Abstract
As one of the major glutathione conjugation enzymes, GSTM1 detoxifies a number of drugs and xenobiotics. Its expression and activity have been shown to correlate both with cancer risks and drug resistance. Through a genome-wide association study, we identified a significant association between HapMap SNP rs366631 and GSTM1 expression. In this study, utilizing lymphoblastoid cell lines derived from International HapMap Consortium CEU and YRI populations, we designed and performed site-specific genotyping assays for both rs366631 and a highly homologous GSTM1 upstream site. Copy number variation (CNV) assays were performed for three different regions of the GSTM1 gene. We demonstrated that HapMap SNP rs366631 is a non-polymorphic site. The false genotyping call arises from sequence homology, a common GSTM1 region deletion and a non-specific genotyping platform used to identify the SNP. However, the HapMap call for rs366631 genotype is an indicator of GSTM1 upstream region deletion. Furthermore, this upstream deletion can be used as a marker of GSTM1 gene deletion. Using a novel GSTM1 CNV assay, we showed a population-specific CNV in this region upstream of the gene. More than 75% of the Caucasian (CEU) samples exhibit GSTM1 deletion and none contain two copies of GSTM1. In contrast, up to 25% of African (YRI) samples were found to have two copies of GSTM1. In conclusion, HapMap rs366631 is a pseudo-SNP that can be used as a GSTM1 deletion marker. Both the pseudo-SNP allele frequency and GSTM1 upstream region CNV show population-specific patterns between CEU and YRI samples.
INTRODUCTION
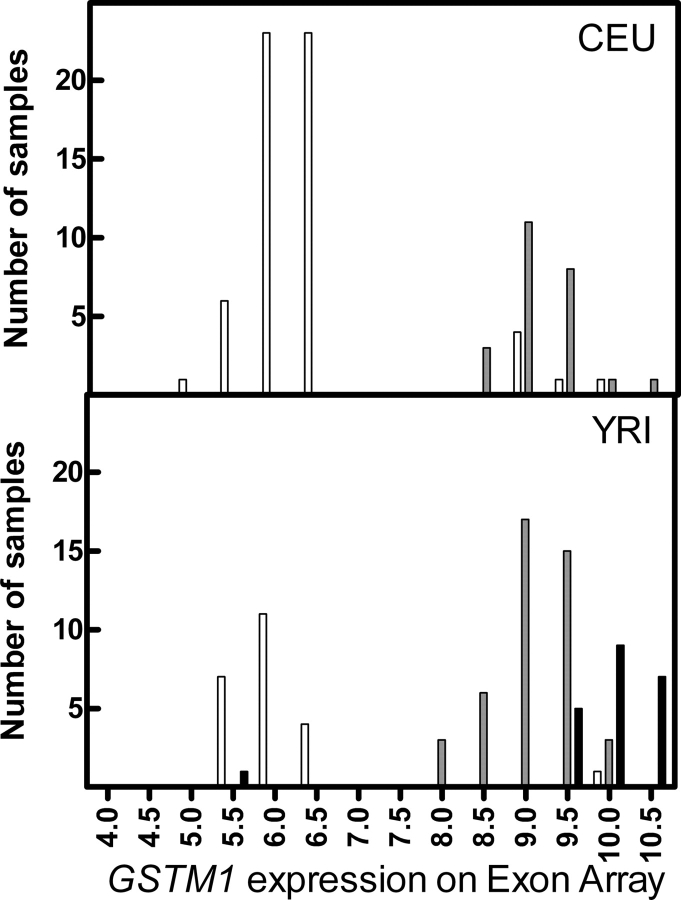
Glutathione S-transferase (GST), a major Phase II family of conjugation enzymes, plays a crucial role in the detoxification of drugs and environmental carcinogens (1). In humans, cytosolic GSTs are divided into eight classes: α (GSTA), µ (GSTM), π (GSTP), θ (GSTT), τ (GSTZ), σ (GSTS), o (GSTO) and κ (GSTK), each of which contains one or more of the homodimeric or heterodimeric isoforms (2). To date, a total of five µ class genes (M1–M5) situated in tandem on chromosome 1p13.3 have been identified (3). GSTM1, a member of GSTM family enzymes, detoxifies a number of electrophilic substances, including carcinogens such as polycyclic aromatic hydrocarbons, ethylene oxide, epoxides and styrene (3). Lack of GSTM1 enzyme may impair metabolic elimination of carcinogenic compounds from the body, thereby increase cancer risk (4), while overexpression of GSTM1 isozyme has been shown to associate with chemotherapeutic resistance (5,6). Interestingly, our laboratory recently found a significant inter-ethnic difference in GSTM1 expression in a set of 176 HapMap lymphoblastoid cell lines (LCLs) derived from individuals of European or African descent (7) (Fig. 1).
Figure 1.
Frequency distribution plot of GSTM1 expression. The total number of samples is plotted against GSTM1 expression obtained through Affymetrix GeneChip® Human Exon 1.0 ST array. Open bars represent samples with HapMap rs366631 TT genotype call; grey bars represent samples with HapMap rs366631 CT genotype call and solid black bars represent samples with HapMap rs366631 CC genotype call.
