Abstract
Because of the extreme diversity in immunoglobulin genes, tolerance mechanisms are necessary to ensure that B cells do not respond to self-antigens. One such tolerance mechanism is called receptor editing. If the B cell receptor (BCR) on an immature B cell recognizes self-antigen, it is down-regulated from the cell surface, and light chain gene rearrangement continues in an attempt to edit the autoreactive specificity. Analysis of a heterozygous mutant mouse in which the NF-κB–dependent IκBα gene was replaced with a lacZ (β-gal) reporter complementary DNA (cDNA; IκBα+/lacZ) suggests a potential role for NF-κB in receptor editing. Sorted β-gal+ pre–B cells showed increased levels of various markers of receptor editing. In IκBα+/lacZ reporter mice expressing either innocuous or self-specific knocked in BCRs, β-gal was preferentially expressed in pre–B cells from the mice with self-specific BCRs. Retroviral-mediated expression of a cDNA encoding an IκBα superrepressor in primary bone marrow cultures resulted in diminished germline κ and rearranged λ transcripts but similar levels of RAG expression as compared with controls. We found that IRF4 transcripts were up-regulated in β-gal+ pre–B cells. Because IRF4 is a target of NF-κB and is required for receptor editing, we suggest that NF-κB could be acting through IRF4 to regulate receptor editing.
B lymphocytes gain the potential to recognize >108 antigens (Cobb et al., 2006) by using a novel genetic mechanism called V(D)J recombination to generate a large repertoire of Ig heavy chain (IgHC) and Ig light chain (IgLC) variable domain exons (Brack et al., 1978; Tonegawa, 1983). Variable domain exons are composed of V, D, and J gene segments (IgHC) or V and J gene segments (IgLC). Successive stages of B cell development are defined by the ordered assembly of Ig genes; the IgHC locus rearranges in pro–B cells, the IgLC locus rearranges in pre–B cells, and the newly synthesized B cell receptor (BCR) is first expressed on the cell surface in immature B cells. V(D)J recombination begins with recognition and cleavage of a pair of recombination signal sequences (RSSs) flanking rearranging gene segments by the V(D)J recombinase composed of the lymphoid-restricted RAG1 and RAG2 proteins (Schatz et al., 1989; Oettinger et al., 1990). After RAG-mediated cleavage, the nonhomologous end-joining machinery repairs the DNA breaks, forming coding joints between the gene segments and signal joints between the two broken RSS ends (Bassing et al., 2002).
Transcription of rearranging gene segments correlates with their developmentally regulated activation for rearrangement (Alt et al., 1987). Mutations that disrupt this “germline” transcription interfere with V(D)J recombination. This has led various workers to examine specific transcription factors for their ability to influence gene rearrangement and B cell development. One such factor, NF-κB, was initially discovered as a result of its ability to bind to a sequence in the Igκ intronic enhancer (Sen and Baltimore, 1986). NF-κB is composed of homo- or heterodimers of five rel family members: RelA (p65), RelB, c-Rel, p50, and p52 (Hayden et al., 2006). Recent evidence suggests that additional proteins may associate with the rel proteins and influence the affinity and specificity of binding (Wan et al., 2007). Inactive NF-κB is sequestered in the cytoplasm bound to an inhibitory protein of the IκB family. Various signaling pathways result in the activation of a kinase that phosphorylates IκBα leading to its degradation. Once released from IκBα, NF-κB can translocate to the nucleus, bind DNA sequences, and regulate transcription. Remarkably, one of the transcriptional targets of NF-κB is IκBα itself, leading to negative-feedback regulation of NF-κB activation (Chiao et al., 1994).
Previous work attempting to elucidate the role of NF-κB in B cell development has lead to contradictory conclusions. Expression of a mutant IκBα “superrepressor” was reported to prevent light chain gene rearrangements in a transformed cell line (Scherer et al., 1996; O'Brien et al., 1997; Bendall et al., 2001). Retrovirus-mediated expression of a similar IκBα superrepressor in primary B cells, however, revealed a different phenotype: a block at the pro–B stage of development as defined by cell surface marker expression (Feng et al., 2004; Jimi et al., 2005) or a complete lack of B cells (Igarashi et al., 2006). This block could be overcome by expression of an antiapoptosis gene (Feng et al., 2004) or by neutralizing TNF-α (Igarashi et al., 2006). Adding to this confusion, targeted disruption NEMO, a protein required in some pathways leading to IκBα degradation, did not seem to alter B cell development until the mature stage (Sasaki et al., 2006).
A potential role for NF-κB in the regulation of IgLC gene rearrangement was reported by workers studying receptor editing, a process in which engagement of the BCR on an immature B cell with self-antigen provokes further recombination in an effort to replace the offending variable exon with an innocuous one (Nemazee, 2006). At least 25% of the primary B cell repertoire is thought to undergo editing (Casellas et al., 2001). These workers found that in vitro cross-linking of BCR on immature B cells leads to an increase in RAG expression and IgLC gene rearrangement that correlates with NF-κB activation and the binding of NF-κB to sites in the RAG locus and the κ intronic enhancer (Verkoczy et al., 2005). Another group, however, showed that targeted mutation of the NF-κB binding site in the κ intronic enhancer had little if any apparent effect on V-to-Jκ rearrangement (Inlay et al., 2004).
In an attempt to clarify the role of NF-κB in B cell development, we took advantage of a targeted mutant mouse that expresses a lacZ complementary DNA (cDNA; encoding β-galactosidase [β-gal]) from the IκBα locus (Beg et al., 1995). Because IκBα is directly regulated by active nuclear NF-κB, we were able to analyze NF-κB activity using a fluorescent β-gal substrate and multiparameter flow cytometry without perturbing its endogenous activity. Our results point to a role for NF-κB in the regulation of receptor editing.
RESULTS
IκBα+/lacZ mice report IκBα and NF-κB activity
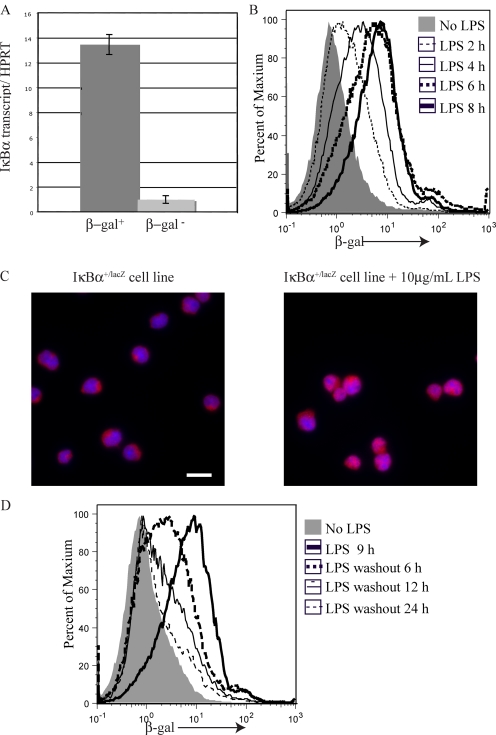
In an attempt to assess NF-κB activity at various stages of B cell development, we analyzed IκBα+/lacZ mice. Although these mice express less IκBα protein than wild-type mice, they display no obvious phenotype (Beg et al., 1995). To confirm that β-gal activity correlates with IκBα expression in IκBα+/lacZ reporter mice, IκBα transcripts were analyzed in RNA purified from pre–B cells sorted for β-gal activity as assayed using the fluorescent substrate fluorescein di-β-d-galactopyranoside (FDG). Using real-time PCR, we determined that the β-gal+ pre–B cells express 13-fold more IκBα messenger RNA (mRNA) as compared with β-gal− cells (Fig. 1 A). The greater amounts of IκBα transcripts in β-gal+ cells confirms that β-gal detected using FDG is a suitable reporter for IκBα expression in IκBα+/lacZ mice.
Figure 1.
