Abstract
Alcohol dependence (AD) is a complex disorder with environmental and genetic origins. The role of two genetic variants in ALDH2 and ADH1B in AD risk has been extensively investigated. This study tested for associations between nine polymorphisms in ALDH2 and 41 in the seven ADH genes, and alcohol-related flushing, alcohol use and dependence symptom scores in 4597 Australian twins. The vast majority (4296) had consumed alcohol in the previous year, with 547 meeting DSM-IIIR criteria for AD. There were study-wide significant associations (P < 2.3 × 10−4) between ADH1B-Arg48His (rs1229984) and flushing and consumption, but only nominally significant associations (P < 0.01) with dependence. Individuals carrying the rs1229984 G-allele (48Arg) reported a lower prevalence of flushing after alcohol (P = 8.2 × 10−7), consumed alcohol on more occasions (P = 2.7 × 10−6), had a higher maximum number of alcoholic drinks in a single day (P = 2.7 × 10−6) and a higher overall alcohol consumption (P = 8.9 × 10−8) in the previous year than those with the less common A-allele (48His). After controlling for rs1229984, an independent association was observed between rs1042026 (ADH1B) and alcohol intake (P = 4.7 × 10−5) and suggestive associations (P < 0.001) between alcohol consumption phenotypes and rs1693482 (ADH1C), rs1230165 (ADH5) and rs3762894 (ADH4). ALDH2 variation was not associated with flushing or alcohol consumption, but was weakly associated with AD measures. These results bridge the gap between DNA sequence variation and alcohol-related behavior, confirming that the ADH1B-Arg48His polymorphism affects both alcohol-related flushing in Europeans and alcohol intake. The absence of study-wide significant effects on AD results from the low P-value required when testing multiple single nucleotide polymorphisms and phenotypes.
INTRODUCTION
Alcohol dependence (AD) is a complex behavioral disorder with both social and biological origins (1–3), characterized by a syndrome of serious problems related to alcohol use. Diagnostic criteria include withdrawal symptoms after cessation of heavy intake, tolerance to alcohol's effects, continued use despite evidence of physical or psychological problems due to drinking, persistent desire to quit drinking or inability to do so, and spending substantial amounts of time drinking or recovering from drinking. Family, twin and adoption studies support the view that genes contribute between 40 and 60% of the variance in AD risk (4–8). Environment also contributes a substantial proportion to the risk of AD, and different phenotypic characteristics add to the AD risk in different families (9–13). The genetic variance in AD risk is likely to reflect gene polymorphism effects on multiple characteristics, which collectively impact on the probability of hazardous or harmful drinking and on the progression to dependence. Some of these characteristics seem to be related to alcohol's metabolism or to its acute effects, while others relate to other types of psychopathology or to personality.
The roles of the biologically relevant alcohol-metabolizing genes, alcohol dehydrogenases (particularly the Class I, low-Km ADHs ADH1A, ADH1B and ADH1C) and aldehyde dehydrogenases (particularly the low-Km mitochondrial ALDH2), in AD risk have been extensively investigated, initially in Asians (14–16) and subsequently in European (17–22) and African (23,24) populations. The significant associations between both ALDH2 and ADH1B variants and AD risk have been explained on the hypothesis that any increase in acetaldehyde production, or reduction in its subsequent elimination, will reduce an individual's vulnerability to alcohol abuse and alcoholism because of the aversive effects associated with elevated blood and tissue acetaldehyde levels (2,25). This has been demonstrated by the measurement of acetaldehyde in the blood after alcohol consumption in people with ALDH2 deficiency, but not in relation to ADH variants.
ALDH2
Acetaldehyde, generated by ethanol oxidation in the liver, is further metabolized to acetate by aldehyde dehydrogenases (ALDH) (26,27). The high-affinity mitochondrial ALDH2 is mainly responsible, but ALDH2 deficiency is common in parts of Asia. The effect of the inactive ALDH2*2 enzyme was first investigated by Wolff (28) who observed racial differences in the facial flushing response during alcohol consumption. The ALDH2*1 (504Glu) allele encodes an active subunit while ALDH2*2 (504Lys) encodes a subunit that is essentially inactive (consequently causing a build-up of acetaldehyde in the blood and other tissues). Hybridization of an ALDH2*2 enzyme subunit with an ALDH2*1 subunit results in the inactivation of the isozyme and an ALDH2-deficient phenotype (1,29,30). Individuals homozygous for the ALDH2*2, or heterozygous, are therefore deficient in the conversion of acetaldehyde to acetate, have high blood acetaldehyde levels after alcohol consumption and suffer from adverse reactions to alcohol, such as severe facial flushing, nausea, headache and tachycardia. The importance of ALDH2 genetic variation in risk for AD has been well established among Asian populations where the flushing response is observed in 57–80% of individuals, and the ALDH2*2 allele frequency ranges from 0.25 to 0.35. Heterozygotes are at reduced risk of AD compared to ALDH2*1 homozygotes, while individuals homozygous for ALDH2*2 have a very low risk for AD (14,15,31). This polymorphism (rs671) is confined to populations from North-East Asia and groups with ancestry from that region.
ADH1B
Both ADH1B and ADH1C exhibit single nucleotide polymorphisms (SNPs) resulting in critical single amino acid exchanges at different sites in the NAD+ coenzyme-binding domain. Accordingly, the enzymes encoded by polymorphic forms of ADH1B and ADH1C display different maximal activities (Vmax) and affinities (Km) for ethanol (1,32,33). This feature has been widely cited as the explanation for the well-validated effects of ADH1B variation (34–36) on alcohol use and dependence.
The β2β2 isozyme resulting from the 48His form of ADH1B Arg48His has a significantly higher Km value for ethanol, and a maximal activity 40-fold that of β1β1 (27). Therefore, ADH1B 48His homozygotes are predicted to produce higher levels of acetaldehyde than any other ADH1B genotype. While facial flushing has been observed in Japanese drinkers either hetero- or homozygous for ADH1B*2 after controlling for ALDH2 genotype (37–39), other reports are negative or equivocal (40–43).
The ADH1C-encoded γ1 and γ2 homodimers also show a difference in their Vmax activities. Since the Vmax of the γ1γ1 isozyme is 2.5-fold higher than γ2γ2, people carrying ADH1C 349Ile have a faster predicted elimination rate for alcohol. Conflicting reports have been published on the association between ADH1C Ile349Val and AD. Although strong LD exists between ADH1B Arg48His and ADH1C Ile349Val (16,18), the genetic polymorphisms of ADH1B, rather than ADH1C, have the stronger association with the development of alcoholism.
Most early studies on the effects of ADH1B variation were performed in China or Japan, where the frequency of the ADH1B 48His allele is higher than elsewhere (44). Turning to studies on population groups outside East Asia, a study on Israeli Jews by Neumark et al. (45) indicated that the ADH1B polymorphism accounted for 20–30% of variation in alcohol intake, and the less common ADH1B 48His served as a protective factor against AD. Similar effects on alcohol consumption have been reported in young (age <33), but not older, Israeli Jews (46). We also found an association between ADH1B 48His and alcohol intake and dependence in our earlier study of Australians of European descent (17). Borras et al. (18) found an association between ADH1B but not ADH1C genotype and AD in Caucasian people. Other studies have observed a small protective function for ADH1C 369Ile in Caucasians (2,47). Some recent studies indicate that both ADH1C and ADH1B independently affect drinking habits in European (48) and Japanese (49) people.
The main gap in our understanding lies between the effects of ADH polymorphisms on enzyme activity and the differences in alcohol use associated with these polymorphisms. Although in vitro ADH activity is affected, the evidence that alcohol metabolism in vivo is affected is sparse and largely (50,51) but not entirely (52) negative. Also, there is no direct evidence that ADH variation produces differences in acetaldehyde generation during alcohol metabolism, nor that the ADH variants lead to differences in the subjective responses (positive or negative) to alcohol use.
To date, the majority of association studies investigating the role of alcohol metabolizing genes in risk for AD have focused on the well-characterized coding variants within ADH1B, ADH1C and ALDH2 and on the phenotype of AD. Three recent studies, however, have systematically analyzed polymorphisms within the seven ADH cluster genes (53) as well as ALDH1A1 and ALDH2 (21) in non-Asian samples. Edenberg et al. (19) observed evidence of association between AD and multiple ADH4 SNPs spanning a single haplotype block within the Collaborative Study on the Genetics of Alcoholism (COGA) cohort of United States families enriched for alcoholism. They did not report association with either the ADH1B Arg48His or ADH1C Ile369Val variants. Luo et al. (20) typed 20 SNPs in ALDHs and ADHs and found evidence for effects of the major ADH1B variants in European- and African-Americans. They also found evidence for differences in ADH4 SNP genotype (but not allele) frequencies between controls and patients with alcohol or cocaine dependence (54). Kuo et al. (21) reported the association between one SNP each in ADH5 and ADH1B and AD in the Irish Affected Sib Pair Study of Alcohol Dependence (IASPSAD) sample composed of severely affected cases. No association was observed with any ALDH1A1 or ALDH2 marker, nor with the ADH1C*1 variant.
We have now genotyped 50 SNPs, spanning the seven ADH genes and ALDH2, in 4597 individuals representative of the general European-Australian population. SNPs were chosen to provide comprehensive coverage of the ADH gene cluster, including the atypical ADH1B*2 (His48) marker, and also of ALDH2. We explore the relationships between variation in the alcohol metabolizing genes and self-reported reactions to alcohol, as well as quantitative alcohol consumption measures and both quantitative and binary AD measures within our Caucasian population.
RESULTS
A total of 4597 twin participants provided blood samples and completed the SSAGA-OZ interview. Two thousand six hundred and eighteen families were included in the analysis. Since parents were not genotyped, only families containing a non-identical sibling pair contributed to the ‘within-family’ tests of association. One thousand one hundred and seventy-seven families included a dizygotic (DZ) pair and hence were potentially informative for both the ‘within’ and the ‘total’ tests of association. Eight hundred and fourteen families included a monozygotic (MZ) pair and hence contributed information to only the ‘total’ test. Similarly, 627 families had just a single offspring and these contributed information to only the ‘total’ test. The mean age of the study population was 43.8 ± 11.5 years, ranging from 26 to 89 years. The mean age of female participants was significantly higher (44.1 versus 42.5) than for males (P = 0.001) (Table 1). While a small percentage (<3%) of the sample had always abstained from alcohol, the majority (∼93%) had consumed alcohol in the previous year, with 7.6 and 5.5% of male and females, respectively, reporting that they drank alcohol on a daily basis. With respect to AD, male participants were approximately four times more likely to meet a lifetime diagnosis of DSM-IIIR dependence than females (25.4 versus 6.1%). However, ∼36% of males and ∼70% of females did not report any symptoms of AD.