Given the functional importance of GSTM1 in cellular protection from environmental and oxidative stress, genetic variations that alter this gene expression or enzyme activity may play an important role in both risk of disease development and cellular sensitivity to drugs. Two polymorphisms of the GSTM1 gene, namely GSTM1*0 and GSTM1*A/GSTM1*B, have been identified. GSTM1*0 is a deleted allele, and the homozygous allele (GSTM1-null genotype) expresses no GSTM1 protein. Most studies of GSTM1 polymorphism have compared the homozygous deletion genotype with the genotypes containing at least one functional allele. Common homozygous gene deletions have been reported for GSTM1 with variable frequency in different ethnic groups (4,8). Alleles GSTM1A and GSTM1B differ by a C > G substitution at base position 534. This C > G substitution results in the substitution of Asn > Lys at amino acid 172 (9); however, this substitution results in no functional difference between the two alleles (10). As a result, GSTM1A and GSTM1B are categorized together as non-null conjugator phenotypes. Numerous studies have shown the positive correlation between GSTM1*0 (homozygous deletion) and increased risk of cancers (11–15) and with better treatment outcomes (16–18).
Utilizing a set of EBV-transformed LCLs from the International HapMap Consortium, our laboratory has recently performed a genome-wide association study between over 2 million HapMap SNPs and global gene expression (19). In an effort to identify relationships between genetic variants and gene expression for a set of important pharmacogenes in humans (7), we identified a previously unknown significant association between a SNP (rs366631) genotype and GSTM1 expression in cell lines derived from individuals with either Caucasian (CEU) or African (YRI) ancestry. rs366631 is located 11 265 bp downstream of GSTM1 gene on chromosome 1 with a population-specific minor allele frequency (0.18 and 0.49 for CEU and YRI, respectively). Further examination revealed that the TT genotype of rs366631 is associated with little or no GSTM1 expression, and the C allele (minor allele) carriers are associated with higher GSTM1 expression in both CEU and YRI samples (Fig. 1). The objective of our current study is to validate this predictor of GSTM1 expression.
RESULTS
Genotyping
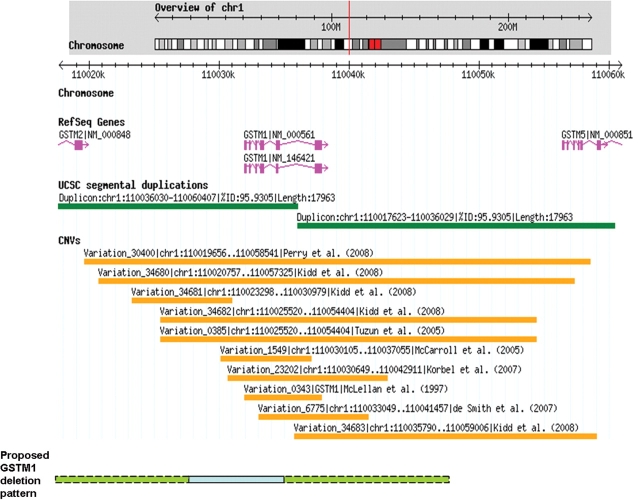
UCSC Genome Brower shows two duplications of more than 1000 bases of non-repeat masked sequence flanking the GSTM1 gene region (Fig. 2). rs366631 is located in one of these duplicons downstream of GSTM1 (http://genome.ucsc.edu). In contrast to a T being the common allele for rs366631, at the corresponding site in the upstream duplicon, a C allele is the common allele. Due to the high homology between these two duplicons, we designed and performed two sequence-specific genotyping assays, which included sequence-specific PCR, single-base extension (SBE) reaction followed by denaturing high-performance liquid chromatography (DHPLC). PCR products from both the rs366631 site and the upstream C site were sequenced and results confirmed that the amplified sequences were specific to the region of interest. We genotyped 30 randomly selected HapMap samples for both the rs366631 site and the upstream C site, and all samples genotyped showed non-polymorphic TT genotype at the rs366631 site. Approximately half of the samples tested showed non-polymorphic amplification of the C allele at the upstream C site and the remainder showed no amplification at the upstream C site (Table 1). The sequencing results were consistent with the SBE-DHPLC results.
Figure 2.
GSTM1 region overview. Information illustrated was obtained from Database of Genomic Variants (http://projects.tcag.ca/variation/ accessed on 10 July 2008; Human genome build 36). The chromosome location of GSTM1 gene, relative location of rs366631 (chromosome 1: 110,053,995, listed as ‘T’) and upstream C site (chromosome 1: 110,029,548, listed as ‘C’), UCSC segmental duplications finding, current CNV findings as well as our proposed novel GSTM1 deletion pattern are shown. The blue segment of proposed deletion pattern has been shown through our three CNV assays, which stretch from upstream C region to exon 7 of GSTM1. The dashed green segment of proposed deletion represents potential deletion region, which was derived from Kerb et al.'s report (20) but has not been confirmed in our study.