IκBα+/lacZ mouse reports IκBα transcription and nuclear NF-κB activity. (A) Real-time RT-PCR analysis of IκBα expression in RNA purified from flow-sorted β-gal+ (dark gray) and β-gal− (light gray) IκBα+/lacZ pre–B cells (B220+CD43lowsIgM/D−). The ratio is shown between IκBα transcript and a control transcript, HPRT, with error bars indicating the standard deviation of triplicate assays. (B) Flow cytometric analysis of β-gal expression in IκBα+/lacZ Ableson cells cultured in the absence (gray shading) or presence of LPS for the indicated lengths of time. (C) Anti-p65 immunofluorescence microscopy was performed on IκBα+/lacZ Ableson cells cultured in the presence (right) or absence (left) of LPS for 2 h. Bar, 10 µm. (D) Flow cytometric analysis of β-gal expression in IκBα+/lacZ Ableson cells cultured in the absence (gray shading) or presence (thick black line) of LPS for 9 h, or after the indicated times after the LPS had been washed out and the cells had been recultured with cycloheximide. Each of the experiments in A–C were repeated a minimum of twice with similar results.
Several groups have shown that IκBα transcription is regulated by NF-κB activity (Gugasyan et al., 2000; Ghosh and Karin, 2002; Hoffmann et al., 2002; Hayden et al., 2006; Hayden and Ghosh, 2008). Therefore, we hypothesized that β-gal activity also reports NF-κB activity in IκBα+/lacZ mice. We confirmed this using an Ableson virus–transformed pro–B cell line made from IκBα+/lacZ mice. After LPS treatment, β-gal was up-regulated (Fig. 1 B) and p65 was relocalized from the cytoplasm to the nucleus in this cell line (Fig. 1 C). β-gal activity was observed in IκBα+/lacZ Ableson cells within 2 h of LPS treatment and the level of β-gal activity increased with longer treatment, reaching a plateau after 8 h (Fig. 1 B). To determine the half-life of β-gal in this system, we treated the IκBα+/lacZ Ableson cell line with LPS for 9 h, washed out the LPS, and recultured the cells in the presence of the protein synthesis inhibitor cycloheximide (Fig. 1 D). Using this approach, we found the half-life of β-gal to be 7.55 h. These experiments demonstrate that known activators of NF-κB can also activate β-gal in IκBα+/lacZ cells and that β-gal is reporting recent NF-κB activity.
Increased light chain gene rearrangements correlate with IκBα expression in pre–B cells
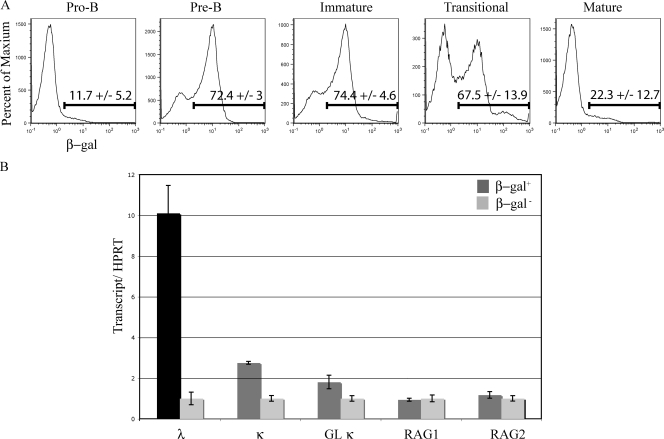
We used multiparameter flow cytometry to analyze β-gal expression in cells isolated from IκBα+/lacZ bone marrow to determine the pattern of IκBα transcription and, thus, NF-κB activity during B cell development. We observed stage-specific β-gal expression; it was low in pro–B cells, peaked at the pre–B stage, remained high in immature B cells, and was low in mature B cells (Fig. 2 A). The IκBα-positive and -negative subsets of pre–B cells were chosen for further analysis. We used real-time RT-PCR to quantify various light chain locus transcripts. A 10-fold greater amount of Vλ1,2-Cλ–rearranged transcript was observed in β-gal–expressing pre–B cells compared with β-gal–negative pre–B cells (Fig. 2 B). Germline κ transcripts are transcripts through the unrearranged κ locus and correlate with the accessibility of the locus to the recombinase. There was a twofold increase in both germline and rearranged κ transcripts in the pre–B cells with β-gal activity (Fig. 2). These data indicate that increased κ accessibility and, presumably, increased light chain gene rearrangement correlate with IκBα expression and NF-κB activity in pre–B cells.
Figure 2.
IκBα expression during B cell development. (A) β-gal expression in an IκBα+/lacZ reporter mouse was analyzed at various stages of B cell development. Cells were gated on forward and side scatter and divided into developmental stages using the cell surface markers B220, CD43, IgM, and IgD (Hardy et al., 1991). The data are representative of three independent experiments as indicated by the percentages and standard deviations accompanying each FACS histogram. (B) cDNA synthesized from sorted β-gal–positive and –negative pre–B cell subpopulations was analyzed for the indicated transcripts. Each transcript level was normalized to HPRT levels to control for any differences in cDNA template and data from each β-gal− sample was arbitrarily set to 1, with error bars indicating range of triplicate assays. The cDNA analysis used pre B cells from a pool of five to six mice and was repeated in two independent experiments.
We analyzed RAG1 and RAG2 transcripts in the β-gal–positive and –negative populations to assess whether differences in expression of these proteins might be responsible for the differences observed in light chain rearrangements. RAG levels were almost identical between the β-gal–positive and –negative pre–B cells (Fig. 2 B). This implies that a mechanism other then differentially regulated RAG expression is responsible for the differences observed in light chain rearrangements in the two pre–B cell subpopulations. To verify that the sorted β-gal pre–B cell population was not contaminated with pro–B cells, which could potentially account for the observed differences in various transcripts, we analyzed V(D)J-rearranged heavy chain gene transcripts in these populations. We found very similar levels of heavy chain transcripts in these two populations (Fig. S2), confirming that they were comparable B cell populations.
Receptor-editing markers correlate with IκBα expression
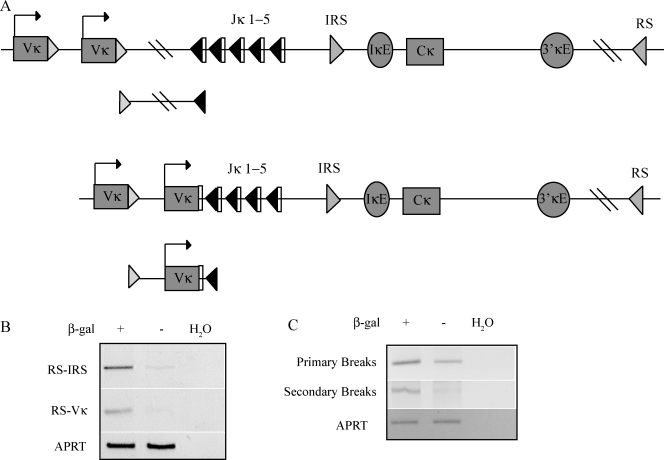
Considering what process might divide the pre–B cell compartment into nonequivalent subpopulations, we proceeded to test the idea that NF-κB might be specifically activated in cells undergoing receptor editing. To test this idea, we examined β-gal–positive and –negative pre–B cells for various markers of receptor editing. Rearrangements involving the RS element, which lies 25 kb 3′ of Cκ, and either a Vκ gene segment or the IRS sequence in the J-C interval are considered hallmarks of receptor editing (Fig. 3 A; Tiegs et al., 1993; Retter and Nemazee, 1998; Vela et al., 2008). Using a PCR assay, we detected more RS-IRS and RS-Vκ rearrangements in the β-gal+ than in the β-gal− pre–B cell population (Fig. 3 B). Thus, RS rearrangements are enriched in pre–B cells expressing IκBα.
Figure 3.