Table 1.
Summary of the sample demographics and alcohol-related traits, by sex
| Male (n = 1478) | Female (n = 2818) | |
|---|---|---|
| Age at time of interview (years) | 42.5 ± 10.9 | 44.1 ± 11.6 |
| Age range (years) | 26–89 | 28–84 |
| Has consumed alcohol in the previous 12 months, n (%) | 1407 (95.2) | 2624 (93.1) |
| Drinking frequency during the past 12 months (days) | 124.6 ± 112.8 | 82.6 ± 105.3 |
| Percentage who drink daily (%) | 8 | 6 |
| Percentage who drink at least weekly (%) | 69 | 46 |
| Number of drinks typically consumed on days when they drank alcohol in the past 12 months | 2.9 ± 2.3 | 1.9 ± 1.5 |
| Percentage of men who consume four or more drinks and women who consume three or more drinks (%) | 28 | 24 |
| Quantity of drinks consumed in previous 12 months (Freq × TypDy) | 406.9 ± 537.8 | 180.7 ± 277.0 |
| Most drinks consumed in a single day in the previous 12 months | 7.6 ± 6.6 | 3.7 ± 3.2 |
| Most drinks ever consumed in a single day | 16.7 ± 11.5 | 7.1 ± 6.0 |
| Has ever flushed or blushed while consuming 1 or 2 drinks of alcohol, n (%) | 241 (16.6) | 887 (31.6) |
| Percentage who always experience flushing (%) | 2.3 | 8.1 |
| Has ever had a negative reaction (flushing, nausea, headaches, palpitations, hives, extreme sleepiness) while consuming one or two drinks of alcohol, n (%) | 575 (39.6) | 1452 (52.0) |
| Percentage who always experience negative reaction (%) | 7.9 | 15.5 |
| DSM-IIIR AD score (number of criteria met) | 2.15 ± 2.12 | 0.80 ± 1.47 |
| Percentage with no symptoms (%) | 34 | 69 |
| Diagnosed with DSM-IIIR AD, n (%) | 375 (25.4) | 172 (6.1) |
Lifetime abstainers are excluded.
Marker information including chromosomal position and minor allele frequencies for the 50 SNPs genotyped in ALDH2 and the seven ADH genes are given in Table 2. Call rates of ≥98% were achieved for all SNPs except rs441 (97.9%), rs3098808 (97.7%), rs737280 (97.3%) and rs968529 (95.4%). The average genotyping rate among all SNPs was 98.8% with samples successfully genotyped on an average for 49 out of 50 SNPs. Two markers (rs1154409 in ADH5 and rs671 in ALDH2) were non-polymorphic in our sample and two SNPs had a minor allele frequency (MAF) <0.05. Discordant genotypes between MZ twins were identified using PEDSTATS and made up 0.01% of the MZ data. Non-polymorphic markers and SNPs that showed significant deviations from Hardy–Weinberg equilibrium (HWE) at a P < 0.001 level in the overall sample (rs284784 in ADH7 and rs3098808 in ADH1C) were excluded from further analysis. The physical locations of and linkage disequilibrium (LD) between the remaining 8 ALDH2 SNPs and 38 ADH SNPs are presented schematically in Figure 1. Substantial LD (|D′| >0.8) was observed between eight ALDH2 SNPs which are consistent with LD data (on a smaller number of people) from the HapMap database for CEPH families of European origin. A gene-centric pattern of LD was observed among the ADH gene markers in which high LD was observed within each gene and lower LD between genes.
Table 2.
ADH and ALDH2 gene marker information
| Gene | Chr | SNP | Chromosome locationa | Gene locationb | Alleles Minorc | Major | MAF | pHWEd | Call rate (%) |
|---|---|---|---|---|---|---|---|---|---|
| METAP1 | 4 | rs1230210 | 100 186 714 | Intron 5 | T | C | 0.273 | 0.39 | 99.2 |
| ADH5 | 4 | rs1230165 | 100 205 396 | 3′-UTR | C | T | 0.181 | 0.79 | 99.1 |
| ADH5 | 4 | rs896992 | 100 221 395 | Intron 4 | G | A | 0.329 | 1.00 | 99.1 |
| ADH5 | 4 | rs1154409 | 100 226 893 | Intron 1 | T | G | 0e | 1.00 | 99.3 |
| 4 | rs2602877 | 100 258 870 | Intergenic | A | T | 0.271 | 0.59 | 99.1 | |
| 4 | rs2602878 | 100 258 976 | Intergenic | A | C | 0.270 | 0.57 | 99.1 | |
| ADH4 | 4 | rs1042364 | 100 264 597 | 3′-UTR | A | G | 0.277 | 1.00 | 99.1 |
| ADH4 | 4 | rs1126672 | 100 266 835 | Exon 8 | T | C | 0.278 | 0.65 | 99.4 |
| ADH4 | 4 | rs1800759 | 100 284 532 | Promoter | A | C | 0.385 | 0.18 | 98.6 |
| ADH4 | 4 | rs4140388 | 100 284 757 | Upstream | C | G | 0.459 | 0.69 | 98.4 |
| 4 | rs3762894 | 100 285 107 | Intergenic | G | A | 0.160 | 0.98 | 98.7 | |
| 4 | rs1984364 | 100 289 806 | Intergenic | G | T | 0.275 | 0.95 | 99.2 | |
| 4 | rs1540053 | 100 301 177 | Intergenic | C | T | 0.235 | 0.95 | 98.6 | |
| ADH6 | 4 | rs2000864 | 100 342 799 | Downstream | T | G | 0.487 | 0.43 | 98.9 |
| ADH6 | 4 | rs4147545 | 100 347 776 | Intron 6 | G | A | 0.320 | 0.24 | 99.4 |
| 4 | rs1230025 | 100 405 399 | Intergenic | T | A | 0.230 | 0.51 | 98.0 | |
| ADH1A | 4 | rs3819197 | 100 419 532 | Intron 8 | A | G | 0.231 | 0.05 | 98.6 |
| ADH1A | 4 | rs2276332 | 100 422 470 | Intron 6 | G | T | 0.086 | 0.68 | 98.9 |
| ADH1B | 4 | rs1042026 | 100 447 489 | 3′-UTR | G | A | 0.279 | 0.92 | 98.6 |
| ADH1B | 4 | rs17033 | 100 447 968 | 3′-UTR | G | A | 0.086 | 0.52 | 99.5 |
| ADH1B | 4 | rs6850217 | 100 452 643 | Intron 6 | A | G | 0.390 | 0.72 | 99.2 |
| ADH1B | 4 | rs2018417 | 100 454 163 | Exon 6 | T | G | 0.040 | 1.00 | 98.9 |
| ADH1B | 4 | rs4147536 | 100 458 135 | Intron 3 | T | G | 0.213 | 0.89 | 99.0 |
| ADH1B | 4 | rs1229984 | 100 458 342 | Exon 3 | A | G | 0.035 | 0.11 | 99.0 |
| ADH1B | 4 | rs1159918 | 100 462 032 | Upstream | T | G | 0.361 | 0.89 | 98.9 |
| 4 | rs2866152 | 100 470 090 | Intergenic | G | C | 0.244 | 0.98 | 99.5 | |
| ADH1C | 4 | rs1789895 | 100 475 752 | Downstream | C | G | 0.323 | 0.60 | 98.9 |
| ADH1C | 4 | rs3098808 | 100 475 847 | Downstream | C | T | 0.114 | <0.0001 | 97.7 |
| ADH1C | 4 | rs1693482 | 100 482 988 | Exon 6 | A | G | 0.410 | 0.84 | 98.7 |
| ADH1C | 4 | rs283416 | 100 490 379 | Intron 1 | T | C | 0.065 | 1.00 | 98.6 |
| 4 | rs1583973 | 100 506 906 | Intergenic | T | C | 0.099 | 0.95 | 98.8 | |
| 4 | rs1826906 | 100 520 071 | Intergenic | C | T | 0.326 | 0.45 | 99.1 | |
| 4 | rs2032350 | 100 533 085 | Intergenic | T | C | 0.198 | 0.08 | 98.5 | |
| 4 | rs1348276 | 100 544 653 | Intergenic | G | T | 0.410 | 0.48 | 98.5 | |
| 4 | rs994772 | 100 546 687 | Intergenic | A | G | 0.106 | 0.87 | 98.8 | |
| ADH7 | 4 | rs284784 | 100 554 897 | Intron 8 | T | G | 0.270 | <0.0001 | 98.4 |
| ADH7 | 4 | rs1154454 | 100 557 365 | Intron 7 | C | T | 0.163 | 1.00 | 99.3 |
| ADH7 | 4 | rs1154458 | 100 559 545 | Intron 6 | G | C | 0.408 | 0.23 | 98.8 |
| ADH7 | 4 | rs971074 | 100 560 884 | Exon 6 | A | G | 0.113 | 0.97 | 99.4 |
| ADH7 | 4 | rs1154461 | 100 561 925 | Intron 5 | C | G | 0.351 | 0.45 | 98.8 |
| 4 | rs1583971 | 100 626 045 | Intergenic | A | T | 0.111 | 0.08 | 99.7 | |
| ALDH2 | 12 | rs737280 | 110 657 696 | Upstream | C | T | 0.272 | 0.04 | 97.3 |
| ALDH2 | 12 | rs886205 | 110 667 147 | 5′-UTR | G | A | 0.170 | 0.20 | 99.0 |
| ALDH2 | 12 | rs2238151 | 110 674 553 | Intron 1 | C | T | 0.322 | 0.07 | 99.0 |
| ALDH2 | 12 | rs4648328 | 110 685 508 | Intron 3 | T | C | 0.165 | 0.10 | 99.1 |
| ALDH2 | 12 | rs441 | 110 691 569 | Intron 6 | C | C | 0.184 | 0.68 | 97.9 |
| ALDH2 | 12 | rs968529 | 110 697 088 | Intron 9 | T | C | 0.069 | 0.19 | 95.4 |
| ALDH2 | 12 | rs4646778 | 110 698 503 | Intron 9 | A | C | 0.165 | 0.17 | 99.1 |
| ALDH2 | 12 | rs671 | 110 704 486 | Exon 12 | A | G | 0e | 1.00 | 99.3 |
| ALDH2 | 12 | rs16941667 | 110 707 133 | Intron 12 | T | C | 0.079 | 0.23 | 99.4 |
Chr, chromosome; HWE, Hardy–Weinberg equilibrium; MAF, minor allele frequency; SNP, single-nucleotide polymorphism; UTR, untranslated region.
aPosition in nucleotides as estimated in dbSNP (Build 127).
bPosition within or near gene.
cThe allele with the lowest frequency.
dpHWE = P-value for the Hardy–Weinberg equilibrium test run in PEDSTATS (71).
ers671 and rs1154409 are non-polymorphic.