Table 1.
Genotyping results for randomly selected 30 HapMap samples at the rs366631 site and the upstream C site as compared with HapMap rs366631 calls
| Ethnicity | Sample id | HapMap calls for rs366631 | SBE assay rs366631 site | SBE assay upstream C site |
|---|---|---|---|---|
| YRI | NA19200 | C/C | T/T | C/C |
| CEU | NA12815 | C/T | T/T | C/C |
| CEU | NA12892 | C/T | T/T | C/C |
| CEU | NA11840 | C/T | T/T | C/C |
| YRI | NA18502 | C/T | T/T | C/C |
| YRI | NA19201 | C/T | T/T | C/C |
| YRI | NA18870 | C/T | T/T | C/C |
| YRI | NA19137 | C/T | T/T | C/C |
| YRI | NA19144 | C/T | T/T | C/C |
| YRI | NA19203 | C/T | T/T | C/C |
| YRI | NA19143 | C/T | T/T | C/C |
| CHB | NA18545 | C/T | T/T | C/C |
| CHB | NA18566 | C/T | T/T | C/C |
| JPT | NA18942 | C/T | T/T | C/C |
| CEU | NA06985 | C/T | T/T | C/C |
| CEU | NA11995 | C/T | T/T | C/C |
| CEU | NA12891 | T/T | T/T | – |
| CEU | NA11993 | T/T | T/T | – |
| CEU | NA11994 | T/T | T/T | – |
| CEU | NA11881 | T/T | T/T | – |
| CEU | NA11882 | T/T | T/T | – |
| YRI | NA19153 | T/T | T/T | – |
| YRI | NA18504 | T/T | T/T | – |
| CHB | NA18526 | T/T | T/T | – |
| CHB | NA18562 | T/T | T/T | – |
| CHB | NA18609 | T/T | T/T | – |
| JPT | NA18945 | T/T | T/T | – |
| JPT | NA18961 | T/T | T/T | – |
| JPT | NA18964 | T/T | T/T | – |
| JPT | NA18967 | T/T | T/T | – |
To insure non-polymorphic TT genotyping calls were not due to the sample selection, we also tested 52 non-HapMap CEPH (Centre d'Etude du Polymorphisme Humain) and an additional 28 HapMap samples that had HapMap rs366631 CC calls, all samples showed TT genotype at the rs366631 site, further suggesting HapMap rs366631 is non-polymorphic.
Upstream C region quantification
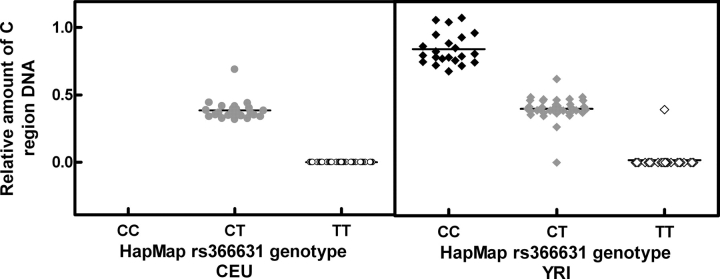
To evaluate the relative abundance of the genomic region that includes the upstream C site, we performed quantitative PCR (qPCR) assays on 176 HapMap CEU and YRI LCLs. We found a significant association between HapMap rs366631 genotyping calls and the relative amount of C region genomic sequence (P = 2 × 10−79, when association was analyzed with all 176 samples using population and sex as covariates). The CC genotype was found to correlate with two copies, CT with one copy and TT with zero copies of the GSTM1 upstream C site region (Fig. 3). Furthermore, the relative amount of the C region genomic sequence was significantly correlated with GSTM1 expression (measured by Affymetrix GeneChip® Human Exon 1.0 ST array, P < 0.0001). Upon evaluating the ‘pseudo genotype’ and C region copy number relationship, we identified an ethnic-specific GSTM1 upstream C region copy number variation (CNV), for which ∼75% of CEU samples showed a complete deletion of the upstream C region, 25% had a single copy and none of the CEU samples contained two copies of C region DNA (Fig. 3). In contrast, 25% of the YRI samples showed deletion, 50% had a single copy and 25% had two copies of the C region. The deletion allele frequency is 0.86 and 0.51 for CEU and YRI, respectively. This was consistent with Hardy–Weinberg equilibrium (P > 0.05) and showed a significant population difference (Fst = 0.15).
Figure 3.
Relationship between HapMap rs366631 genotype and relative amounts of upstream C region in both CEU and YRI samples. Circles represent CEU samples; diamonds represent YRI samples. P-values for association between genotype and upstream CNV are P = 2 × 10−52 and P = 2 × 10−37 for CEU and YRI, respectively.