Markers of receptor editing are increased in IκBα-expressing cells. (A) The top diagram shows that the κ locus is in its germline configuration with the primary break intermediate shown below. The bottom diagram is the κ locus after a Vκ has rearranged to Jκ1, with the secondary break intermediate shown below. (B) Agarose gel analysis of PCR assays for RS rearrangements to either IRS (RS-IRS) or Vκ (RS-Vκ) in DNA from pre–B cells sorted for β-gal expression. PCR amplification of the APRT locus was used as a template control, and H20 indicates PCR reactions without template. (C) LM-PCR assays to detect primary and secondary double-stranded DNA RSS break intermediates in DNA from sorted β-gal–positive and –negative pre–B cells. An ethidium-stained agarose gel analysis of LM-PCR products is shown. Pooled bone marrow from five to six mice was used for each of two repetitions of this experiment yielding similar results.
The relative amounts of primary and secondary κ rearrangement reaction intermediates can also help identify cell populations undergoing receptor editing. Primary κ rearrangements occur on germline alleles. After this initial Vκ-to-Jκ rearrangement, an upstream Vκ can rearrange to a downstream Jκ if the primary rearrangement is nonproductive, if it is unable to pair with the preexisting IgHC, or if it contributes to a self-specific BCR. These are termed secondary rearrangements and are increased in cells undergoing receptor editing. Ligation-mediated PCR (LM-PCR; Schlissel et al., 1993) was used to detect reaction intermediates associated with primary and secondary Igκ rearrangements in DNA purified from β-gal–positive and –negative pre–B cells. In both populations, similar amounts of primary Igκ rearrangement intermediates were detected, but secondary rearrangement intermediates were highly enriched in the β-gal+ subpopulation (Fig. 3 C). Thus, secondary Igκ rearrangements, which are enhanced in receptor editing, correlate with IκBα expression.
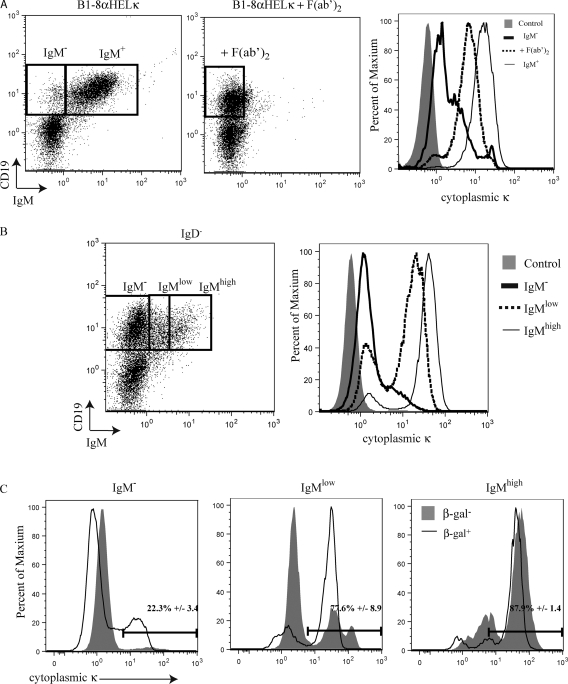
The BCR is down-regulated from the cell surface during receptor editing (Tze et al., 2000). One feature that distinguishes cells undergoing receptor editing from proper pre–B cells is cytoplasmic κ expression (Pelanda et al., 1997). To examine this feature of receptor editing, we cultured bone marrow from an innocuous BCR knockin mouse (B1-8 α-HEL-κ, see subsequent section), in the presence or absence of anti-IgM F(ab′)2 fragments, that mimic a signal inducing receptor editing (Hertz and Nemazee, 1997). As expected, the cultures where IgM was cross-linked down-regulated IgM off the cell surface, resulting in levels of cytoplasmic Igκ greater than those seen in sIgM− cells from unperturbed cultures (Fig. 4 A). The level of cytoplasmic Ig κ, however, is less than that found on sIgM+ cells cultured in the absence of F(ab′)2 fragments (Fig. 4 A).
Figure 4.
Cytoplasmic κ expression correlates with IκBα expression. (A) B1-8 α-HEL-κ bone marrow was cultured in IL-7 with or without cross-linking with anti-IgM F(ab′)2 to imitate a receptor-editing signal. Cells were surface stained with anti-CD19 and anti-IgM antibodies and a fourfold excess of a biotinylated anti-Igκ antibody to block surface Igκ. Cells were fixed, permeabilized, and stained with anti-Igκ antibody. The dot plots show CD19 versus IgM. The histogram displays cytoplasmic κ staining with the shaded gray representing isotype control staining, the thick black line representing IgM− cells without cross-linking, the dashed line representing cells cross-linked with anti-IgM F(ab′)2, and the thin black line representing IgM+ cells without cross-linking. This experiment was repeated three times. (B) Bone marrow from a wild-type mouse was stained as described in A. The dot plots show CD19 and IgM staining with gates defining IgM−, IgMlow, and IgMhigh populations. The histograms display cytoplasmic κ expression. The shaded gray indicates isotype control, the thick black line indicates IgM−, the dashed line indicates IgMlow, and the thin black line indicates IgMhigh. This experiment was performed three times with bone marrow from different individual mice. (C) IκBα+/lacZ bone marrow was sorted for β-gal expression and subsequently stained as described in A. The histograms display cytoplasmic κ expression in β-gal–sorted cells gated on the IgM−, IgMlow, and IgMhigh populations. β-gal–positive cells are indicated by the black lines and β-gal–negative cells are shown in gray shading. The numbers indicate the percentage of cytoplasmic κ–positive β-gal–positive cells, with mean values and standard error generated from analyses of three different mice.
We proceeded to analyze wild-type mice for cytoplasmic κ expression in three different B cell populations: CD19+IgM−, IgMlow, and IgMhigh cells (Fig. 4 B). We found that only a small percentage of CD19+IgM− cells express cytoplasmic κ (13%; Fig. 4 B). We suggest that this cytoplasmic Igκ+ subpopulation of sIgM− cells is undergoing receptor editing. We then analyzed IκBα+/lacZ bone marrow. In this same CD19+IgM− population, we detected cytoplasmic Igκ in β-gal+ cells but not in β-gal− cells (Fig. 4 C). This result implies that, in the pro–/pre–B cell compartment, cells undergoing receptor editing are β-gal+. In wild-type mice, the majority of IgMlow and IgMhigh cells express cytoplasmic Igκ (74 and 87%, respectively; Fig. 4 B). IgMlow cells are either early immature cells (in the process of up-regulating IgM expression) or cells undergoing receptor editing (in the process of down-regulating surface IgM). Most IgMlow cells undergoing receptor editing will be cytoplasmic Igκ positive because these cells once expressed this protein on the cell surface and are now in the process of down-regulating it or deleting the offending Igκ variable exon. Newly generated innocuous immature B cells, which are also IgMlow, will not have synthesized high levels of Igκ protein, so these cells do not necessarily express high levels of cytoplasmic Igκ. High levels of cytoplasmic Igκ were expressed in the majority of the β-gal+IgMlow cells but were only expressed in ∼30% of β-gal−IgMlow cells (Fig. 4 C). This suggests that the β-gal+IgMlow cells are receptor-editing cells and β-gal−IgMlow cells may be newly generated immature B cells. The majority of IgMhigh cells express cytoplasmic Igκ, regardless of β-gal expression levels (Fig. 4 C). This is not surprising, as cells with high IgM expression are continuously synthesizing immunoglobulin, accounting for the cytoplasmic Igκ expression. Thus, expression of cytoplasmic Igκ is associated with IκBα expression in populations of cells where receptor-editing signals are present.