Figure 1.
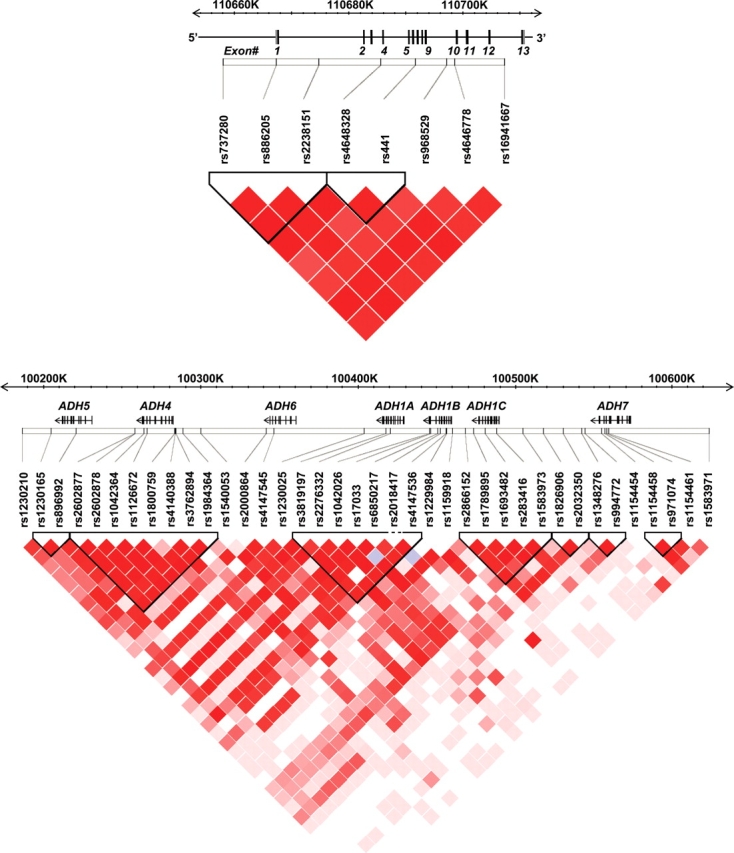
LD between the SNPs genotyped in ALDH2 and seven ADH genes: (A) the gene structure of ALDH2 is shown with exons numbered (1–13) and relative exon size denoted by the width of the vertical bars. Pairwise marker–marker LD (shown below the gene structure) was generated using Haploview 4.0 (75). Regions of low to high LD (D′) are represented by white to red shading, respectively; (B) gene location, structure and LD (D′) between the SNPs genotyped in the ADH cluster.
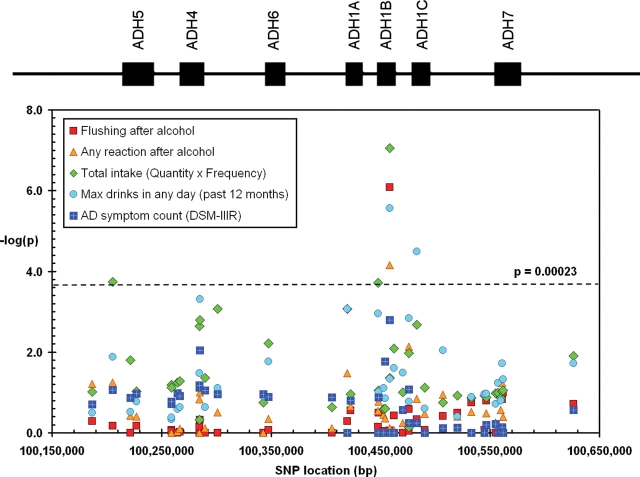
Results of the association testing are summarized in Figure 2 for ADH, and given in full in Table 3.
Figure 2.
Total association of SNPs across the ADH gene cluster with quantitative AD variables, alcohol consumption variables and self-reported alcohol reactions. The location of the seven ADH genes is shown at the top of the figure. Results are plotted as –log10(P-value) against the physical location of each SNP on chromosome 4. See Table 1 for further description of the variables.
Table 3.
Allelic associations between SNPs in ADH and ALDH2 genes, and phenotypes associated with alcohol reactions, alcohol use and AD
| Genes and SNPs |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene(s) | Phenotypes (P-values) Usual quantity (no. of drinks) | Max drinks in any day past 12 months | Max drinks on any day lifetime | Frequency of alcohol use | Total intake (Quantity × frequency) | Flushing after alcohol | Any reaction after alcohol | AD symptom score (DSM-IIIR) |
| ADH (chromosome 4) | |||||||||
| rs1230210 | METAP1 | 0.24 | 0.32 | 0.79 | 0.26 | 0.097 | 0.51 | 0.061 | 0.2 |
| rs1230165 | ADH5 | 0.061 | 0.0133 | 0.047 | 0.00035 | 0.00018 | 0.68 | 0.056 | 0.086 |
| rs896992 | ADH5 | 0.53 | 0.3 | 1 | 0.03 | 0.016 | 1 | 0.37 | 0.137 |
| rs2602877 | ADH4 | 1 | 0.46 | 0.64 | 0.071 | 0.064 | 0.85 | 1 | 0.19 |
| rs2602878 | ADH4 | 0.75 | 0.42 | 0.72 | 0.075 | 0.076 | 0.89 | 1 | 0.17 |
| rs1042364 | ADH4 | 0.57 | 0.26 | 0.44 | 0.098 | 0.056 | 1 | 0.93 | 0.105 |
| rs1126672 | ADH4 | 0.5 | 0.23 | 0.59 | 0.08 | 0.052 | 0.94 | 0.79 | 0.124 |
| rs1800759 | ADH4 | 0.15 | 0.033 | 0.0075 | 0.0055 | 0.0023 | 1 | 0.148 | 0.075 |
| rs4140388 | ADH4 | 0.62 | 0.072 | 1 | 0.53 | 0.48 | 0.71 | 1 | 0.067 |
| rs3762894 | ADH4 | 0.00078 | 0.00048 | 0.0139 | 0.02 | 0.0016 | 0.47 | 0.1 | 0.009 |
| rs1984364 | ADH4 | 0.46 | 0.23 | 0.48 | 0.069 | 0.044 | 1 | 0.76 | 0.089 |
| rs1540053 | NA | 0.126 | 0.077 | 0.61 | 0.0049 | 0.00084 | 1 | 0.31 | 0.109 |
| rs2000864 | ADH6 | 0.19 | 0.125 | 1 | 0.33 | 0.18 | 1 | 1 | 0.11 |
| rs4147545 | ADH6 | 0.004 | 0.017 | 0.49 | 0.066 | 0.006 | 0.86 | 0.45 | 0.129 |
| rs1230025 | NA | 0.21 | 0.133 | 0.72 | 0.35 | 0.23 | 1 | 0.76 | 0.132 |
| rs3819197 | ADH1A | 0.00033 | 0.00085 | 0.076 | 0.0069 | 0.00086 | 0.52 | 0.033 | 1 |
| rs2276332 | ADH1A | 0.38 | 0.16 | 0.01 | 0.075 | 0.11 | 0.28 | 0.22 | 0.16 |
| rs1042026 | ADH1B | 0.00067 | 0.00111 | 0.046 | 0.0023 | 0.00019 | 0.7 | 0.29 | 1 |
| rs17033 | ADH1B | 0.33 | 0.096 | 0.0041 | 0.079 | 0.088 | 0.32 | 0.17 | 0.132 |
| rs6850217 | ADH1B | 0.17 | 0.078 | 0.24 | 1 | 0.26 | 0.91 | 0.35 | 1 |
| rs2018417 | ADH1B | 0.43 | 0.139 | 0.031 | 0.28 | 0.25 | 1 | 0.44 | 0.017 |
| rs4147536 | ADH1B | 0.066 | 0.046 | 0.46 | 0.133 | 0.043 | 0.88 | 0.76 | 1 |
| rs1229984 | ADH1B | 0.0005 | 2.7E−06 | 0.022 | 2.7E−06 | 8.9E−08 | 8.2E−07 | 0.000069 | 0.0016 |
| rs1159918 | ADH1B | 0.084 | 0.025 | 0.26 | 0.025 | 0.0081 | 0.37 | 0.86 | 1 |
| rs2866152 | ADH1C | 0.18 | 0.032 | 0.16 | 0.19 | 0.1 | 1 | 0.57 | 0.27 |
| rs1789895 | ADH1C | 0.0058 | 0.00143 | 0.0132 | 0.114 | 0.0106 | 0.26 | 0.0074 | 0.57 |
| rs1693482 | ADH1C | 0.00045 | 0.000032 | 0.0062 | 0.098 | 0.0021 | 0.46 | 0.142 | 0.57 |
| rs283416 | ADH1C | 0.63 | 0.25 | 0.28 | 0.11 | 0.075 | 0.85 | 0.34 | 1 |
| rs1583973 | NA | 0.057 | 0.0091 | 0.32 | 0.31 | 0.18 | 0.38 | 0.113 | 0.77 |
| rs1826906 | NA | 1 | 0.39 | 0.28 | 0.097 | 0.119 | 0.32 | 0.39 | 0.74 |
| rs2032350 | NA | 0.16 | 0.128 | 0.39 | 0.25 | 0.136 | 0.18 | 0.3 | 1 |
| rs1348276 | ADH7 | 0.28 | 0.109 | 0.34 | 0.18 | 0.119 | 0.86 | 1 | 1 |
| rs994772 | ADH7 | 0.7 | 0.106 | 0.46 | 0.27 | 0.133 | 0.16 | 0.33 | 0.64 |
| rs1154454 | ADH7 | 1 | 0.058 | 1 | 0.24 | 0.102 | 0.91 | 0.63 | 1 |
| rs1154458 | ADH7 | 0.39 | 0.16 | 0.34 | 0.6 | 0.137 | 1 | 0.27 | 0.96 |
| rs971074 | ADH7 | 0.077 | 0.019 | 0.22 | 0.36 | 0.093 | 0.149 | 0.058 | 0.73 |
| rs1154461 | ADH7 | 0.26 | 0.047 | 0.18 | 0.097 | 0.09 | 0.101 | 0.4 | 1 |
| rs1583971 | NA | 0.09 | 0.019 | 0.22 | 0.043 | 0.0124 | 0.19 | 0.25 | 0.27 |
| ALDH2 (chromosome 12) | |||||||||
| rs737280 | NA | 0.23 | 0.112 | 0.021 | 0.15 | 0.21 | 0.81 | 0.7 | 0.0033 |
| rs886205 | ALDH2 | 0.49 | 0.8 | 0.45 | 0.11 | 0.47 | 0.35 | 0.85 | 0.22 |
| rs2238151 | ALDH2 | 0.15 | 0.05 | 0.0061 | 0.2 | 0.096 | 0.8 | 0.42 | 0.00095 |
| rs4648328 | ALDH2 | 0.45 | 1 | 0.38 | 0.23 | 1 | 0.54 | 1 | 0.138 |
| rs441 | ALDH2 | 0.44 | 0.38 | 0.041 | 0.47 | 0.61 | 0.9 | 0.39 | 0.079 |
| rs968529 | ALDH2 | 0.4 | 1 | 0.5 | 0.126 | 0.23 | 0.9 | 1 | 0.19 |
| rs4646778 | ALDH2 | 0.48 | 1 | 0.44 | 0.17 | 0.62 | 0.43 | 0.95 | 0.17 |
| rs16941667 | ALDH2 | 0.28 | 0.69 | 1 | 0.18 | 0.66 | 0.84 | 1 | 1 |
NA, SNP in intragenic region.