GSTM1 region CNV
This upstream C region has been reported to be within GSTM1 gene deletion by Kerb et al. (20). To study whether the relative abundance of upstream C region quantity is part of the GSTM1 gene deletion, we performed two additional GSTM1 CNV assays and compared results to those obtained from our upstream C region assay (Table 2). A total of 14 samples were randomly selected from all three rs366631 genotyping groups. The upstream C region CNV results are in 100% agreement with those obtained through Applied Biosystems (ABI) TaqMan GSTM1 copy number assay (evaluating exon 1 region) and through an in-house GSTM1 copy number assay (evaluating intron 6/exon 7 region). Given the different genomic regions these assays have evaluated, our findings support that the upstream C region is a component of the GSTM1 gene deletion.
Table 2.
GSTM1 CNV assay comparison on 14 HapMap LCL samples
| Sample ids | C Site qPCR | ABI assay | In-house assay | HapMap rs366631 genotype call |
|---|---|---|---|---|
| 18500 | 0.78 | 0.75 | 0.83 | CC |
| 18517 | 0.75 | 0.82 | 0.85 | CC |
| 18863 | 0.81 | 1.12 | 0.78 | CC |
| 19142 | 0.87 | 0.79 | 0.94 | CC |
| 11840 | 0.40 | 0.40 | 0.45 | CT |
| 12146 | 0.36 | 0.37 | 0.62 | CT |
| 12760 | 0.32 | 0.39 | 0.39 | CT |
| 19139 | 0.46 | 0.40 | 0.39 | CT |
| 19140 | 0.45 | 0.41 | 0.39 | CT |
| 19194 | 0.38 | 0.42 | 0.45 | CT |
| 10860 | 0 | NA | 0 | TT |
| 12234 | 0 | NA | 0 | TT |
| 18503 | 0 | NA | 0 | TT |
| 19154 | 0 | NA | 0 | TT |
C site qPCR assay quantifies the GSTM1 upstream genomic region.
ABI assay quantifies the GSTM1 5′-UTR and exon 1 region.
In-house assay quantifies the GSTM1 intron 6 and exon 7 region.
DISCUSSION
In this study, we evaluated in greater depth the relationship between the HapMap reported SNP, rs366631, and GSTM1 gene expression. Our site-specific SBE-DHPLC assay designed to genotype rs366631, demonstrated that rs366631 is a non-polymorphic site. Given the high homology between the duplicon where rs366631 is located and the duplicon upstream of GSTM1 gene, it is possible that HapMap genotyping calls were evaluating both sites. We therefore designed an additional site-specific SBE-DHPLC assay for the upstream C site. All amplified C site PCR products showed a CC genotype; however, ∼50% were not amplified. When comparing our site-specific genotyping results with HapMap rs366631 calls, we reasoned that non-amplification of the C site was consistent with a TT call for rs366631; while a CT call for rs366631 was likely resulting from the presence of one copy of the upstream C site and a CC call for rs366631 corresponding to two copies.
The platform used to make the genotype calls for HapMap rs366631 was Illumina HapMap-BeadArrays. The Illumina HapMap-BeadArray utilized GoldenGate™ Assay design, which contains a set of three oligonucleotides. Two of them are 5′ assay oligos designed to hybridize upstream of the SNP, with the 3′ base of each being allele-specific. The third oligo is located 3′ end of the two allele-specific oligos and is locus-specific (http://www.ncbi.nlm.nih.gov.proxy.uchicago.edu/SNP/snp_viewTable.cgi?method_id=915). Given the high homology for two duplicons flanking GSTM1 region, this design may non-specifically produce signals from both duplicons. Therefore, SNP calls are made when, in fact, two non-polymorphic homologous sites are evaluated using a single assay. Furthermore, literature supports homozygous GSTM1 deletion frequencies of 20–80% in various ethnic populations (4,8). Considering both the genotyping platform design and the potential inclusion of the upstream C site in the common GSTM1 deletion, it became clear that when the GSTM1 region is deleted, a TT call is made; however, we were puzzled that CC calls were made when both C and T sites were present. One possibility is that the Illumina HapMap BeadArray oligos were designed to capture the upstream C site nucleotide and as a consequence also non-specifically captured the downstream T site nucleotide at a decreased efficiency. After quantifying the relative prevalence of the upstream C region, a significant association was found between HapMap rs366631 genotyping calls and the relative amount of C site genomic sequence. When two copies of C site genomic regions exist, a CC call is made. When only one copy of C site region is present, a CT call is made. When there was a homozygous deletion of this region, TT genotype was called. These findings support the possibility that the relative abundance of the upstream C region is responsible for HapMap genotype calls. However, other possible explanation may also result in the wrong genotyping calls (e.g. genotype calling algorithm forces the call of the three possible genotypes). Without an evaluation of the raw genotype intensity data that is not currently in the public domain, we can only speculate the cause. Interestingly, rs366631 is listed as a marker on both Illumina Human 1M-duo (http://www.illumina.com/pages.ilmn?ID=261&source=home) and Human 610-quad beadchips (http://www.illumina.com/pages.ilmn?ID=248). However, the genotype results on this site are read as a ‘no call’ (M. Kibriya, H. Ahsan, C. Brown and K. White, personal communication). In addition, the rs366631 SNP genotype is not currently reported in the PhaseIII HapMap data release (http://www.hapmap.org/cgi-perl/gbrowse/hapmap3_B36/).