Developing B cells expressing an innocuous BCR do not express appreciable levels of IκBα and have a reduction in nuclear p65 and ribosomal protein S3 (RPS3)
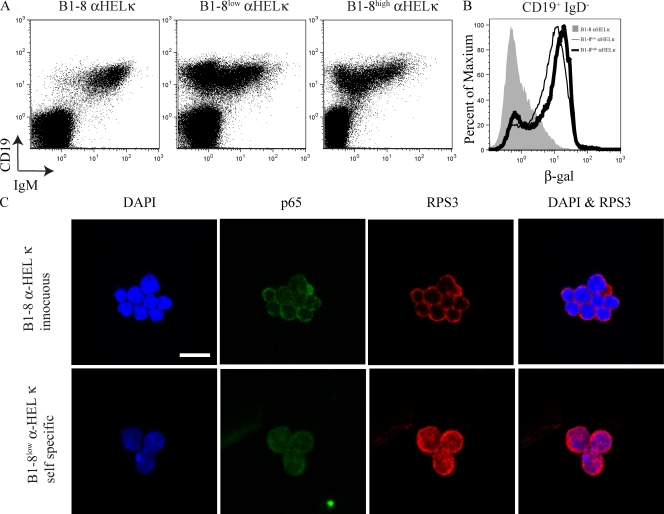
To further test the idea that NF-κB is involved in receptor editing, IκBα+/lacZ mice were crossed to various BCR knockin mice. The B1-8 heavy chain, when paired with the anti–HEL-κ light chain (α-HEL-κ), produces an innocuous BCR (Casellas et al., 2001). Specific mutations in the B1-8 heavy chain result in autoreactivity when paired with α-HEL-κ. Thus, B1-8high α-HEL-κ and B1-8low α-HEL-κ mice express mutated B1-8 heavy chains and self-specific BCRs (Casellas et al., 2001). There are few pro–/pre–B cells in B1-8 α-HEL-κ because these mice express an innocuous BCR and rapidly generate immature and mature B cells (Fig. 5 A). B1-8low α-HEL-κ and B1-8high α-HEL-κ B cells also rush through pro– and pre–B cell development, but because the knocked-in BCR in these cells is self-specific, it is down-regulated at the immature stage during receptor editing. Therefore, the cells in the pro–/pre–B gate in B1-8low α-HEL-κ and B1-8high α-HEL-κ mice are almost all undergoing receptor editing. IκBα+/lacZ B1-8 α-HEL-κ mice, with an innocuous BCR, express almost no β-gal in pro–B, pre–B, and immature B cells (CD19+IgD− gate). In contrast, ∼70% of either IκBα+/lacZ B1-8low α-HEL-κ and IκBα+/lacZ B1-8high α-HEL-κ mice express β-gal in the pro–B, pre–B, and immature B cell gate (Fig. 5 B). Thus, when cells do not undergo receptor editing, the IκBα reporter is not expressed. However, in two different mice where a majority of B cells undergo receptor editing, the majority of B cells express the IκBα reporter.
Figure 5.
Cells undergoing receptor editing show increased IκBα levels as well as increased nuclear NF-κB and RPS3. (A) Bone marrow was isolated from IκBα+/lacZ B1-8 α-HEL-κ, IκBα+/lacZB1-8high α-HEL-κ, and IκBα+/lacZB1-8low α-HEL-κ mice. Cells were loaded with FDG and stained with anti-CD19, anti-CD43, anti-IgM, and anti-IgD antibodies. Cells were first gated on IgD−. CD19 verses IgM staining is displayed. (B) β-gal activity in the CD19+IgD− gate (pro–B, pre–B, and immature B cells) is displayed. IκBα+/lacZ B1-8 α-HEL-κ is shaded gray, IκBα+/lacZB1-8low α-HEL-κ is the thin black line, and IκBα+/lacZB1-8high α-HEL-κ the thick black line. A and B are representative of at least three different mice individually analyzed for each genotype. (C) Immunofluorescence microscopy detecting p65 and RPS3 in B1-8 α-HEL-κ, B1-8low α-HEL-κ, and B1-8high α-HEL-κ pro–B, pre–B, and immature B cells. The data shown is representative of two independent sorts on each occasion scoring between 250 and 400 cells. Bar, 10 µm. Fig. S3 displays quantitative data from a repetition of this experiment.
RPS3, a component of the 40S ribosome subunit, was recently found to interact with p65, increase the activity and binding affinity of NF-κB, and influence the binding site preferences of the NF-κB complex (Wan et al., 2007). Various data implicate RPS3 in the regulation of IκBα and the κ locus. Chromatin immunoprecipitation assays show that this ribosomal protein binds to the IκBα promoter. In addition, a small interfering RNA directed against RPS3 inhibits IκBα expression and a RPS3-specific short hairpin RNA reduces κ expression. As with p65, RPS3 is localized in the cytoplasm but enters the nucleus when it is participating in transcriptional regulation (Wan et al., 2007). We used immunofluorescence microscopy to determine the localization of RPS3 and p65 in a mixture of pro–B, pre–B, and immature B cells (B220+IgD−) from B1-8 α-HEL-κ and B1-8low α-HEL-κ bone marrow. Interestingly, the B1-8 α-HEL-κ cells contained mostly cytoplasmic RPS3 expression (only 17% of these cells scored positive for nuclear RPS3), whereas the majority (68% of the cells scored) of B1-8low α-HEL-κ cells contained nuclear RPS3 expression (Fig. 5 C and Fig. S3). Although there was more nuclear p65 in the B1-8low α-HEL-κ cells compared with the B1-8 α-HEL-κ cells, the difference is not as striking as RPS3 (Fig. S3). These data imply that RPS3 could be acting to regulate NF-κB during receptor editing.
Inhibition of NF-κB inhibits light chain rearrangements
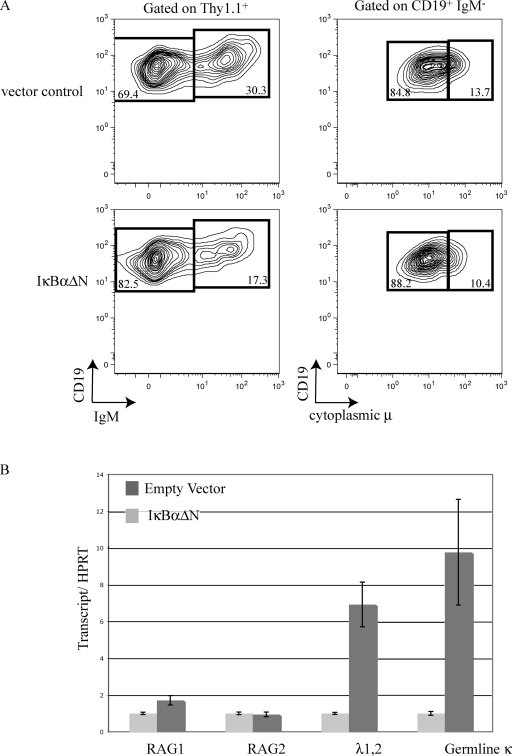
To determine if the differences observed between light chain locus transcription and rearrangement in the IκBα-positive and -negative populations were dependent on NF-κB activity, we took advantage of a retrovirus expressing an IκBα superrepressor (IκBαΔN). IκBαΔN is missing amino acids 1–36 of the N terminus and cannot be phosphorylated or ubiquitinated. Therefore, it remains bound to NF-κB dimers and inhibits activation of the classical NF-κB pathway (Scherer et al., 1996). Pro–B cells in IL-7–dependent wild-type bone marrow cultures were infected with an IκBαΔN-expressing retrovirus or an empty vector control. We failed to detect a block in pro–/pre–B development in IκBαΔN-expressing cells using the surface CD19 expression and cytoplasmic μ (Fig. 6 A). There was, however, a twofold reduction in IgM+ cells among those expressing IκBαΔN (Fig. 6 A). RNA was purified and transcript levels in the IκBαΔN-infected pro–/pre–B cells were compared with empty vector–infected pro–/pre–B cells. RAG expression was comparable in these two populations (Fig. 6 B). In the IκBαΔN-infected pro–/pre–B cells, a large decrease in Vλ1,2-Cλ–rearranged transcripts was observed (7-fold), as well as a decrease in germline κ transcripts (10-fold; Fig. 6 B). These results indicate that B cells with diminished NF-κB activity have diminished λ light chain gene rearrangements and κ accessibility. This confirms our results from the IκBα+/lacZ reporter mice that NF-κB influences light chain status without changing RAG expression.