Alcohol reactions
None of the tested SNPs in ALDH2 were associated with flushing or with the more broadly defined reactions to alcohol. As expected, we found no occurrences of ALDH2*2 among our subjects.
Self-reported flushing after consumption of small amounts of alcohol was significantly associated (P = 8.2 × 10−7) with the A-allele of rs1229984 (ADH1B Arg48His). This allele also conferred a higher likelihood of experiencing any negative reaction (P = 6.9 × 10−5). The direction of effect is consistent with published findings that the ADH1B His48 (A-allele) variant is protective against AD. The prevalence of flushing or other reactions by rs122984 genotype is shown in Table 4.
Table 4.
Effects of rs1229984 (ADH1B Arg48His) genotype on proportions of participants reporting flushing or other alcohol reactions, alcohol consumption (number of drinks in the past year), symptom score for DSM-IIIR AD and DSM-IIIR AD
| Male |
Female |
|||||
|---|---|---|---|---|---|---|
| GG (RR) | AG (RH) | AA (HH) | GG (RR) | AG (RH) | AA (HH) | |
| Genotypes, n (%) | 1381 (93) | 107 (7) | 2 (0.1) | 2699 (94) | 157 (6) | 9 (0.3) |
| Flushing, n (%) | ||||||
| Never | 1123 (84) | 83 (78) | 1 (50) | 1782 (69) | 81 (53) | 2 (25) |
| Sometimes | 188 (14) | 19 (18) | 1 (50) | 600 (23) | 53 (35) | 3 (38) |
| Always | 28 (2) | 5 (5) | 0 (0) | 206 (8) | 18 (12) | 3 (38) |
| Any negative reaction, n (%) | ||||||
| Never | 815 (61) | 60 (56) | 0 (0) | 1251 (48) | 52 (34) | 2 (25) |
| Sometimes | 423 (32) | 38 (36) | 1 (50) | 944 (37) | 67 (44) | 3 (38) |
| Always | 103 (8) | 9 (8) | 1 (50) | 396 (15) | 35 (23) | 3 (38) |
| Yearly drinks (median) | 208 | 150 | 12 | 60 | 30 | 3 |
| DSM-IIIR symptom count, n (% of column total) | ||||||
| 0 | 446 (33) | 48 (45) | 1 (50) | 1791 (69) | 119 (77) | 8 (100) |
| 1 | 144 (11) | 10 (9) | 0 (0) | 170 (6) | 10 (6) | 0 (0) |
| 2 | 205 (15) | 12 (11) | 1 (50) | 307 (12) | 11 (7) | 0 (0) |
| 3 | 202 (15) | 19 (18) | 0 (0) | 183 (7) | 9 (6) | 0 (0) |
| 4 | 144 (11) | 7 (7) | 0 (0) | 89 (3) | 3 (2) | 0 (0) |
| 5 | 106 (8) | 4 (4) | 0 (0) | 38 (1) | 1 (1) | 0 (0) |
| >5 | 99 (7) | 7 (7) | 0 (0) | 36 (1) | 2 (1) | 0 (0) |
| DSM-IIIR AD (%) | ||||||
| Negative | 989 (74) | 89 (83) | 2 (100) | 2442 (94) | 150 (97) | 8 (100) |
| Positive | 352 (26) | 18 (17) | 0 (0) | 166 (6) | 5 (3) | 0 (0) |
Genotypes are shown as the base on the reverse strand of DNA (G or A) and as the corresponding amino acid (R = Arginine, H = Histidine). Note that some phenotypic information was missing so that the number of subjects cross-tabulated by phenotype and genotype may not sum to the total of subjects with valid genotypes.
Alcohol consumption
No significant linkage findings were observed across the polymorphisms in the ADH gene cluster or ALDH2. Total association results for quantitative measures of alcohol consumption are presented in Figure 2 and Table 3.
For ALDH2, we only detected nominal association (P = 0.006) with maximum drinks, where individuals carrying two T-alleles of rs2238151 (located in intron 1) reported consuming a higher maximum number of drinks in a single day (5.24 drinks) than heterozygous T/C (4.98) or homozygous C (4.49) individuals. However, this signal was not significant after taking account of multiple testing (with a threshold level of significance set at 2.3 × 10−4, see Materials and Methods).
For ADH, the strongest evidence of association was found for the non-synonymous SNP rs1229984 located in exon 3 of ADH1B (ADH1B Arg48His). Within the past 12 months, individuals carrying the rs1229984 G-allele (the more common arginine 48 variant, Arg48) reported consuming higher quantities of alcohol on a typical day when drinking (P = 0.0005), drank alcohol on more occasions (P = 2.7 × 10−6), reported a higher maximum number of alcoholic beverages consumed in a single day in the past year (P = 2.7 × 10−6) and had a higher overall consumption of alcohol (P = 8.9 × 10−8) than individuals with the A-allele (His48). The frequency distributions of overall alcohol intake by rs1229984 genotype in men and women are shown in Figure 3, and the medians are shown in Table 4.
Figure 3.
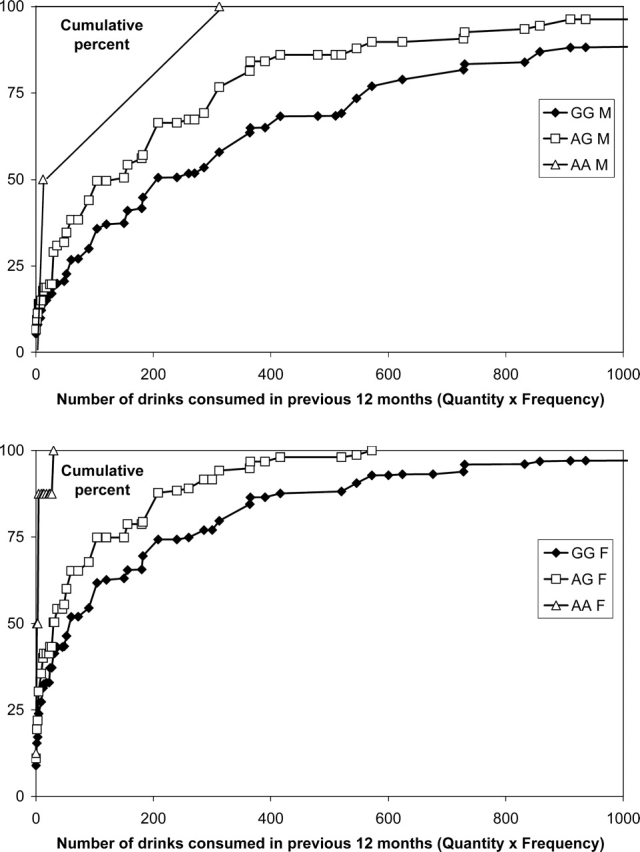
Cumulative frequency distribution plots for number of drinks in the previous 12 months (estimated from self-reported frequency and quantity) by rs1229984 (ADH1B Arg48His) genotypes, in (A) men and (B) women (GG = 48Arg homozygotes, AG = heterozygotes, AA = 48His homozygotes).
Study participants with 0, 1 and 2 A-alleles for rs1229984 reported consuming, on average, a maximum of 5.1, 4.1 and 1.9 drinks in a single day in the previous year, respectively, and this trait has previously been shown to be highly correlated with alcohol use disorders (55). Similarly, the number of DSM-IIIR AD symptoms met were 1.25, 1.08 and 0.18 symptoms for 0, 1 and 2 A-alleles, respectively.
Among the remaining ADH markers analyzed, rs1693482 (the non-synonymous Arg272Gln polymorphism in ADH1C) was the only SNP significantly associated with alcohol consumption after correcting for multiple testing (P =3.2 × 10−5, with the maximum number of drinks consumed in a single day in the past year). SNPs in ADH1A (rs3819197), ADH1B (rs1042026), ADH4 (rs3762894) and ADH5 (rs1230165) were associated to a smaller degree (P < 0.001) with one or more consumption variables tested (Table 3). To determine whether LD with rs1229984 accounted for the association signals across this region, we analyzed multiple SNPs jointly for four variables (total quantity, frequency, maximum number of drinks in past 12 months and flushing) by regression analysis with rs1229984 genotype included. There was evidence for residual association at rs1042026 (which is in moderate LD with rs1229984; D′ = 0.67; r2 = 0.01) for the total quantity variable (P = 4.7 × 10−5) with the A-allele conferring higher alcohol intake over the previous 12 months, indicating there may be two independent signals in ADH1B. Alternatively, both SNPs may be in incomplete LD with a single causal variant. The significant association observed between rs1693482 (ADH1C Arg272Gln) and maximum number of drinks in past 12 months was mostly explained by rs1229984 (D′ = 0.84; r2 = 0.02), but with nominally significant residual association detected (P = 0.0007) after the effect of rs1229984 was accounted for. Smaller residual associations were also observed between rs1230165 in ADH5 and frequency (P = 0.001) and rs37262894 in ADH4 (P = 0.0004) and maximum number of drinks in the past 12 months. For the flushing trait, the large effect at rs1229984 explained all of the other signals in the ADH region.