In addition, we found a significant correlation between GSTM1 genomic deletion (measured by qPCR using primers designed to the upstream C region) and GSTM1 expression. This was consistent with previous findings by French et al. that GSTM1-null genotype corresponds to lower GSTM1 expression when compared with non-null genotype (21). Furthermore, McLellan et al. have shown that duplication of GSTM1 gene may cause ultra rapid GSTM1 enzyme activity (22). Interestingly, in our study, all CEU HapMap samples exhibit at least partial deletion at the GSTM1 upstream C region; while ∼25% of YRI samples have two copies of the C region. This ethnic-specific genomic deletion is a likely explanation for the observed population difference in GSTM1 expression between the CEU and YRI samples. GSTM1 deletion polymorphism has generally been described as either null (homozygous deletion) or non-null. Although significant inter-ethnic differences in GSTM1 homozygous deletion have been reported (4,8), our study is the first to show ethnic differences in the number of expressed GSTM1 alleles between samples with Caucasian ancestry and those with African ancestry, which corresponds to differences in GSTM1 expression. A considerable functional difference between carriers of two versus one active GSTM1 allele has been suggested (20). Given the important role GSTM1 in disease risk and drug response, the functional significance of this ethnic-specific GSTM1 genetic CNV should be further explored. Finally, although the low GSTM1 allele frequency in CEU samples may justify studies using null versus non-null GSTM1 genotype analysis, when studying the genetic contribution of GSTM1 in populations that have fairly high GSTM1 allele frequency such as the Yorubans, a more quantitative analysis of GSTM1 allele is warranted.
Two additional GSTM1 CNV assays were performed and compared with the upstream C region CNV assay. We found complete agreement between these assays that interrogated different sections of the GSTM1 genomic region in randomly selected 14 HapMap LCL samples. When two copies of the upstream region are present, this encompasses two copies of exon 1 and intron 6/exon 7 DNA; but if there is no copies of the upstream region, exon 1 and intron 6/exon 7 DNA are not present. Upon evaluating the Database of Genomic Variants (http://projects.tcag.ca/variation/ accessed on 10 July 2008; Human genome build 36), nine patterns of GSTM1 region deletions are currently listed. Three of these patterns contain large deleted regions encompassing both the T (GSTM1 downstream) and C (GSTM1 upstream) sites (23–25); while the other four are smaller deletion regions which include neither T nor C site (22,26–28). In addition, one deletion pattern included only the T site (23). The final pattern included only the C site in the deletion region (23), which is in agreement with our findings. In addition, although not listed in the Database of Genomic Variants, Kerb et al. have reported an approximately 18 500 bp deletion, which includes the upstream C region, but not the downstream T region (20). When combining three CNV assays, our data suggest the potential deletion region extends at least from the upstream C region through the exon 7 region of GSTM1, a region longer than that reported by Kidd et al. (23) and within the deletion region described by Kerb et al. (20). These data suggest either potential individual specific CNV patterns due to sample selection (Kidd et al. studied eight individuals and we evaluated 14 individuals with one sample that overlapped between the two studies), or a potential new pattern of GSTM1 region CNV that warrants further study.
In summary, through site-specific genotyping, we demonstrated that HapMap SNP rs366631 is a non-polymorphic site. The sequence homology, the common GSTM1 region deletion, and non-specific genotyping platform usage are likely contributing to the false genotyping call. Although referred to as HapMap rs366631, this genotype is an indicator of GSTM1 upstream region deletion. Furthermore, this upstream deletion can be used as a marker of GSTM1 gene deletion. Lastly, we developed a new GSTM1 CNV assay that interrogates upstream of GSTM1 and demonstrates a population-specific CNV in this region.
MATERIALS AND METHODS
DNA isolation
EBV-transformed B-LCLs were purchased from the Coriell Institute for Medical Research (Camden, NJ). Cell lines were maintained and diluted as described previously (29). DNA was isolated from 87 HapMap CEU, 89 YRI, 8 CHB (Chinese from Beijing), 8 JPT (Japanese from Tokyo) LCLs and 52 non-HapMap CEPH LCLs. For each cell line, a total of 5 × 106 cells were pelleted, washed in ice-cold PBS and centrifuged to remove PBS. All cell pellets were flash-frozen and stored at −80°C until DNA isolation. Total DNA was extracted using the 5Prime kit (5Prime Inc., Fisher Scientific, Pittsburgh, PA) following the manufacturer's protocol. DNA quality assessment and quantification were conducted using the optical spectrometry 260/280 nm ratio.
Genotyping
For specificity, two region-specific PCR amplification primer sets were designed with each one specifically amplifying one duplicon to produce 151 bp PCR products (Table 3). Both the rs366631 site and upstream C site genotyping were performed on a set of randomly selected 30 HapMap samples which consist of 10 CEU, 10 YRI, 5 CHB and 5 JPT. In addition, rs366631 genotyping was performed on 52 non-HapMap CEPH samples and 28 HapMap samples (22 YRI, 3 CHB and 3 JPT) that had CC genotyping calls. Sample IDs can be found in the Supplementary Material.