Figure 6.
B cell development in IκBαΔN-infected IL-7 bone marrow cultures. (A) Bone marrow IL-7 cultures were infected with an IκBαΔN retrovirus or an empty vector control. 3 d after infection, cells were surface stained with Thy1.1 (a marker of viral infection), CD19, and IgM antibodies. The cells were fixed, permeabilized, stained with IgM antibody, and then analyzed by flow cytometry. The top shows the empty vector control infection and the bottom shows IκBαΔN-infected cells. The left displays CD19 versus IgM gated on Thy1.1+ cells. The right displays CD19 verses cytoplasmic μ gated on CD19+IgM−. (B) Real-time RT-PCR analysis of the indicated transcripts using RNA purified from Thy1.1-sorted IκBαΔN or empty vector retrovirally transduced pro–/pre–B cells. All transcripts were normalized to HPRT transcription and data from each IκBαΔN sample was arbitrarily set to 1, with error bars indicating range of triplicate assays. These experiments were repeated twice using bone marrow pooled from six mice.
IκBα expression correlates with IRF4 expression but not with expression of Bcl-2 family members
To investigate potential targets of regulation by NF-κB that might play a role in receptor editing, transcript levels of multiple NF-κB target genes were analyzed in RNA purified from sorted β-gal–positive and –negative pre–B cells. Bcl-2 family members are antiapoptosis genes and targets of NF-κB (Catz and Johnson, 2001). Mice that contain self-specific BCR transgenes, as well as a Bcl-2 transgene, display enhanced receptor editing (Lang et al., 1997), suggesting that survival regulated by this gene may be involved in receptor editing. We used RT-PCR to compare levels of transcripts from the antiapoptosis genes Bcl-2, Bcl-X, and Mcl-1 in β-gal–positive and –negative pre–B cell cDNA. There was no increase in these antiapoptosis genes in the β-gal–positive cells (Fig. S1). These data indicate that increased survival, mediated by these antiapoptosis genes, is unlikely to be responsible for the increased receptor editing observed in the β-gal–positive cells.
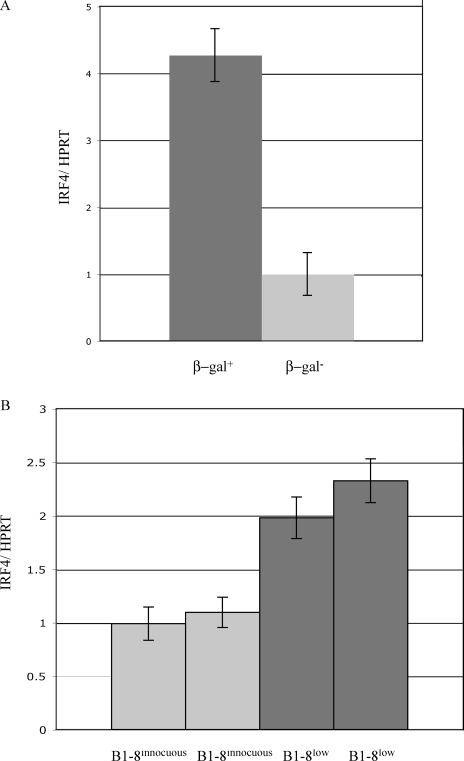
IRF4 is a target of NF-κB that has been previously shown to be important in both pre–B cell development and receptor editing. (Lu et al., 2003; Muljo and Schlissel, 2003; Ma et al., 2006; Saito et al., 2007; Johnson et al., 2008; Pathak et al., 2008). We used real-time RT-PCR to quantify IRF4 transcripts in pro–/pre–B cells sorted based on β-gal activity. The signal was normalized to hypoxanthine-guanine phosphoribosyl transferase (HPRT) to account for differences in template quality. We observed a fourfold increase in IRF4 transcripts in β-gal+ pre–B cells (Fig. 7 A). A twofold increase was observed in IRF4 expression in B1-8low α-HEL-κ CD19+IgD− cells compared with B1-8 α-HEL-κ CD19+IgD− cells (Fig. 7 B). IRF4 has binding sites in the 3′κ enhancer as well as in both λ enhancers (Pongubala et al., 1992; Eisenbeis et al., 1995). This increase in IRF4 in the β-gal–positive population implies that NF-κB could be acting through IRF4 to regulate receptor editing.
Figure 7.
Expression of NF-κB target genes in IκBαΔ-transduced pre–B cells. (A) cDNA from sorted β-gal–positive and –negative pre–B cells was analyzed by real time RT-PCR for IRF4 transcripts and the data were normalized to HPRT transcripts to control for any differences in cDNA template. β-gal–negative cell result was arbitrarily set to 1, with error bars indicating range of triplicate assays. (B) cDNA from CD19+IgD− cells from B1-8 α-HEL-κ and B1-8low α-HEL-κ mice was analyzed by real time RT-PCR for IRF4 transcripts and the data were normalized to HPRT, with error bars indicating range of triplicate assays. These results are representative of two independent repetitions of this experiment using pools of sorted bone marrow cells from five to six mice.
DISCUSSION
Receptor editing is stimulated in immature B cells by BCR recognition of autoantigen, which results in down-modulation of BCR from the cell surface. BCR engagement is known to activate NF-κB. Using an IκBα-lacZ knockin reporter allele, we detected a subpopulation of pre–B cells that contains active nuclear NF-κB and found that this same population expresses markers of receptor editing. We propose that this correlation between NF-κB activity and receptor editing may indicate a functional role for this transcription factor in a key mechanism of self-tolerance.
Previous results have suggested various roles for NF-κB during B cell development, ranging from involvement in light chain gene rearrangements (Scherer et al., 1996; O'Brien et al., 1997; Bendall et al., 2001) to being dispensable for B cell development (Igarashi et al., 2006; Sasaki et al., 2006). Because receptor editing involves a specific subset of light chain gene rearrangements, our data imply that NF-κB could be involved in light chain rearrangements without being required for B cell development, which is consistent with seemingly contradictory results. In addition, the role of NF-κB in receptor editing could be either direct or indirect, and its activation in pre–B cells might be via the classical or nonclassical pathway or, perhaps, even a novel pathway. Suggestive of the latter possibility, Derudder et al. (2009) have found that NEMO, IKK1, and IKK2 are all dispensable for receptor editing. Further experiments will be required to determine whether alternative pathways of NF-κB activation exist in developing B cells.
Much of the data presented in this paper are correlative; nuclear NF-κB activity correlates with β-gal expression, which in turn correlates with numerous markers of receptor editing in phenotypic pre–B cells including elevated levels of Igλ rearrangement and transcription, increased cytoplasmic Igκ expression, increased Jκ locus RS rearrangements, and increased Jκ RSS replacement breaks. We also observed a several-fold increase in IRF-4 mRNA levels, which is consistent with prior studies showing that IRF-4 plays a role in Igκ and Igλ locus activation enhancers (Pongubala et al., 1992; Eisenbeis et al., 1995) and is involved in receptor editing (Pathak et al., 2008). We provided a critical test of these correlations by examining NF-κB activity in two knockin models of receptor editing, in each case finding that nuclear NF-κB activity is elevated as compared with cells expressing a knocked-in innocuous receptor.
In addition, we found that overexpression of a cDNA encoding an IκBαΔN mutant suppressed the generation of sIgM+ immature B cells and diminished levels of germline Igκ and rearranged Igλ transcripts in a short-term primary cell culture system. The results of this perturbation of NF-κB activity are consistent with a causal role for this factor in receptor editing.