Analysis of the subset of the data known to have European ancestry (defined as having all four grandparents with known European ancestry) gave results which were consistent with those seen in the full sample—restricting analysis to this subsample halved the sample size but the effect size estimates were similar (data not shown). Similarly, family based ‘within’ association tests yielded results consistent with those seen in the main analysis (Supplementary Material, Table S)—for these ‘within’ analyses, the effective sample size was substantially smaller because only families with two DZ individuals contribute (since parents were not genotyped, singleton and MZ families contribute no information to the ‘within’ test). These analyses rejected a role for the population stratification in the effects observed. A small proportion of subjects had reported their religion as ‘Jewish’ in a previous questionnaire, and in view of the reported higher prevalence of the rs1229984 48His allele in this group the allele frequency was estimated. Out of 33 such subjects, 4 were 48His homozygotes and 16 were heterozygotes, giving a MAF of 0.36. However, further analysis of the alcohol-related phenotypes controlling for Jewish ancestry left the strength of the associations unchanged.
Alcohol dependence
Total association results for the quantitative measure of AD are presented in Figure 2 and Table 3. For ALDH2, the most significant findings (P < 0.001) in the total association analysis were detected with the ALDH2 SNP rs2238151 (intron 1) where T-allele conferred a higher DSM-IIIR score. The T-allele of rs737280 (ALDH2) also conferred higher DSM-IIIR AD scores, but the effect was less significant (P ≈ 0.003). No study-wide significant association was noted between the AD measures and the SNPs genotyped in the ADH gene cluster. However, rs1229984 did show nominal association with the DSM-IIIR score (P = 0.0016).
Subsequent binary analyses of DSM-IIIR AD diagnosis were run, both on the overall data and in DZ families to permit within-family tests of association to be performed. The purpose of these analyses was to be able to directly compare our findings with those of other groups who only analysed the dichotomous diagnoses of AD. The number of affected (case) individuals in the entire sample was 959, with a reduced number of cases (540) within DZ families only. No study-wide significant association was observed with AD diagnosis in the DZ family or MZ case–control analyses (smallest P-value 0.01 before correction for multiple testing).
DISCUSSION
The object of this study was to examine a connected series of phenotypes on a pathway to alcoholism, and to test for allelic associations with multiple polymorphisms across alcohol dehydrogenases (ADH1A, ADH1B, ADH1C, ADH4, ADH5, ADH6 and ADH7) and the high-affinity aldehyde dehydrogenase (ALDH2). Although there have been many genetic association studies on these genes, they have either concentrated on a single phenotype (usually AD), or on a narrow range of genetic markers, or both. There have been suggestions that the polymorphisms studied have not been causative, or that they account for only part of the effects located within these genes; there have been questions about the applicability of results across populations; and a greater integration of information about gene and enzyme variation, the subjective effects of alcohol, habits of alcohol use and symptoms of dependence have been needed. We have found that the effects of the much-studied ADH1B Arg48His polymorphism can be traced through these alcohol-related phenotypes, and that the most easily detected effects are on the phenotypes near the start of the proposed sequence of events. On the whole, our results confirm the prevailing theory, but with some additions and with some features which require further consideration.
ALDH2
None of the ALDH2 SNP-phenotype associations reached the study-wide level of significance of 2.3 × 10−4. However, it would be unwise to dismiss the possibility of a true effect of variation in ALDH2 in Europeans; precautions against false-positive results can lead to false negatives even with large sample size. A number of phenotypes related to AD, particularly the symptom score for dependence, were associated with rs2238151 at P ≈ 0.001 and with rs737280 at P < 0.01 (Table 3). In a previous study, on a smaller group who had participated in our Alcohol Challenge Twin Study, we found that these two SNPs were associated with significant differences in alcohol metabolism (22). We postulated that this could be due to a variant which decreased the activity of ALDH2, to a lesser extent than the Asian rs671 polymorphism, and predicted that people homozygous for C at rs2238151 and/or for G at rs737280 should have a lower prevalence of AD and a lower symptom score. That was not shown among the 376 people typed for ALDH2 SNPs in our previous study, but with the larger number of subjects typed in this study we note that the T allele at rs2238151 showed higher AD symptom scores, with P-values around 0.001 and in the predicted direction. Further studies or meta-analysis on ALDH2 variants in Europeans seem justified. However, neither of these two SNPs showed associations with self-reported flushing or with alcohol consumption so the mechanism of any effect on dependence is unclear.
ADH
Our main finding is that the variation at rs1229984, the well-known ADH1B Arg48His polymorphism, is associated with flushing or other reactions to alcohol as well as with alcohol consumption in Europeans. Self-reported unpleasant reactions to alcohol are known to occur in a substantial proportion of people, particularly women [see Table 1, and (56)], but our earlier attempts to test whether ADH1B variation affected these reactions (57) were inconclusive. With larger numbers in the present study, the ADH1B 48His allele, which has a frequency of around 5% in this group, is highly significantly associated with more frequent alcohol reactions and lower alcohol intake. The effect of this polymorphism on reported alcohol intake is summarized in Figure 3 and Table 4; for both men and women, the median alcohol intake was notably lower in heterozygotes than in 48Arg homozygotes, and in the small number of 48His homozygotes alcohol consumption was very low.
However, this polymorphism is not significantly associated, at least at our study-wide significance level, with AD or AD symptom count. Practically, all previous studies, in Asia and in Europe, have found that the 48His allele is protective against AD. For Europeans, the relative risk of AD for heterozygotes against 48Arg homozygotes has been estimated at 0.47 with 95% confidence intervals of 0.29 to 0.76 (36). Our estimated relative risk for DSM-IIIR AD in heterozygotes compared to 48Arg homozygotes, from the current data, is 0.57 for men and 0.49 for women. There is no evidence of heterogeneity and the pooled odds ratio estimate is 0.53 with 95% confidence intervals 0.32–0.88. The point to note is that our results are compatible with the expected size of the effect on AD risk. As noted above in relation to ALDH2 variation, failure to meet a stringent significance level does not exclude an effect; and the AD symptom score phenotype showed association with rs1229984 at the P < 0.002 level. On balance, we consider that rs1229984 does affect AD risk but the nature of our study (with subjects drawn from the general population), the low MAF for this polymorphism, and the stringent study-wide significance level arising from the examination of multiple SNPs and phenotypes, contribute to our inability to demonstrate this unequivocally.
A number of previous reports have considered whether variation at ADH1B Arg48His affects the alcohol flush reaction or alcohol reactions in general, both in Asians (38–43,58) and in Europeans. The results are very mixed; in Europeans a measure of ‘level of response’ to alcohol was affected by this polymorphism (59) but alcohol-induced flushing was not (60). In Asians also there are about equal numbers of positive and negative reports, and some suggestion of gene–gene interaction in that ADH1B genotype may only affect flushing in ALDH2 heterozygotes. However, our results on this point are strong, and we conclude that ADH1B Arg48His does affect the immediate response to alcohol consumption.
No other polymorphisms in either ADH or ALDH2 had significant or suggestive associations with alcohol reactions. However, many ADH SNPs showed associations with drinking behavior (Fig. 3 and Table 3), some because of LD with rs1229984. When the effect of rs1229984 was regressed out, there was an independent association between rs1042026 in ADH1B and the overall quantity phenotype (P = 4.7 × 10−5), with the A-allele conferring higher alcohol intake on average over the previous 12 months. Smaller associations between rs1230165 in ADH5 and frequency of alcohol use, and between both rs37262894 (in ADH4) and rs1693482 (ADH1C Arg272Gln) and the maximum number of drinks consumed in 1 day in the previous 12 months were also observed. No SNPs producing stronger effects than rs1229984, or explaining the effects at rs1229984 through LD with a distinct causative variant, were found. However, there were indications of independent effects in ADH1B, ADH1C, ADH4 and ADH5 which did not reach our required significance level, but which still need to be confirmed or refuted.
Comparable data are available from three other studies with SNP coverage of the ADH region and one of ADH4 only, which focused on associations with AD. Using a conventional test for allelic association, Luo et al. (20) found significant results for rs1229984 (Arg48His) in European-Americans and for rs2066702 (Arg369Cys) in African-Americans, both in ADH1B. Using an analysis based on deviation from HWE, there were indications that variation in ADH1A, ADH1B, ADH4, ADH5 and ADH7 affected dependence risk (20,54). Significant associations between AD in European-Americans and SNPs in ADH4, ADH1A and ADH1B were found in another ADH-cluster-wide study by the Collaborative Study on the Genetics of Alcoholism (19). While no association was detected between rs1229984 and AD in European-Americans, in African-Americans rs2066702 was significantly (P < 0.05) associated with AD. The most significant associations were in the ADH1A-ADH1B region or in ADH4, depending on the AD criteria used. It is interesting to note that out of 110 SNPs tested on 2139 people, three were significant at the 0.01 level and a further 18 had P-values between 0.05 and 0.01; we can infer that the effects on AD risk are either small or they are associated (as we found) with polymorphisms with low minor allele frequencies. A further study with recruitment in Ireland (21) found associations at P < 0.01 between AD and SNPs in ADH1B and ADH5, and at P < 0.05 in ADH1A, ADH1B, ADH1B and ADH7. Significant association between AD and ADH4 SNPs was also reported in Brazilian subjects (61). Overall, the genes coding for Class I ADHs with high affinity for ethanol are the ones which most frequently show significant effects on dependence risk, but ADH4 has also been implicated in several studies.
From our results and the published literature, we can now propose that rs1229984, or ADH1B Arg48His, affects the subjective experiences associated with alcohol use, specifically through flushing or similar reactions, even in Europeans homozygous for the active form of ALDH2. This affects alcohol use, which in turn affects AD risk. Other polymorphisms in ADH genes may also affect AD through similar mechanisms, but no strong evidence of this is yet available. The main anomaly in this narrative is that rs1229984 probably does not affect the rate of alcohol metabolism, at least so far as it can be measured from changes in blood and breath alcohol concentrations (51). It still remains possible that this variant affects the steady-state concentration of acetaldehyde within the hepatocytes, which is where acetaldehyde is produced, and that this affects the release of vasoactive compounds into the circulation and causes alcohol-induced flushing. Measurement of acetaldehyde concentrations in the liver of humans seems beyond our reach, but measurements of histamine or other vasoactive molecules in the circulation might allow testing of this concept; and transgenic mice expressing rs1229984 might allow more invasive studies of the effects of this polymorphism on alcohol and acetaldehyde metabolism.
