. 2005;579:2868–2872. doi: 10.1016/j.febslet.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 28.Richardson J. P., Wang M., Clarke J. H., Patel K. J., Irvine R. F. Genomic tagging of endogenous type IIβ phosphatidylinositol 5-phosphate 4-kinase in DT40 cells reveals a nuclear localisation. Cell. Signal. 2007;19:1309–1314. doi: 10.1016/j.cellsig.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammond G. R., Dove S. K., Nicol A., Pinxteren J. A., Zicha D., Schiavo G. Elimination of plasma membrane phosphatidylinositol (4,5)-bisphosphate is required for exocytosis from mast cells. J. Cell Sci. 2006;119:2084–2094. doi: 10.1242/jcs.02912. [DOI] [PubMed] [Google Scholar]
- 30.Pagano R. E., Sepanski M. A., Martin O. C. Molecular trapping of a fluorescent ceramide analogue at the Golgi apparatus of fixed cells: interaction with endogenous lipids provides a trans-Golgi marker for both light and electron microscopy. J. Cell Biol. 1989;109:2067–2079. doi: 10.1083/jcb.109.5.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watt S. A., Kular G., Fleming I. N., Downes C. P., Lucocq J. M. Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem. J. 2002;363:657–666. doi: 10.1042/0264-6021:3630657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran D., Gascard P., Berthon B., Fukami K., Takenawa T., Giraud F., Claret M. Cellular distribution of polyphosphoinositides in rat hepatocytes. Cell. Signal. 1993;5:565–581. doi: 10.1016/0898-6568(93)90052-n. [DOI] [PubMed] [Google Scholar]
- 33.Stauffer T. P., Ahn S., Meyer T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- 34.Várnai P., Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]-inositol-labeled phosphoinositide pools. J. Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terasaki M. Localization of endoplasmic reticulum in living and glutaraldehyde-fixed cells with fluorescent dyes. Cell. 1984;38:101–108. doi: 10.1016/0092-8674(84)90530-0. [DOI] [PubMed] [Google Scholar]
- 36.Yokogawa T., Nagata S., Nishio Y., Tsutsumi T., Ihara S., Shirai R., Morita K., Umeda M., Shirai Y., Saitoh N., Fukui Y. Evidence that 3′-phosphorylated polyphosphoinositides are generated at the nuclear surface: use of immunostaining technique with monoclonal antibodies specific for PI(3,4)P2. FEBS Lett. 2000;473:222–226. doi: 10.1016/s0014-5793(00)01535-0. [DOI] [PubMed] [Google Scholar]
- 37.Yip S. C., Eddy R. J., Branch A. M., Pang H., Wu H., Yan Y., Drees B. E., Neilsen P. O., Condeelis J., Backer J. M. Quantification of PtdIns(3,4,5)P3 dynamics in EGF-stimulated carcinoma cells: a comparison of PH-domain-mediated methods with immunological methods. Biochem. J. 2008;411:441–448. doi: 10.1042/BJ20071179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laux T., Fukami K., Thelen M., Golub T., Frey D., Caroni P. GAP43, MARCKS, and CAP23 modulate PI(4,5)P2 at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J. Cell Biol. 2000;149:1455–1472. doi: 10.1083/jcb.149.7.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Micheva K. D., Holz R. W., Smith S. J. Regulation of presynaptic phosphatidylinositol 4,5-biphosphate by neuronal activity. J. Cell Biol. 2001;154:355–368. doi: 10.1083/jcb.200102098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma V. P., Desmarais V., Sumners C., Shaw G., Narang A. Immunostaining evidence for PI(4,5)P2 localization at the leading edge of chemoattractant-stimulated HL-60 cells. J. Leukocyte Biol. 2008;84:440–447. doi: 10.1189/jlb.0907636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weixel K. M., Blumental-Perry A., Watkins S. C., Aridor M., Weisz O. A. Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J. Biol. Chem. 2005;280:10501–10508. doi: 10.1074/jbc.M414304200. [DOI] [PubMed] [Google Scholar]