It is possible that the activation of NF-κB in a population of cells undergoing receptor editing is serving a purpose other than the induction of editing. For example, NF-κB is known to regulate the expression of various antiapoptotic genes including Bcl-2, Bcl-xL, and Mcl-1. Thus, it is possible that NF-κB's role in editing may be to promote a sufficient period of cell survival to allow for ongoing V(D)J recombination activity to result in the generation of a self-tolerant BCR. To test this idea, we compared the levels of antiapoptosis gene mRNA in sorted β-gal+ and β-gal− pre–B cells. We found no significant differences in expression levels between these cell populations (Fig. S1).
A series of elegant studies from the Behrens laboratory has led to the suggestion that receptor editing is associated with the loss of IgM from the surface of editing cells (Schram et al., 2008). Others have found that cytoplasmic Igκ expression is elevated after BCR down-regulation in cells undergoing receptor editing (Pelanda et al., 1997). These results are consistent with our observation that a high fraction of β-gal+ pre–B cells express cytoplasmic Igκ chain (Fig. 4). We found that cytoplasmic Igκ levels increase in the population of CD19+IgM− cells that results when immature B cells are treated with cross-linking anti-IgM antibodies, an in vitro mimic of the receptor-editing signal. Schram et al. (2008) showed that this loss of surface BCR in editing cells was associated with increased levels of transcription of both IκBα and Bcl-x, which are known NF-κB target genes.
RS rearrangements are considered a hallmark of receptor editing (Retter and Nemazee, 1998; Vela et al., 2008). Large increases in these rearrangements are observed in receptor-editing mouse models (Chen et al., 1997; Pelanda et al., 1997; Ait-Azzouzene et al., 2005). Mice lacking the RS sequence display decreased receptor editing and fewer B cells expressing a λ light chain (Vela et al., 2008). An increase in RS rearrangement to both IRS and Vκ gene segments correlates with β-gal expression in our experiments. The increase in RS rearrangement indicates that a substantial number of cells in the β-gal–positive pre–B cell population have inactivated the κ locus, providing further evidence that this population is undergoing receptor editing.
In both experimental systems used to study NF-κB activity, analysis of IκBα+/lacZ pre–B cells, and retroviral infection of cultured B cells with IκBαΔN, the change in light chain status was not accompanied by a change in RAG expression. This data implies that an increase in RAGs is not responsible for receptor-editing rearrangements. There are two hypotheses regarding RAG expression during receptor editing: either an increase or a persistence of RAG expression upon recognition of an autoantigen (Jankovic et al., 2004; Verkoczy et al., 2005). We interpret our data as being consistent with the idea that RAG expression is maintained, not necessarily increased, during receptor editing.
RPS3 interacts with p65, increases the activity and binding affinity of NF-κB, and influences the binding sites of the NF-κB complex (Wan et al., 2007). Using an immunofluorescence microscopy assay, we found increased nuclear RPS3 in pro–B, pre–B, and immature B cells from receptor-editing model mice (B1-8low α-HEL-κ) compared with the same population of cells from mice with innocuous BCRs (B1-8 α-HEL-κ). Preliminary data suggests that knocking down RPS3 causes a defect in receptor editing (unpublished data). We infer from this data that RPS3 is influencing binding sites of NF-κB during receptor editing and we are currently examining this issue in greater detail.
Our results implicate NF-κB in the regulation of receptor editing. These data imply that NF-κB is not required for B cell development to take place but, instead, contributes to a key process that prevents the expression of self specific BCRs.
MATERIALS AND METHODS
Mice.
B1-8, B1-8high, B1-8low, and Igκ α-HEL mice were a gift from M. Nussenzweig (Rockefeller University, New York, NY). IκBα+/lacZ mice were a gift from B. Sha (University of California, Berkeley, Berkeley, CA). All wild-type mice used were C57BL/6 (The Jackson Laboratory). Animal experimentation was approved by the University of California, Berkeley Animal Care and Use Committee.
Cell culture.
Cells were cultured in RPMI 1640 medium supplemented with 10% (vol/vol) fetal calf serum, 100 µg/ml penicillin, 100 µg/ml streptomycin, and 50 µM β-mercaptoethanol and grown at 37°C in 5% CO2. 10 µg/ml LPS was added to specified cultures for various lengths of time. For the LPS washout experiments, cells were washed twice in PBS and then recultured in RPMI as before with the addition of 50 µg/ml cycloheximide.
For primary cell cultures or retroviral infection, bone marrow was isolated from 4–6-wk-old mice. The cells were passed through a 40-µm filter to create a single cell suspension. Red blood cells were depleted by ACK lysis and cells were filtered again. Cells were overlayed on irradiated S17 stromal cells that were plated 1 d prior. 100 U/ml rIL-7 (R&D Systems) was added to the culture. Goat anti–mouse IgM F(ab′)2 fragments (Jackson ImmunoResearch Laboratories) were used at a final concentration of 10 µg/ml.
Retroviral infection of bone marrow cultures.
Retroviral plasmids were incubated with lipofectamine 2000 (Invitrogen) and added to Phoenix cells. 5 h after transfection, 10% fetal calf serum was added to the Opti-MEM media. 2 d after culture, the viral supernatant was removed and put through a 0.45-µM syringe filter and added directly to the bone marrow cells supplemented with 100 U/ml IL-7 and 4 µg/ml polybrene. The cells and viral supernatant were spun at 2,400 rpm for 90 min at room temperature. After 4–6 h, the cells were pelleted and resuspended in media and overlayed on irradiated S17 cells that had been plated 1 d prior.
FDG loading.
Cells were resuspended at a concentration of three million cells/50 µl RPMI media. FDG (Invitrogen) was diluted to 2 mM in H2O. 50 μL of 2-mM FDG was added to 50 µl of cells and incubated at 37°C for exactly 1 min. 1.5 ml RPMI media was added to stop the reaction.
Real-time PCR.
All taqman primers used fam/tamra chemistry. The primers used were the following: HPRT forward, 5′-CTGGTGAAAAGGACCTCTCG-3′; HPRT reverse, 5′-TGAAGTACTCATTATAGTCAAGGGCA-3′; HPRT TM probe, 5′-TGTTGGATACAGGCCAGACTTTGTTGGAT-3′; κGT 5′, 5′-GGACGTTCGGTGGAGGC-3′; κGT 3′, 5′-GGAAGATGGATACAGTTGGTGCA-3′; κGT probe, 5′-CCAAGCTGGAAATCAAACGCTGAT-3′; Irf4 sense, 5′-GAAGCCTTGGCGCTCTCA-3′; Irf4 antisense, 5′-TCACGAGGATGTCCCGGTAA-3′; Irf4 probe, 5′-CTGCCGGCTGCATATCTGCCTGT-3′; RAG1 sense, 5′-CATTCTAGCACTCTGGCCGG-3′; RAG1 antisense, 5′-TCATCGGGGCAGAACTGAA-3′; RAG1 probe, 5′-AAGGTAGCTTAGCCAACATGGCTGCCTC-3′; RAG2 sense, 5′-TTAATTCCTGGCTTGGCCG-3′; RAG2 antisense, 5′-TTCCTGCTTGTGGATGTGAAAT-3′; and RAG2 probe, 5′-AGGGATAAGCAGCCCCTCTGGCC-3′. Real time PCR was also performed using Evergreen chemistry. The following primers were used: Vλ1,2, 5′-TGGAGACAAGGCTGCCCTCACCATCACAG-3′; Vλ3, 5′-TGGTGCTGATCGCTACCTTAGCATTTCCA-3′; Cλ RT, 5′-GAGCTCYTCAGRGGAAGGTGGAAACABGGT-3′; IκBα intron forward, 5′-GCAATCATCCACGAAGAGAAGC-3′; IκBα intron reverse, 5′-CGTTGACATCAGCACCCAAAG-3′; Cκ, 5′-GTCCTGATCAGTCCAACTGTTCAGG-3′; and VκS, 5′-CCGAATTCGSTTCAGTGGCAGTGGRTCRGGRAC-3′.