MATERIALS AND METHODS
Participants
Participants were recruited for a 1992–1995 telephone interview based twin study conducted at the Queensland Institute of Medical Research (QIMR) (7,62). This interview was based upon an Australian modified version (SSAGA-OZ) of the Semi-Structured Assessment for the Genetics of Alcoholism instrument designed for genetic studies of alcoholism. The SSAGA is a psychiatric interview that retrospectively assesses physical, psychological and social manifestations of AD along with several other psychiatric disorders and has undergone both reliability and validity testing (63,64). It included questions on alcohol-related flushing or other unpleasant reactions to alcohol, quantity and frequency of alcohol use over the last 12 months, and about the maximum number of drinks in a single day in the last 12 months and ever. A total of 4597 subjects (34.6% males) from 2618 families, comprising 814 (583 female and 231 male) MZ pairs, 1177 (482 female, 198 male and 497 opposite sex) DZ pairs and 627 twins (38.8% male) whose co-twin did not participate, were included in genetic analysis. The participants were predominantly of Northern European ancestry, from information they provided on the place of birth and ethnicity of their grandparents. Fifty-one percent of the sample had ancestry information on all four grandparents. Of these, 88% had all European grandparents (with 89% of these Northern European, N = 1873 twins), a further 1% indicated ‘adopted’, and 8% indicated ‘Australian’ ancestry. Most of the ‘Australian’ ancestry individuals are thought to have fully European ancestry (all grandparents born in Australia but with European ancestry), whereas others have one or more aboriginal grandparents. The final 3% had one or more Asian or African grandparent. They were aged 26–89 years (mean age was 43.8 ± 11.5 years) at the time of testing. Subjects gave written informed consent and provided blood samples from which DNA was isolated using standard protocols. Genetic studies were approved by the QIMR Human Research Ethics committee. Zygosity of same-sex twin pairs was assessed using a combination of self-report only (32% of same-sex pairs), three blood groups (ABO, MNS and Rh), data from genome-wide microsatellite markers and a set of nine polymorphic DNA microsatellite markers (AmpF1STR Profiler Plus Amplification Kit, Applied Biosystems, Foster City, CA, USA). The zygosity assignment from the nine-marker set is highly accurate [probability of correct assignment >99.99% (65)] and we estimate that the overall accuracy of zygosity assignment in this cohort is >99%.
Phenotypes
Assessment of current alcohol use, reactions to alcohol and lifetime history of AD were from the adapted SSAGA interview data. A small number of participants (2.3% of men and 3.1% of women) reported they had never had even one alcoholic drink, and their alcohol-related phenotypes were set to missing. Participants who reported never drinking more than three drinks in a single day were allowed to skip detailed questions from the SSAGA interview on drinking behavior. Reactions to alcohol use were self-reported (62). Alcohol consumption measures were frequency and quantity of alcohol consumption, maximum number of alcoholic drinks consumed in a single day ever and within the past 12 months. The total number of drinks taken in the past year was estimated from the reported quantity and frequency categories. AD was diagnosed by a computer algorithm based on the criteria of the Third Revised (DSM-IIIR) Diagnostic and Statistical Manual of the American Psychiatric Association (66). In addition to the dichotomous definition of AD, a quantitative measure based on reported AD symptoms was calculated (67).
Genotyping
Eleven SNPs flanking or within ALDH2 locus were selected on the basis of (i) previous work completed by our group (68) in which six SNPs within or flanking ALDH2 were genotyped in a partially overlapping Australian twin sample that completed an alcohol challenge test (69); and (ii) data available at the time from the International HapMap Project public database (http://www.hapmap.org/). Two SNPs failed during the assay design or provided unreliable genotype data and were excluded. rs671 was monomorphic. The locations of the eight remaining SNPs typed in the study are shown in Figure 1 and SNP information, including the observed MAF, is given in Table 2.
LD data from another study in which 104 SNPs across the ADH gene cluster were genotyped (Birley et al., submitted for publication), indicated that a reduced set of 43 haplotype-tagging SNPs would provide appropriate coverage (r2≥ 0.80) of the gene cluster. Additional tagging SNPs plus SNPs selected a priori based on existing research, specifically ADH1B Arg48His (rs1229984) and ADH1C Ile350Val (rs698), were typed giving a total of 51 SNPs. Ten SNPs provided unreliable genotype data and were excluded. One of these was rs698 (ADH1C Ile350Val) but it is in near-complete LD (D′ = 0.99 (70)) with rs1693482 (ADH1C Arg272Gln). The locations of the 41 remaining SNPs typed are shown in Figure 1 and SNP information is described in Table 2.
Assays were designed using the Sequenom MassARRAY Assay Design (version 3.0) software (Sequenom Inc., San Diego, CA, USA). Genotyping was carried out in standard 384-well plates with 12.5 ng genomic DNA used per sample. We used a modified Sequenom protocol where half reaction volumes were used in each of the PCR, SAP and iPLEX stages giving a total reaction volume of 5.5 µl. The iPLEX reaction products were desalted by diluting samples with 18 µl of water and 3 µl SpectroCLEAN resin (Sequenom) and then were spotted on a SpectroChip (Sequenom), processed and analyzed on a Compact MALDI-TOF Mass Spectrometer by MassARRAY Workstation software (version 3.3) (Sequenom). Allele calls for each 384-well plate were reviewed using the cluster tool in the SpectroTyper software (Sequenom) to evaluate assay quality. Genotype error checking, sample identity and zygosity assessment and HWE analyses were completed in PEDSTATS (71).
Statistical analyses
Quantitative trait analysis was performed in Merlin (72) and QTDT (73,74), with eight traits examined. The alcohol-related quantitative traits were first transformed to normality using a piecewise normal transformation and were corrected for sex and age effects by fitting covariates in the regression model. MZ twin status was included in Merlin and QTDT analyses by adding zygosity status to the data file. Tests of total association modeling multipoint linkage were performed in Merlin; non-independent transmissions were corrected for by modeling multipoint linkage in a maximum likelihood framework. SNPs in high LD were clustered together for linkage modeling such that clusters included pairs of SNPs for which pairwise r2 exceeded 0.05 (all intervening markers were included in the cluster). Secondary analyses robust to population stratification were conducted using the orthogonal ‘within’ test in QTDT. Since parents were not genotyped, this analysis only included families with two DZ offspring. To account for the fact that transmissions were not independent in the DZ families due to the presence of linkage in DZ pairs, a permutation procedure (10 000 permutations) was used to correct the P-values for the orthogonal test. The permutation procedure was also used to estimate the correlation within the set of phenotypes and within the set of genotypes (Lind et al., in preparation).
Binary trait analysis was performed for comparison with the above quantitative trait analysis and also for comparison with the work of other groups. Association of DSM-IIIR AD was examined using the UNPHASED statistical package in two stages. First, for DZ families, a pedigree disequilibrium (PDT) style test was used, in the PDTPHASE module, to test for over-transmission of allele to offspring (a ‘within’ family test that should be robust to any population stratification effects). Second, for MZ families, pairs with discordant phenotypes were set to phenotype unknown while pairs with concordant phenotypes were not changed. Subsequently, one MZ twin per family was included for each trait analyzed in COCAPHASE. Since no parents were genotyped, each MZ twin was treated as unrelated and a case–control test was implemented.
Pair-wise marker–marker LD was assessed using the D′ and r2 statistic in Haploview 3.31 (75). Many of the traits studied were correlated and there was substantial LD across ALDH2 and the ADH gene cluster (see Fig. 1A and B). As a result, the effective number of statistical tests done was less than the actual number of tests. Using permutation to take into account the correlated phenotypes and correlated genotypes, a P-value less than 2.3 × 10−4 is required for study wide significance.