Table 3.
PCR and SBE primer design for rs366631, upstream C site and an endogenous control
| Region of interest | PCR primer sets | SBE primer |
|---|---|---|
| rs366631 GSTM1 downstream | F: 5′-TGG CCT CAC AGT GAA TTC TC-3′ | 5′-CAT TAT GGA TTA AGA GTT TAT GAT T-3′ |
| R: 5′-TCT TTC ACA GCT CAA GAC CC-3′ | ||
| GSTM1 upstream C site | F: 5′-TGG CCT CAC AGT GAA CTC TT-3′ | 5′-CAT TAT GGA TTA AGA GTT TAT GAT T-3′ |
| R: 5′-TCT TTC ACA GCT CAA GAC GG-3′ | ||
| huB2M | F: 5′-TGGCACCTGCTGAGATACTG-3′ | none |
| R: 5′-GGATGCTAGGACAGCAGGAC-3′ |
SBE, single-base pair extension. Bold represent nucleotides specific for the region of interest.
PCR reactions were set up in a total volume of 15 µl containing 2.5 mM MgCl2, 50 µM each dNTP, 125 nM forward and reverse primers, 0.625 U AmpliTaq Gold (Applied Biosystems, Foster City, CA, USA) and 30 ng genomic DNA. Reactions were denatured initially at 95°C for 15 min followed by 40 cycles of 95°C for 15 s, 58°C for 15 s and 72°C for 30 s and a final extension at 72°C for 10 min. PCR products were cleaned-up with Shrimp Alkaline Phosphatase and Exonuclease I at 37°C for 45 min. To confirm region-specific amplification, PCR products were sequenced as described later.
Genotyping was conducted through SBE reactions followed by DHPLC. A volume of 7.2 µl of purified PCR product was added for SBE reaction with 1 µM SBE primer, 250 µM each ddNTP and 1.5 U Thermo Sequenase (GE Healthcare) in a 12 µl volume. SBE reactions were run under the following conditions: 96°C for 2 min, followed by 60 cycles at 96°C for 30 s, 55°C for 30 s and 60°C for 30 s. SBE products were denatured and separated on a WAVE 3500HT DHPLC System, and analyzed by Navigator software (Transgenomic, Inc.).
Sequencing
The sequencing reactions were carried out in a total volume of 10 µl containing 0.5 or 1 µl purified PCR product, 400 nM forward or reverse primer, 2 µl Big Dye terminator v3.1 (Applied Biosystems) and 1× Big Dye dilution buffer. The reactions were denatured initially at 95°C for 5 min then cycled at 95°C for 10 s, 50°C for 5 s and 60°C for 4 min for 25 cycles of the standard sequencing conditions. Same T or C site amplification primer sets were used for both forward and reverse direction sequencing. Sequencing was performed on an ABI 3100 Sequencer (Applied Biosystems). The sequence chromatograms were analyzed using the Sequencher 4.5 program.
Upstream C region quantification
Prevalence of the C site genomic region was quantified in 176 HapMap CEU and YRI LCLs. We performed SYBR green qPCR assays using the same primer sets designed for upstream C site amplification described earlier and a primer set for an endogenous control (huB2M) (Table 3) on the Applied Biosystems 7500 real-time PCR system. Reactions were carried out in 25 µl volumes consisting of 12.5 μl 2× SYBR Green PCR Master mix (Applied Biosystems), 1.5 µl primer mix (final of 300 nM for each forward and reverse primers) and 40 ng DNA. The thermocycler parameters were: 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 15 s/63°C for 1 min. Each cycle threshold (Ct) value obtained for C region was normalized using huB2M independently. A standard curve was constructed using a pool of two LCL samples (GM15144 and GM15324) that were known to have two copies of GSTM1. The relative upstream C region expression in our LCL samples was obtained by standard curve fitting. Each experiment was conducted a minimum of two times and samples were run in triplicate for each experiment.
A quantitative transmission disequilibrium test (QTDT) was performed to identify association between HapMap rs366631 genotype and qPCR results for GSTM1 upstream C region using QTDT software (http://www.sph.umich.edu/csg/abecasis/QTDT) (30). QTDT tests were performed within each population with sex as a covariate as well as in the combined CEU and YRI populations with both sex and population as covariates. P < 0.05 was considered statistically significant.