PCR primers.
PCR primers used were the following: RS reverse #1, 5′-GGACATCTACTGACAGGTTATCACAGGTC-3′; IRS forward #1, 5′-ATGACTGCTTGCCATGTAGATACCATGG-3′; VκS, 5′-CCGAATTCGSTTCAGTGGCAGTGGRTCRGGRAC-3′; APRT 747 forward, 5′-TGCTAGACCAACCCGCACCCAGAAG-3′; APRT 964 reverse, 5′-TCGTGACCGCACCTGAACAGCAC-3′; CH, 5′-ATGCAGATCTCTGTTTTTGCCTCC-3′; VH558, 5′-CGAGCTCTCCARCACAGCCTWCATGCARCTCARC-3′; VH7183, 5′-CGGTACCAAGAASAMCCTGTWCCTGCAAATGASC-3′; and VHQ52, 5′-CGGTACCAGACTGARCATCASCAAGGACAAYTCC-3′.
LM-PCR for primary and secondary κ break intermediates.
LM-PCR was performed as previously described (Schlissel et al., 1993). Linker-ligated genomic DNA was analyzed by PCR using the linker primer BW-H (5′-CCGGGAGATCTGAATTCCAC-3′) and the ko5 primer (5′-GCCCAAGCGCTTCCACGCATGCTTGGAG-3′) to assay for primary breaks or a degenerate Vκ primer (5′-CCGAATTCGSTTCAGTGGCAGTGGRTCRGGRAC-3′) to detect secondary breaks. A touchdown PCR program was used: 94°C for 1 min, 19 cycles of 92°C for 30 s, and then 70°C for 40 s, with the temperature dropped by 0.5°C for each successive cycle. This was followed by 19 cycles of 92°C for 30 s and then 60°C for 40 s with 1 s added each successive cycle.
Immunofluorescence microscopy.
In the experiment shown in Fig. 1, cells were resuspended in PBS and spun onto a slide using a cytospin centrifuge. Cells were fixed in 4% paraformaldehyde, washed in PBS, and then incubated in blocking solution (PBS, 0.5% fetal calf serum, 0.5% normal rat serum, 0.2% Triton X-100, and 3% bovine serum albumin). The blocking buffer was removed and the cells were incubated with the primary antibody p65 (sc-372; Santa Cruz Biotechnology, Inc.). The slides were washed in PBS containing 0.1% Triton X-100 and then PBS. Cells were next incubated with secondary antibody, α–rabbit Cy3 (Jackson ImmunoResearch Laboratories), and then washed as for the primary antibody. Antifading solution containing 500 ng/ml DAPI was place on each cell spot. In the experiment shown in Fig. 5, staining was performed as previously described (Wan et al., 2007).
Surface and cytoplasmic staining for flow cytometry.
Antibodies used to stain early B cell development in conjunction with FDG loading were the following: IgM-PE (Southern Biotechnology), B220-PE Texas red (Invitrogen), CD43-biotin (clone S7; BD), and streptavidin-Cy5 (BD). Alternative antibodies used to stain for B cell development were the following: IgD-PE (Southern Biotechnology), CD43-biotin (clone S7; BD), streptavidin- Red613 (Invitrogen), IgM-Cy5 (eBioscience), andCD19-Cy7 (BD).
For cytoplasmic Igκ staining, cells were first surface stained using CD19-Cy7 (BD), IgM-Cy5 (eBioscience), IgD-PE (Southern Biotechnology), and a fourfold excess of κ-biotin (BD), and then fixed, permeabilized in BSA/PBS with 0.1% saponin, and resuspended in BSA/PBS with 0.1% saponin with κ-FITC (BD).
Online supplemental material.
Fig. S1 shows the lack of correlation between NF-κB activity and antiapoptosis gene expression. Fig. S2 is an analysis of rearranged heavy chain gene transcripts in β-gal–sorted pre–B cells. Fig. S3 is a quantitative analysis of the immunofluorescence microscopy presented in Fig. 5 C. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20082815/DC1.
Acknowledgments
The authors wish to acknowledge Bill Sha (University of California, Berkeley, CA) for providing the IκBα+/lacZ mice, as well as important advice during the conduct of this project, and Michel Nussenzweig (Rockefeller University) for sharing various Ig knockin mice.
This work was supported by a grant from the National Institutes of Health (RO1 HL48702) to M.S. Schlissel.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used: β-gal, β-galactosidase; BCR, B cell receptor; cDNA, complementary DNA; FDG, fluorescein di-β-d-galactopyranoside; HPRT, hypoxanthine-guanine phosphoribosyl transferase; IgHC, Ig heavy chain; IgLC, Ig light chain; LM-PCR, ligation-mediated PCR; mRNA, messenger RNA; RPS3, ribosomal protein S3; RSS, recombination signal sequence.
References
- Ait-Azzouzene D., Verkoczy L., Peters J., Gavin A., Skog P., Vela J.L., Nemazee D. 2005. An immunoglobulin Cκ-reactive single chain antibody fusion protein induces tolerance through receptor editing in a normal polyclonal immune system.J. Exp. Med. 201:817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F.W., Blackwell T.K., Yancopoulos G.D. 1987. Development of the primary antibody repertoire.Science. 238:1079–1087 [DOI] [PubMed] [Google Scholar]
- Bassing C.H., Swat W., Alt F.W. 2002. The mechanism and regulation of chromosomal V(D)J recombination.Cell. 109:S45–S55 [DOI] [PubMed] [Google Scholar]
- Beg A.A., Sha W.C., Bronson R.T., Baltimore D. 1995. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice.Genes Dev. 9:2736–2746 [DOI] [PubMed] [Google Scholar]
- Bendall H.H., Sikes M.L., Oltz E.M. 2001. Transcription factor NF-kappa B regulates Ig lambda light chain gene rearrangement.J. Immunol. 167:264–269 [DOI] [PubMed] [Google Scholar]
- Brack C., Hirama M., Lenhard-Schuller R., Tonegawa S. 1978. A complete immunoglobulin gene is created by somatic recombination.Cell. 15:1–14 [DOI] [PubMed] [Google Scholar]
- Casellas R., Shih T.A., Kleinewietfeld M., Rakonjac J., Nemazee D., Rajewsky K., Nussenzweig M.C. 2001. Contribution of receptor editing to the antibody repertoire.Science. 291:1541–1544 [DOI] [PubMed] [Google Scholar]
- Catz S.D., Johnson J.L. 2001. Transcriptional regulation of bcl-2 by nuclear factor kappa B and its significance in prostate cancer.Oncogene. 20:7342–7351 [DOI] [PubMed] [Google Scholar]
- Chen C., Prak E.L., Weigert M. 1997. Editing disease-associated autoantibodies.Immunity. 6:97–105 [DOI] [PubMed] [Google Scholar]
- Chiao P.J., Miyamoto S., Verma I.M. 1994. Autoregulation of I kappa B alpha activity.Proc. Natl. Acad. Sci. USA. 91:28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb R.M., Oestreich K.J., Osipovich O.A., Oltz E.M. 2006. Accessibility control of V(D)J recombination.Adv. Immunol. 91:45–109 [DOI] [PubMed] [Google Scholar]
- Derudder E., Cadera E.J., Vahl J.C., Wang J., Fox C.J., Zha S., van Loo G., Pasparakis M., Schlissel M.S., Schmidt-Supprian M., Rajewsky K. 2009. Development of immunoglobulin lambda-chain-positive B cells, but not editing of immunoglobulin kappa-chain, depends on NF-kappaB signals.Nat. Immunol. 10:647–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbeis C.F., Singh H., Storb U. 1995. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator.Genes Dev. 9:1377–1387 [DOI] [PubMed] [Google Scholar]
- Feng B., Cheng S., Pear W.S., Liou H.C. 2004. NF-kB inhibitor blocks B cell development at two checkpoints.Med. Immunol. 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Karin M. 2002. Missing pieces in the NF-kappaB puzzle.Cell. 109:S81–S96 [DOI] [PubMed] [Google Scholar]
- Gugasyan R., Grumont R., Grossmann M., Nakamura Y., Pohl T., Nesic D., Gerondakis S. 2000. Rel/NF-kappaB transcription factors: key mediators of B-cell activation.Immunol. Rev. 176:134–140 [DOI] [PubMed] [Google Scholar]