FUNDING
This research was funded by the National Institute on Alcohol Abuse and Alcoholism (NIAAA): sample ascertainment, phenotyping and blood collection by grants AA07535, AA07728, AA11998, AA13320 and AA13321, genotyping by grants (AA013326, AA013326 and AA014041). S.M. is the recipient of a Career Development Award from the Australian National Health and Medical Research Council (grant number 496674).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the twins for their long-term co-operation; Genetic Epidemiology Laboratory staff for sample processing and DNA extraction; Dixie Statham for coordinating the SSAGA Study; and the clinical staff and many research interviewers for data collection.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bosron W.F., Li T.K. Genetic polymorphism of human liver alcohol and aldehyde dehydrogenases, and their relationship to alcohol metabolism and alcoholism. Hepatology. 1986;6:502–510. doi: 10.1002/hep.1840060330. [DOI] [PubMed] [Google Scholar]
- 2.Thomasson H.R., Crabb D.W., Edenberg H.J., Li T.K., Hwu H.G., Chen C.C., Yeh E.K., Yin S.J. Low frequency of the ADH2*2 allele among Atayal natives of Taiwan with alcohol use disorders. Alcohol Clin. Exp. Res. 1994;18:640–643. doi: 10.1111/j.1530-0277.1994.tb00923.x. [DOI] [PubMed] [Google Scholar]
- 3.Dick D.M., Bierut L.J. The genetics of alcohol dependence. Curr. Psychiatry Rep. 2006;8:151–157. doi: 10.1007/s11920-006-0015-1. [DOI] [PubMed] [Google Scholar]
- 4.Heath A.C., Madden P.A., Bucholz K.K., Dinwiddie S.H., Slutske W.S., Bierut L.J., Rohrbaugh J.W., Statham D.J., Dunne M.P., Whitfield J.B., et al. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychol. Med. 1999;29:1069–1081. doi: 10.1017/s0033291799008909. [DOI] [PubMed] [Google Scholar]
- 5.Kendler K.S., Heath A.C., Neale M.C., Kessler R.C., Eaves L.J. Alcoholism and major depression in women. A twin study of the causes of comorbidity. Arch. Gen. Psychiatry. 1993;50:690–698. doi: 10.1001/archpsyc.1993.01820210024003. [DOI] [PubMed] [Google Scholar]
- 6.Pickens R.W., Svikis D.S., McGue M., Lykken D.T., Heston L.L., Clayton P.J. Heterogeneity in the inheritance of alcoholism. A study of male and female twins. Arch. Gen. Psychiatry. 1991;48:19–28. doi: 10.1001/archpsyc.1991.01810250021002. [DOI] [PubMed] [Google Scholar]
- 7.Heath A.C., Bucholz K.K., Madden P.A., Dinwiddie S.H., Slutske W.S., Bierut L.J., Statham D.J., Dunne M.P., Whitfield J.B., Martin N.G. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol. Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- 8.Knopik V.S., Heath A.C., Madden P.A., Bucholz K.K., Slutske W.S., Nelson E.C., Statham D., Whitfield J.B., Martin N.G. Genetic effects on alcohol dependence risk: re-evaluating the importance of psychiatric and other heritable risk factors. Psychol. Med. 2004;34:1519–1530. doi: 10.1017/s0033291704002922. [DOI] [PubMed] [Google Scholar]
- 9.Burmeister M. Basic concepts in the study of diseases with complex genetics. Biol. Psychiatry. 1999;45:522–532. doi: 10.1016/s0006-3223(98)00316-3. [DOI] [PubMed] [Google Scholar]
- 10.Hyman S.E. Introduction to the complex genetics of mental disorders. Biol. Psychiatry. 1999;45:518–521. doi: 10.1016/s0006-3223(98)00332-1. [DOI] [PubMed] [Google Scholar]
- 11.Schuckit M.A., Smith T.L. The relationships of a family history of alcohol dependence, a low level of response to alcohol and six domains of life functioning to the development of alcohol use disorders. J. Stud. Alcohol. 2000;61:827–835. doi: 10.15288/jsa.2000.61.827. [DOI] [PubMed] [Google Scholar]
- 12.Heath A.C., Madden P.A., Bucholz K.K., Dinwiddie S.H., Slutske W.S., Bierut L.J., Rohrbaugh J.W., Statham D.J., Dunne M.P., Whitfield J.B., et al. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychol. Med. 1999;29:1069–1081. doi: 10.1017/s0033291799008909. [DOI] [PubMed] [Google Scholar]
- 13.Slutske W.S., Heath A.C., Dinwiddie S.H., Madden P.A., Bucholz K.K., Dunne M.P., Statham D.J., Martin N.G. Common genetic risk factors for conduct disorder and alcohol dependence. J. Abnorm. Psychol. 1998;107:363–374. doi: 10.1037//0021-843x.107.3.363. [DOI] [PubMed] [Google Scholar]
- 14.Chen C.C., Lu R.B., Chen Y.C., Wang M.F., Chang Y.C., Li T.K., Yin S.J. Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. Am. J. Hum. Genet. 1999;65:795–807. doi: 10.1086/302540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen W.J., Loh E.W., Hsu Y.P., Chen C.C., Yu J.M., Cheng A.T. Alcohol-metabolising genes and alcoholism among Taiwanese Han men: independent effect of ADH2, ADH3 and ALDH2. Br. J. Psychiatry. 1996;168:762–767. doi: 10.1192/bjp.168.6.762. [DOI] [PubMed] [Google Scholar]
- 16.Osier M., Pakstis A.J., Kidd J.R., Lee J.F., Yin S.J., Ko H.C., Edenberg H.J., Lu R.B., Kidd K.K. Linkage disequilibrium at the ADH2 and ADH3 loci and risk of alcoholism. Am. J. Hum. Genet. 1999;64:1147–1157. doi: 10.1086/302317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitfield J.B., Nightingale B.N., Bucholz K.K., Madden P.A., Heath A.C., Martin N.G. ADH genotypes and alcohol use and dependence in Europeans. Alcohol Clin. Exp. Res. 1998;22:1463–1469. [PubMed] [Google Scholar]
- 18.Borras E., Coutelle C., Rosell A., Fernandez-Muixi F., Broch M., Crosas B., Hjelmqvist L., Lorenzo A., Gutierrez C., Santos M., et al. Genetic polymorphism of alcohol dehydrogenase in Europeans: the ADH2*2 allele decreases the risk for alcoholism and is associated with ADH3*1. Hepatology. 2000;31:984–989. doi: 10.1053/he.2000.5978. [DOI] [PubMed] [Google Scholar]
- 19.Edenberg H.J., Xuei X., Chen H.J., Tian H., Wetherill L.F., Dick D.M., Almasy L., Bierut L., Bucholz K.K., Goate A., et al. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum. Mol. Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- 20.Luo X., Kranzler H.R., Zuo L., Wang S., Schork N.J., Gelernter J. Diplotype trend regression analysis of the ADH gene cluster and the ALDH2 gene: multiple significant associations with alcohol dependence. Am. J. Hum. Genet. 2006;78:973–987. doi: 10.1086/504113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo P.H., Kalsi G., Prescott C.A., Hodgkinson C.A., Goldman D., van den Oord E.J., Alexander J., Jiang C., Sullivan P.F., Patterson D.G., et al. Association of ADH and ALDH genes with alcohol dependence in the Irish Affected Sib Pair Study of alcohol dependence (IASPSAD) sample. Alcohol Clin. Exp. Res. 2008;32:785–795. doi: 10.1111/j.1530-0277.2008.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickson P.A., James M.R., Heath A.C., Montgomery G.W., Martin N.G., Whitfield J.B., Birley A.J. Effects of variation at the ALDH2 locus on alcohol metabolism, sensitivity, consumption, and dependence in Europeans. Alcohol Clin. Exp. Res. 2006;30:1093–1100. doi: 10.1111/j.1530-0277.2006.00128.x. [DOI] [PubMed] [Google Scholar]
- 23.Ehlers C.L., Gilder D.A., Harris L., Carr L. Association of the ADH2*3 allele with a negative family history of alcoholism in African American young adults. Alcohol Clin. Exp. Res. 2001;25:1773–1777. [PubMed] [Google Scholar]
- 24.Ehlers C.L., Montane-Jaime K., Moore S., Shafe S., Joseph R., Carr L.G. Association of the ADHIB*3 allele with alcohol-related phenotypes in Trinidad. Alcohol Clin. Exp. Res. 2007;31:216–220. doi: 10.1111/j.1530-0277.2006.00298.x. [DOI] [PubMed] [Google Scholar]
- 25.Wall T.L. Genetic associations of alcohol and aldehyde dehydrogenase with alcohol dependence and their mechanisms of action. Ther. Drug Monit. 2005;27:700–703. doi: 10.1097/01.ftd.0000179840.78762.33. [DOI] [PubMed] [Google Scholar]
- 26.Crabb D.W., Edenberg H.J., Thomasson H.R., Li T.K. Genetic factors that reduce the risk in developing alcoholism in animals and humans. In: Begleiter H., Kissin B., editors. The Genetics of Alcoholism. Oxford University Press,; 1995. pp. 202–220. [Google Scholar]
- 27.Agarwal D.P., Goedde H.W. Alcohol Metabolism, Alcohol Intolerance and Alcoholism. Berlin: Springer-Verlag; 1990. [Google Scholar]
- 28.Wolff P.H. Ethnic differences in alcohol sensitivity. Science. 1972;175:449–450. doi: 10.1126/science.175.4020.449. [DOI] [PubMed] [Google Scholar]
- 29.Xiao Q., Weiner H., Johnston T., Crabb D.W. The aldehyde dehydrogenase ALDH2*2 allele exhibits dominance over ALDH2*1 in transduced HeLa cells. J. Clin. Invest. 1995;96:2180–2186. doi: 10.1172/JCI118272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao Q., Weiner H., Crabb D.W. The mutation in the mitochondrial aldehyde dehydrogenase (ALDH2) gene responsible for alcohol-induced flushing increases turnover of the enzyme tetramers in a dominant fashion. J. Clin. Invest. 1996;98:2027–2032. doi: 10.1172/JCI119007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W.J., Loh E.W., Hsu Y.P., Cheng A.T. Alcohol dehydrogenase and aldehyde dehydrogenase genotypes and alcoholism among Taiwanese aborigines. Biol. Psychiatry. 1997;41:703–709. doi: 10.1016/S0006-3223(96)00072-8. [DOI] [PubMed] [Google Scholar]
- 32.Hoog J.O., Heden L.O., Larsson K., Jornvall H., von Bahr-Lindstrom H. The gamma 1 and gamma 2 subunits of human liver alcohol dehydrogenase. cDNA structures, two amino acid replacements, and compatibility with changes in the enzymatic properties. Eur. J. Biochem. 1986;159:215–218. doi: 10.1111/j.1432-1033.1986.tb09855.x. [DOI] [PubMed] [Google Scholar]
- 33.Yin S.J. Alcohol dehydrogenase: enzymology and metabolism. Alcohol Alcohol. Suppl. 1994;2:113–119. [PubMed] [Google Scholar]
- 34.Higuchi S., Matsushita S., Murayama M., Takagi S., Hayashida M. Alcohol and aldehyde dehydrogenase polymorphisms and the risk for alcoholism. Am. J. Psychiatry. 1995;152:1219–1221. doi: 10.1176/ajp.152.8.1219. [DOI] [PubMed] [Google Scholar]
- 35.Yin S.J., Agarwal D.P., Agarwal D.P., Seitz H.K. In Alcohol in Health and Disease. New York: Marcel Dekker, Inc.; 2001. Functional polymorphism of alcohol and aldehyde dehydrogenases; pp. 1–26. [Google Scholar]