- 42.Blumental-Perry A., Haney C. J., Weixel K. M., Watkins S. C., Weisz O. A., Aridor M. Phosphatidylinositol 4-phosphate formation at ER exit sites regulates ER export. Dev. Cell. 2006;11:671–682. doi: 10.1016/j.devcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Nelson D. S., Alvarez C., Gao Y. S., Garcia-Mata R., Fialkowski E., Sztul E. The membrane transport factor TAP/p115 cycles between the Golgi and earlier secretory compartments and contains distinct domains required for its localization and function. J. Cell Biol. 1998;143:319–331. doi: 10.1083/jcb.143.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shorter J., Beard M. B., Seemann J., Dirac-Svejstrup A. B., Warren G. Sequential tethering of Golgins and catalysis of SNAREpin assembly by the vesicle-tethering protein p115. J. Cell Biol. 2002;157:45–62. doi: 10.1083/jcb.200112127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas C. L., Steel J., Prestwich G. D., Schiavo G. Generation of phosphatidylinositol-specific antibodies and their characterization. Biochem. Soc. Trans. 1999;27:648–652. doi: 10.1042/bst0270648. [DOI] [PubMed] [Google Scholar]
- 46.Osborne S. L., Thomas C. L., Gschmeissner S., Schiavo G. Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J. Cell Sci. 2001;114:2501–2511. doi: 10.1242/jcs.114.13.2501. [DOI] [PubMed] [Google Scholar]
- 47.Garcia P., Gupta R., Shah S., Morris A. J., Rudge S. A., Scarlata S., Petrova V., McLaughlin S., Rebecchi M. J. The pleckstrin homology domain of phospholipase C-δ1 binds with high affinity to phosphatidylinositol 4,5-bisphosphate in bilayer membranes. Biochemistry. 1995;34:16228–16234. doi: 10.1021/bi00049a039. [DOI] [PubMed] [Google Scholar]
- 48.Lemmon M. A., Ferguson K. M., O'Brien R., Sigler P. B., Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. U.S.A. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narayan K., Lemmon M. A. Determining selectivity of phosphoinositide-binding domains. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei Y. J., Sun H. Q., Yamamoto M., Wlodarski P., Kunii K., Martinez M., Barylko B., Albanesi J. P., Yin H. L. Type II phosphatidylinositol 4-kinase β is a cytosolic and peripheral membrane protein that is recruited to the plasma membrane and activated by Rac-GTP. J. Biol. Chem. 2002;277:46586–46593. doi: 10.1074/jbc.M206860200. [DOI] [PubMed] [Google Scholar]
- 51.Godi A., Pertile P., Meyers R., Marra P., Di Tullio G., Iurisci C., Luini A., Corda D., De Matteis M. A. ARF mediates recruitment of PtdIns-4-OH kinase-β and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- 52.Asano T., Mochizuki Y., Matsumoto K., Takenawa T., Endo T. Pharbin, a novel inositol polyphosphate 5-phosphatase, induces dendritic appearances in fibroblasts. Biochem. Biophys. Res. Commun. 1999;261:188–195. doi: 10.1006/bbrc.1999.0998. [DOI] [PubMed] [Google Scholar]
- 53.Kong A. M., Speed C. J., O'Malley C. J., Layton M. J., Meehan T., Loveland K. L., Cheema S., Ooms L. M., Mitchell C. A. Cloning and characterization of a 72-kDa inositol-polyphosphate 5-phosphatase localized to the Golgi network. J. Biol. Chem. 2000;275:24052–24064. doi: 10.1074/jbc.M000874200. [DOI] [PubMed] [Google Scholar]
- 54.Nakanishi S., Catt K. J., Balla T. A wortmannin-sensitive phosphatidylinositol 4-kinase that regulates hormone-sensitive pools of inositolphospholipids. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5317–5321. doi: 10.1073/pnas.92.12.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willars G. B., Nahorski S. R., Challiss R. A. Differential regulation of muscarinic acetylcholine receptor-sensitive polyphosphoinositide pools and consequences for signaling in human neuroblastoma cells. J. Biol. Chem. 1998;273:5037–5046. doi: 10.1074/jbc.273.9.5037. [DOI] [PubMed] [Google Scholar]
- 56.Blagoveshchenskaya A., Cheong F. Y., Rohde H. M., Glover G., Knödler A., Nicolson T., Boehmelt G., Mayinger P. Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. J. Cell Biol. 2008;180:803–812. doi: 10.1083/jcb.200708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.