GSTM1 region CNV
To study whether the relative abundance of the upstream C region could be used as a GSTM1 gene deletion marker, we compared upstream C region qPCR assay to two other GSTM1 CNV assays. One is commercially available from Applied Biosystem (ABI) that is designed to interrogate the 5′-UTR and exon 1 of GSTM1; the other is our custom-designed CNV assay that interrogates intron 6 and exon 7 region of GSTM1. Assays were performed on 14 samples (four with CC, six with CT and four with TT HapMap rs366631 genotyping calls). For the ABI assay, Assay ID: Hs02575461_cn was used. The total reaction volume was 25 µl which consisted of 13.6 μl 2× PCR Master mix (Applied Biosystems), 1.4 µl primer mix (primer sets and TaqMan probe) along with 12.5 ng DNA. TaqMan RNase P control reagents kit was used as an endogenous control (Part #: 4316844). The in-house SYBR PCR reaction assay was conducted in a total volume of 25 µl that contained 12.5 μl 2× SYBR Green PCR Master mix, 1.5 µl primers mix (F: 5′-CTT CAC GTG TTA TGG AGG TTC-3′; R: 5′-TTG GGC TCA AAT ATA CGG TGG-3′) and 20 ng of DNA. This assay amplifies 111 bp amplicon between the intron 6 and exon 7 of GSTM1. The in-house assay results were normalized using huB2M. Both assays were performed on the Applied Biosystems 7500 real-time PCR system. The thermocycler parameters were: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s/60°C for 1 min for ABI assay; 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s/ 61.5°C for 1 min for the in-house assay.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG Online.
FUNDING
This Pharmacogenetics of Anticancer Agents Research (PAAR) Group (http://pharmacogenetics.org) study was supported by NIH/NIGMS grant UO1GM61393 and UO1GM61374 (http://pharmgkb.org/). This study was also supported by P50 CA125183 University of Chicago Breast Cancer SPORE grant. R.S.H. receives support from NIH grant 5 T32 GM 007019-30.
ACKNOWLEDGEMENTS
The authors are grateful for excellent technical support provided by Shannon M. Delaney and Wasim K. Bleibel.
Conflict of Interest statement. Dr Dolan has presented seminars at meetings funded by Affymetrix, Inc.
REFERENCES
- 1.Keen J., Jakoby W. Glutathione transferases. Catalysis of nucleophilic reactions of glutathione. J. Biol. Chem. 1978;253:5654–5657. [PubMed] [Google Scholar]
- 2.Hayes J., Flanagan J., Jowsey I. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 3.Takahiko K., Yuko Y., Mayumi T., Masatoshi W. Genetic polymorphisms of human cytosol glutathione S-transferases and prostate cancer. Pharmacogenomics. 2008;9:93–104. doi: 10.2217/14622416.9.1.93. [DOI] [PubMed] [Google Scholar]
- 4.Rebbeck T. Molecular epidemiology of the human glutathione S-transferase genotypes GSTM1 and GSTT1 in cancer susceptibility. Cancer Epidemiol. Biomarkers Prev. 1997;6:733–743. [PubMed] [Google Scholar]
- 5.McIlwain C., Townsend D., Tew K. Glutathione S-transferase polymorphisms: cancer incidence and therapy. Oncogene. 2006;25:1639–1648. doi: 10.1038/sj.onc.1209373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith G., Stanley L., Sim E., Strange R., Wolf C. Metabolic polymorphisms and cancer susceptibility. Cancer Surv. 1995;25:27–65. [PubMed] [Google Scholar]
- 7.Huang R., Duan S., Kistner E., Zhang W., Bleibel W., Cox N., Dolan M. Identification of genetic variants and gene expression relationships associated with pharmacogenes in humans. Pharmacogenet. Genomics. 2008;18:545–549. doi: 10.1097/FPC.0b013e3282fe1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moyer A., Salavaggione O., Hebbring S., Moon I., Hildebrandt M., Eckloff B., Schaid D., Wieben E., Weinshilboum R. Glutathione S-transferase T1 and M1: gene sequence variation and functional genomics. Clin. Cancer Res. 2007;13:7207–7216. doi: 10.1158/1078-0432.CCR-07-0635. [DOI] [PubMed] [Google Scholar]
- 9.Seidegard J., Vorachek W., Pero R., Pearson W. Hereditary differences in the expression of the human glutathione transferase active on trans-stilben oxide are due to a gene deletion. Proc. Natl. Acad. Sci. USA. 1998;85:7203–7207. doi: 10.1073/pnas.85.19.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Widersten M., Pearson W., Engstrom A., Mannervik B. Heterologous expression of the allelic variant µ-class glutathione transferase µ and ψ. Biochem. J. 1991;276:519–524. doi: 10.1042/bj2760519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlsten C., Sagoo G., Frodsham A., Burke W., Higgins J. Glutathione S-transferase M1 (GSTM1) polymorphisms and lung cancer: a literature-based systematic HuGE review and meta-analysis. Am. J. Epidemiol. 2008;167:759–774. doi: 10.1093/aje/kwm383. [DOI] [PubMed] [Google Scholar]
- 12.Murta-Nascimento C., Schmitz-Dräger B., Zeegers M., Steineck G., Kogevinas M., Real F., Malats N. Epidemiology of urinary bladder cancer: from tumor development to patient's death. World J. Urol. 2007;25:285–295. doi: 10.1007/s00345-007-0168-5. [DOI] [PubMed] [Google Scholar]