- 36.Whitfield J.B. Alcohol dehydrogenase and alcohol dependence: variation in genotype-associated risk between populations. Am. J. Hum. Genet. 2002;71:1247–1250. doi: 10.1086/344287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeshita T., Mao X.Q., Morimoto K. The contribution of polymorphism in the alcohol dehydrogenase beta subunit to alcohol sensitivity in a Japanese population. Hum. Genet. 1996;97:409–413. doi: 10.1007/BF02267057. [DOI] [PubMed] [Google Scholar]
- 38.Chen W.J., Chen C.C., Yu J.M., Cheng A.T. Self-reported flushing and genotypes of ALDH2, ADH2 and ADH3 among Taiwanese Han. Alcohol Clin. Exp. Res. 1998;22:1048–1052. [PubMed] [Google Scholar]
- 39.Matsuo K., Wakai K., Hirose K., Ito H., Saito T., Tajima K. Alcohol dehydrogenase 2 His47Arg polymorphism influences drinking habit independently of aldehyde dehydrogenase 2 Glu487Lys polymorphism: analysis of 2,299 Japanese subjects. Cancer Epidemiol. Biomarkers Prev. 2006;15:1009–1013. doi: 10.1158/1055-9965.EPI-05-0911. [DOI] [PubMed] [Google Scholar]
- 40.Shibuya A., Yasunami M., Yoshida A. Genotype of alcohol dehydrogenase and aldehyde dehydrogenase loci in Japanese alcohol flushers and nonflushers. Hum. Genet. 1989;82:14–16. doi: 10.1007/BF00288263. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka F., Shiratori Y., Yokosuka O., Imazeki F., Tsukada Y., Omata M. Polymorphism of alcohol-metabolizing genes affects drinking behavior and alcoholic liver disease in Japanese men. Alcohol Clin. Exp. Res. 1997;21:596–601. [PubMed] [Google Scholar]
- 42.Takeshita T., Yang X., Morimoto K. Association of the ADH2 genotypes with skin responses after ethanol exposure in Japanese male university students. Alcohol Clin. Exp. Res. 2001;25:1264–1269. [PubMed] [Google Scholar]
- 43.Cook T.A., Luczak S.E., Shea S.H., Ehlers C.L., Carr L.G., Wall T.L. Associations of ALDH2 and ADH1B genotypes with response to alcohol in Asian Americans. J. Stud. Alcohol. 2005;66:196–204. doi: 10.15288/jsa.2005.66.196. [DOI] [PubMed] [Google Scholar]
- 44.Osier M.V., Pakstis A.J., Soodyall H., Comas D., Goldman D., Odunsi A., Okonofua F., Parnas J., Schulz L.O., Bertranpetit J., et al. A global perspective on genetic variation at the ADH genes reveals unusual patterns of linkage disequilibrium and diversity. Am. J. Hum. Genet. 2002;71:84–99. doi: 10.1086/341290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neumark Y.D., Friedlander Y., Thomasson H.R., Li T.K. Association of the ADH2*2 allele with reduced ethanol consumption in Jewish men in Israel: a pilot study. J Stud. Alcohol. 1998;59:133–139. doi: 10.15288/jsa.1998.59.133. [DOI] [PubMed] [Google Scholar]
- 46.Spivak B., Frisch A., Maman Z., Aharonovich E., Alderson D., Carr L.G., Weizman A., Hasin D. Effect of ADH1B genotype on alcohol consumption in young Israeli Jews. Alcohol Clin. Exp. Res. 2007;31:1297–1301. doi: 10.1111/j.1530-0277.2007.00438.x. [DOI] [PubMed] [Google Scholar]
- 47.Chao Y.C., Liou S.R., Chung Y.Y., Tang H.S., Hsu C.T., Li T.K., Yin S.J. Polymorphism of alcohol and aldehyde dehydrogenase genes and alcoholic cirrhosis in Chinese patients. Hepatology. 1994;19:360–366. [PubMed] [Google Scholar]
- 48.Tolstrup J.S., Nordestgaard B.G., Rasmussen S., Tybjaerg-Hansen A., Gronbaek M. Alcoholism and alcohol drinking habits predicted from alcohol dehydrogenase genes. Pharmacogenomics J. 2008;8:220–227. doi: 10.1038/sj.tpj.6500471. [DOI] [PubMed] [Google Scholar]
- 49.Matsuo K., Hiraki A., Hirose K., Ito H., Suzuki T., Wakai K., Tajima K. Impact of the alcohol-dehydrogenase (ADH) 1C and ADH1B polymorphisms on drinking behavior in nonalcoholic Japanese. Hum. Mutat. 2007;28:506–510. doi: 10.1002/humu.20477. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto K., Ueno Y., Mizoi Y., Tatsuno Y. Genetic polymorphism of alcohol and aldehyde dehydrogenase and the effects on alcohol metabolism. Arukoru Kenkyuto Yakubutsu Ison. 1993;28:13–25. [PubMed] [Google Scholar]
- 51.Whitfield J.B., Zhu G., Duffy D.L., Birley A.J., Madden P.A., Heath A.C., Martin N.G. Variation in alcohol pharmacokinetics as a risk factor for alcohol dependence. Alcohol Clin. Exp. Res. 2001;25:1257–1263. [PubMed] [Google Scholar]
- 52.Neumark Y.D., Friedlander Y., Durst R., Leitersdorf E., Jaffe D., Ramchandani V.A., O'Connor S., Carr L.G., Li T.K. Alcohol dehydrogenase polymorphisms influence alcohol-elimination rates in a male Jewish population. Alcohol Clin. Exp. Res. 2004;28:10–14. doi: 10.1097/01.ALC.0000108667.79219.4D. [DOI] [PubMed] [Google Scholar]
- 53.Edenberg H.J., Xuei X., Chen H.J., Tian H., Wetherill L.F., Dick D.M., Almasy L., Bierut L., Bucholz K.K., Goate A., et al. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum. Mol. Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- 54.Luo X., Kranzler H.R., Zuo L., Lappalainen J., Yang B.Z., Gelernter J. ADH4 gene variation is associated with alcohol dependence and drug dependence in European Americans: results from HWD tests and case-control association studies. Neuropsychopharmacology. 2006;31:1085–1095. doi: 10.1038/sj.npp.1300925. [DOI] [PubMed] [Google Scholar]
- 55.Saccone N.L., Kwon J.M., Corbett J., Goate A., Rochberg N., Edenberg H.J., Foroud T., Li T.K., Begleiter H., Reich T., et al. A genome screen of maximum number of drinks as an alcoholism phenotype. Am. J. Med. Genet. 2000;96:632–637. doi: 10.1002/1096-8628(20001009)96:5<632::aid-ajmg8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 56.Whitfield J.B. Acute reactions to alcohol. Addiction Biol. 1997;2:377–386. doi: 10.1080/13556219772435. [DOI] [PubMed] [Google Scholar]
- 57.Whitfield J.B., Martin N.G. Alcohol reactions in subjects of European descent: effects on alcohol use and on physical and psychomotor responses to alcohol. Alcohol Clin. Exp. Res. 1996;20:81–86. doi: 10.1111/j.1530-0277.1996.tb01048.x. [DOI] [PubMed] [Google Scholar]
- 58.Takeshita T., Mao X.Q., Morimoto K. The contribution of polymorphism in the alcohol dehydrogenase beta subunit to alcohol sensitivity in a Japanese population. Hum. Genet. 1996;97:409–413. doi: 10.1007/BF02267057. [DOI] [PubMed] [Google Scholar]
- 59.Duranceaux N.C., Schuckit M.A., Eng M.Y., Robinson S.K., Carr L.G., Wall T.L. Associations of variations in alcohol dehydrogenase genes with the level of response to alcohol in non-Asians. Alcohol Clin. Exp. Res. 2006;30:1470–1478. doi: 10.1111/j.1530-0277.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- 60.Shea S.H., Wall T.L., Carr L.G., Li T.K. ADH2 and alcohol-related phenotypes in Ashkenazic Jewish American college students. Behav. Genet. 2001;31:231–239. doi: 10.1023/a:1010261713092. [DOI] [PubMed] [Google Scholar]
- 61.Guindalini C., Scivoletto S., Ferreira R.G., Breen G., Zilberman M., Peluso M.A., Zatz M. Association of genetic variants in alcohol dehydrogenase 4 with alcohol dependence in Brazilian patients. Am. J. Psychiatry. 2005;162:1005–1007. doi: 10.1176/appi.ajp.162.5.1005. [DOI] [PubMed] [Google Scholar]
- 62.Slutske W.S., Heath A.C., Madden P.A., Bucholz K.K., Dinwiddie S.H., Dunne M.P., Statham D.S., Whitfield J.B., Martin N.G. Is alcohol-related flushing a protective factor for alcoholism in Caucasians? Alcohol Clin. Exp. Res. 1995;19:582–592. doi: 10.1111/j.1530-0277.1995.tb01552.x. [DOI] [PubMed] [Google Scholar]
- 63.Bucholz K.K., Cadoret R., Cloninger C.R., Dinwiddie S.H., Hesselbrock V.M., Nurnberger J.I., Jr, Reich T., Schmidt I., Schuckit M.A. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J. Stud. Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- 64.Hesselbrock M., Easton C., Bucholz K.K., Schuckit M., Hesselbrock V. A validity study of the SSAGA—a comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- 65.Nyholt D.R. On the probability of dizygotic twins being concordant for two alleles at multiple polymorphic loci. Twin. Res. Hum. Genet. 2006;9:194–197. doi: 10.1375/183242706776382383. [DOI] [PubMed] [Google Scholar]
- 66.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 3rd edn. Washington, DC: APA; 1987. [Google Scholar]
- 67.Hansell N.K., Agrawal A., Whitfield J.B., Morley K.I., Zhu G., Lind P.A., Pergadia M.L., Madden P.A.F., Todd R.D., Heath A.C., et al. Long-term stability and heritability of telephone interview measures of alcohol consumption and dependence. Twin. Res. Hum. Genet. 2008;11:287–305. doi: 10.1375/twin.11.3.287. [DOI] [PubMed] [Google Scholar]
- 68.Dickson P.A., James M.R., Heath A.C., Montgomery G.W., Martin N.G., Whitfield J.B., Birley A.J. Effects of variation at the ALDH2 locus on alcohol metabolism, sensitivity, consumption, and dependence in Europeans. Alcohol Clin. Exp. Res. 2006;30:1093–1100. doi: 10.1111/j.1530-0277.2006.00128.x. [DOI] [PubMed] [Google Scholar]
- 69.Martin N.G., Oakeshott J.G., Gibson J.B., Starmer G.A., Perl J., Wilks A.V. A twin study of psychomotor and physiological responses to an acute dose of alcohol. Behav. Genet. 1985;15:305–347. doi: 10.1007/BF01070893. [DOI] [PubMed] [Google Scholar]
- 70.Djousse L., Levy D., Herbert A.G., Wilson P.W., D'Agostino R.B., Cupples L.A., Karamohamed S., Ellison R.C. Influence of alcohol dehydrogenase 1C polymorphism on the alcohol-cardiovascular disease association (from the Framingham Offspring Study) Am. J. Cardiol. 2005;96:227–232. doi: 10.1016/j.amjcard.2005.03.050. [DOI] [PubMed] [Google Scholar]
- 71.Wigginton J.E., Abecasis G.R. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 72.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 73.Abecasis G.R., Cardon L.R., Cookson W.O. A general test of association for quantitative traits in nuclear families. Am. J. Hum. Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abecasis G.R., Cookson W.O., Cardon L.R. Pedigree tests of transmission disequilibrium. Eur. J. Hum. Genet. 2000;8:545–551. doi: 10.1038/sj.ejhg.5200494. [DOI] [PubMed] [Google Scholar]
- 75.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.