- Hardy R.R., Carmack C.E., Shinton S.A., Kemp J.D., Hayakawa K. 1991. Resolution and characterization of pro-B and pre—pro-B cell stages in normal mouse bone marrow.J. Exp. Med. 173:1213–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M.S., Ghosh S. 2008. Shared principles in NF-kappaB signaling.Cell. 132:344–362 [DOI] [PubMed] [Google Scholar]
- Hayden M.S., West A.P., Ghosh S. 2006. NF-kappaB and the immune response.Oncogene. 25:6758–6780 [DOI] [PubMed] [Google Scholar]
- Hertz M., Nemazee D. 1997. BCR ligation induces receptor editing in IgM+IgD- bone marrow B cells in vitro.Immunity. 6:429–436 [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Levchenko A., Scott M.L., Baltimore D. 2002. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation.Science. 298:1241–1245 [DOI] [PubMed] [Google Scholar]
- Igarashi H., Baba Y., Nagai Y., Jimi E., Ghosh S., Kincade P.W. 2006. NF-kappaB is dispensable for normal lymphocyte development in bone marrow but required for protection of progenitors from TNFalpha.Int. Immunol. 18:653–659 [DOI] [PubMed] [Google Scholar]
- Inlay M.A., Tian H., Lin T., Xu Y. 2004. Important roles for E protein binding sites within the immunoglobulin κ chain intronic enhancer in activating VκJκ rearrangement.J. Exp. Med. 200:1205–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic M., Casellas R., Yannoutsos N., Wardemann H., Nussenzweig M.C. 2004. RAGs and regulation of autoantibodies.Annu. Rev. Immunol. 22:485–501 [DOI] [PubMed] [Google Scholar]
- Jimi E., Phillips R.J., Rincon M., Voll R., Karasuyama H., Flavell R., Ghosh S. 2005. Activation of NF-kappaB promotes the transition of large, CD43+ pre-B cells to small, CD43- pre-B cells.Int. Immunol. 17:815–825 [DOI] [PubMed] [Google Scholar]
- Johnson K., Hashimshony T., Sawai C.M., Pongubala J.M., Skok J.A., Aifantis I., Singh H. 2008. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling.Immunity. 28:335–345 [DOI] [PubMed] [Google Scholar]
- Lang J., Arnold B., Hammerling G., Harris A.W., Korsmeyer S., Russell D., Strasser A., Nemazee D. 1997. Enforced Bcl-2 expression inhibits antigen-mediated clonal elimination of peripheral B cells in an antigen dose–dependent manner and promotes receptor editing in autoreactive, immature B cells.J. Exp. Med. 186:1513–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Medina K.L., Lancki D.W., Singh H. 2003. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development.Genes Dev. 17:1703–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Turetsky A., Trinh L., Lu R. 2006. IFN regulatory factor 4 and 8 promote Ig light chain kappa locus activation in pre-B cell development.J. Immunol. 177:7898–7904 [DOI] [PubMed] [Google Scholar]
- Muljo S.A., Schlissel M.S. 2003. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines.Nat. Immunol. 4:31–37 [DOI] [PubMed] [Google Scholar]
- Nemazee D. 2006. Receptor editing in lymphocyte development and central tolerance.Nat. Rev. Immunol. 6:728–740 [DOI] [PubMed] [Google Scholar]
- O'Brien D.P., Oltz E.M., Van Ness B.G. 1997. Coordinate transcription and V(D)J recombination of the kappa immunoglobulin light-chain locus: NF-kappaB-dependent and -independent pathways of activation.Mol. Cell. Biol. 17:3477–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettinger M.A., Schatz D.G., Gorka C., Baltimore D. 1990. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination.Science. 248:1517–1523 [DOI] [PubMed] [Google Scholar]
- Pathak S., Ma S., Trinh L., Lu R. 2008. A role for interferon regulatory factor 4 in receptor editing.Mol. Cell. Biol. 28:2815–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelanda R., Schwers S., Sonoda E., Torres R.M., Nemazee D., Rajewsky K. 1997. Receptor editing in a transgenic mouse model: site, efficiency, and role in B cell tolerance and antibody diversification.Immunity. 7:765–775 [DOI] [PubMed] [Google Scholar]
- Pongubala J.M., Nagulapalli S., Klemsz M.J., McKercher S.R., Maki R.A., Atchison M.L. 1992. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3′ enhancer activity.Mol. Cell. Biol. 12:368–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retter M.W., Nemazee D. 1998. Receptor editing occurs frequently during normal B cell development.J. Exp. Med. 188:1231–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M., Gao J., Basso K., Kitagawa Y., Smith P.M., Bhagat G., Pernis A., Pasqualucci L., Dalla-Favera R. 2007. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma.Cancer Cell. 12:280–292 [DOI] [PubMed] [Google Scholar]
- Sasaki Y., Derudder E., Hobeika E., Pelanda R., Reth M., Rajewsky K., Schmidt-Supprian M. 2006. Canonical NF-kappaB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation.Immunity. 24:729–739 [DOI] [PubMed] [Google Scholar]
- Schatz D.G., Oettinger M.A., Baltimore D. 1989. The V(D)J recombination activating gene, RAG-1.Cell. 59:1035–1048 [DOI] [PubMed] [Google Scholar]
- Scherer D.C., Brockman J.A., Bendall H.H., Zhang G.M., Ballard D.W., Oltz E.M. 1996. Corepression of RelA and c-rel inhibits immunoglobulin kappa gene transcription and rearrangement in precursor B lymphocytes.Immunity. 5:563–574 [DOI] [PubMed] [Google Scholar]
- Schlissel M., Constantinescu A., Morrow T., Baxter M., Peng A. 1993. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated.Genes Dev. 7:2520–2532 [DOI] [PubMed] [Google Scholar]
- Schram B.R., Tze L.E., Ramsey L.B., Liu J., Najera L., Vegoe A.L., Hardy R.R., Hippen K.L., Farrar M.A., Behrens T.W. 2008. B cell receptor basal signaling regulates antigen-induced Ig light chain rearrangements.J. Immunol. 180:4728–4741 [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. 1986. Multiple nuclear factors interact with the immunoglobulin enhancer sequences.Cell. 46:705–716 [DOI] [PubMed] [Google Scholar]
- Tiegs S.L., Russell D.M., Nemazee D. 1993. Receptor editing in self-reactive bone marrow B cells.J. Exp. Med. 177:1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. 1983. Somatic generation of antibody diversity.Nature. 302:575–581 [DOI] [PubMed] [Google Scholar]
- Tze L.E., Baness E.A., Hippen K.L., Behrens T.W. 2000. Ig light chain receptor editing in anergic B cells.J. Immunol. 165:6796–6802 [DOI] [PubMed] [Google Scholar]
- Vela J.L., Ait-Azzouzene D., Duong B.H., Ota T., Nemazee D. 2008. Rearrangement of mouse immunoglobulin kappa deleting element recombining sequence promotes immune tolerance and lambda B cell production.Immunity. 28:161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkoczy L., Ait-Azzouzene D., Skog P., Martensson A., Lang J., Duong B., Nemazee D. 2005. A role for nuclear factor kappa B/rel transcription factors in the regulation of the recombinase activator genes.Immunity. 22:519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F., Anderson D.E., Barnitz R.A., Snow A., Bidere N., Zheng L., Hegde V., Lam L.T., Staudt L.M., Levens D., et al. 2007. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation.Cell. 131:927–939 [DOI] [PubMed] [Google Scholar]