- 13.Reszka E., Czekaj P., Adamska J., Wasowicz W. Relevance of glutathione S-transferase M1 and cytochrome P450 1A1 genetic polymorphisms to the development of head and neck cancers. Clin. Chem. Lab. Med. 2008;46:1090–1096. doi: 10.1515/CCLM.2008.227. [DOI] [PubMed] [Google Scholar]
- 14.Saxena A., Dhillon V., Raish M., Asim M., Rehman S., Shukla N., Deo S., Ara A., Husain S. Detection and relevance of germline genetic polymorphisms in glutathione S-transferases (GSTs) in breast cancer patients from northern Indian population. Breast Cancer Res. Treat. 2008 doi: 10.1007/s10549-008-0098-y. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 15.Ye Z., Song H. Glutathione S-transferase polymorphisms (GSTM1, GSTP1 and GSTT1) and the risk of acute leukaemia: a systematic review and meta-analysis. Eur. J. Cancer. 2005;41:980–989. doi: 10.1016/j.ejca.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 16.Müller P., Asher N., Heled M., Cohen S., Risch A., Rund D. Polymorphisms in transporter and phase II metabolism genes as potential modifiers of the predisposition to and treatment outcome of de novo acute myeloid leukemia in Israeli ethnic groups. Leuk. Res. 2008;32:919–929. doi: 10.1016/j.leukres.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Nagle C., Chenevix-Trench G., Spurdle A., Webb P. The role of glutathione-S-transferase polymorphisms in ovarian cancer survival. Eur. J. Cancer. 2007;43:283–290. doi: 10.1016/j.ejca.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Ott K., Lordick F., Becker K., Ulm K., Siewert J., Höfler H., Keller G. Glutathione-S-transferase P1, T1 and M1 genetic polymorphisms in neoadjuvant-treated locally advanced gastric cancer: GSTM1-present genotype is associated with better prognosis in completely resected patients. Int. J. Colorectal Dis. 2008;23:773–782. doi: 10.1007/s00384-008-0490-4. [DOI] [PubMed] [Google Scholar]
- 19.Duan S., Huang R.S., Zhang W., Bleibel W.K., Roe C., Clark T., Chen T., Schweitzer A., Blume J., Cox N., et al. Genetic architecture of transcript-level variation in humans. Am. J. Hum. Genet. 2008;82:1101–1113. doi: 10.1016/j.ajhg.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerb R., Brockmöller J., Sachse C., Roots I. Detection of the GSTM1*0 allele by long polymerase chain reaction. Pharmacogenetics. 1999;9:89–94. [PubMed] [Google Scholar]
- 21.French D., Wilkinson M.R., Yang W., Chaisemartin L.d., Cook E.H., Das S., Ratain M.J., Evans W.E., Downing J.R., Pui C.-H., et al. Global gene expression as a function of germline genetic variation. Hum. Mol. Genet. 2005;14:1621–1629. doi: 10.1093/hmg/ddi170. [DOI] [PubMed] [Google Scholar]
- 22.McLellan R., Oscarson M., Alexandrie A., Seidegård J., Evans D., Rannug A., Ingelman-Sundberg M. Characterization of a human glutathione S-transferase mu cluster containing a duplicated GSTM1 gene that causes ultrarapid enzyme activity. Mol. Pharmacol. 1997;52:958–965. doi: 10.1124/mol.52.6.958. [DOI] [PubMed] [Google Scholar]
- 23.Kidd J., Cooper G., Donahue W., Hayden H., Sampas N., Graves T., Hansen N., Teague B., Alkan C., Antonacci F., et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry G., Ben-Dor A., Tsalenko A., Sampas N., Rodriguez-Revenga L., Tran C., Scheffer A., Steinfeld I., Tsang P., Yamada N., et al. The fine-scale and complex architecture of human copy-number variation. Am. J. Hum. Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuzun E., Sharp A., Bailey J., Kaul R., Morrison V., Pertz L., Haugen E., Hayden H., Albertson D., Pinkel D., et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 26.de Smith A., Tsalenko A., Sampas N., Scheffer A., Yamada N., Tsang P., Ben-Dor A., Yakhini Z., Ellis R., Bruhn L., et al. Array CGH analysis of copy number variation identifies 1284 new genes variant in healthy white males: implications for association studies of complex diseases. Hum. Mol. Genet. 2007;16:2783–2794. doi: 10.1093/hmg/ddm208. [DOI] [PubMed] [Google Scholar]
- 27.Korbel J., Urban A., Affourtit J., Godwin B., Grubert F., Simons J., Kim P., Palejev D., Carriero N., Du L., et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarroll S., Hadnott T., Perry G., Sabeti P., Zody M., Barrett J., Dallaire S., Gabriel S., Lee C., Daly M., et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 29.Huang R.S., Kistner E.O., Bleibel W.K., Shukla S.J., Dolan M.E. Effect of population and gender on chemotherapeutic agent-induced cytotoxicity. Mol. Cancer Ther. 2007;6:31–36. doi: 10.1158/1535-7163.MCT-06-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abecasis G., Cardon L., Cookson W. A general test of association for quantitative traits in nuclear families. Am. J. Hum. Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.