Abstract
Persistent infection with hepatitis C virus (HCV) is among the leading causes of chronic liver disease. Previous studies suggested that genetic variation in hypervariable region 1 (HVR1) of the second envelope protein, possibly in response to host immune pressure, influences the outcome of HCV infection. In the present study, a chimpanzee transfected intrahepatically with RNA transcripts of an infectious HCV clone (pCV-H77C) from which HVR1 was deleted became infected; the ΔHVR1 virus was subsequently transmitted to a second chimpanzee. Infection with ΔHVR1 virus resulted in persistent infection in the former chimpanzee and in acute resolving infection in the latter chimpanzee. Both chimpanzees developed hepatitis. The ΔHVR1 virus initially replicated to low titers, but virus titer increased significantly after mutations appeared in the viral genome. Thus, wild-type HCV without HVR1 was apparently attenuated, suggesting a functional role of HVR1. However, our data indicate that HVR1 is not essential for the viability of HCV, the resolution of infection, or the progression to chronicity.
Hepatitis C virus (HCV) is a small, enveloped positive strand RNA virus (Flaviviridae family) that is an important cause of chronic liver disease worldwide (1–3). Three to four million people in the United States and more that 100 million people globally are infected with HCV. A key feature of HCV is its capacity to persist in most infected individuals. About 80% of acutely infected individuals become chronically infected, and some develop chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma.
In an infected individual, HCV circulates as a heterogeneous population (quasispecies). The envelope glycoproteins (E1 and E2) exhibit the highest degree of genetic heterogeneity, especially in the 81 nucleotides comprising the hypervariable region 1 (HVR1) located at the N terminus of E2 (4–6). This region has been extensively used to characterize the quasispecies nature of HCV (7–10). It evolves rapidly in infected individuals, suggesting that it is under strong immune pressure, and there is evidence that the virus is able to escape the host immune response by accumulating mutations in the HVR1 region (11–14). Consistent with these observations, the HVR1 has been shown to contain at least one neutralization epitope (15, 16). In vitro, antibodies to HVR1 can inhibit the binding of recombinant E2 (17) or of virus (18) to cells, and rabbit hyperimmune serum generated to the carboxyl-terminal 21 amino acids of HVR1 can neutralize the infectivity of HCV for chimpanzees (15, 16). In addition, a correlation between the early appearance of anti-HVR1 and resolution of HCV infection in acutely infected patients was reported (19–21). A recent study provided evidence that the evolutionary dynamics of the HVR1 quasispecies during the acute phase of hepatitis C can predict the outcome of the infection (22). Also, it has been suggested that the HVR1 might be a decoy antigen, diverting the host immune response away from more conserved neutralization epitopes of HCV (23) or that variants of HVR1 might act as T cell receptor antagonists for HCV-specific CD4+ or CD8+ T cells (24–26). These studies all assume a central role of HVR1 in the natural history of HCV.
Despite the high degree of genetic variability of HVR1, detailed sequence analysis has revealed that this region actually may be structurally constrained. It contains highly conserved amino acid positions, and even at variable positions, amino acid substitutions do not follow a random pattern (10, 27, 28). Furthermore, hydrophobicity plots are similar for highly divergent HVR1 sequences (J.B., unpublished data). It remains possible, therefore, that the HVR1 of HCV must maintain a defined structure to preserve virus viability, as was described for the V3 loop of HIV gp120 (29, 30). Deletion of the V3 loop does not cause appreciable misfolding of gp120, but it is lethal for the virus because V3 has a critical role in the binding of the virus to its principal receptor, CD4 (29, 30).
The generation of infectious cDNA clones has made it possible to study HCV with recombinant DNA technology. Because there is not a reproducible cell culture system for HCV, infectivity studies of molecular clones must be performed in chimpanzees (31–36). In the present study, we inoculated chimpanzees with HCV lacking HVR1 and studied whether this hypervariable region plays an essential role for the natural history of this important human pathogen.
Materials and Methods
Construction of Plasmids.
Two HCV expression vectors were constructed using pDisplay (Invitrogen) as described previously (37). Plasmid pE1E2surf-715 encoded the E1 protein of HCV followed by a truncated form of the E2 protein (amino acids 192–715). Plasmid pE1E2surf-715(ΔHVR1) encoded the E1 protein (amino acids 192–383) and a truncated E2 protein lacking the HVR1 region (amino acids 411–715). The sequence of both clones was confirmed.
Construction of a deletion mutant of pCV-H77C lacking the HVR1 was performed by digestion of the expression vector pE1E2surf(ΔHVR1) with MunI and cloning the resulting fragment into the MunI-digested HCV cDNA clone H77C. A clone containing the correct insert was selected and retransformed, and large-scale plasmid DNA was prepared as previously described (32). The complete sequence of the HVR1 deletion mutant [pH77C(ΔHVR1)] was identical to pCV-H77C except for the absence of the first 81 nucleotides (positions 1491 to 1571 of H77C) of E2.
In Vitro Protein Synthesis and Processing.
Expression vectors pE1E2surf-715 and pE1E2surf-715(ΔHVR1) were assayed for in vitro protein synthesis and processing. Reactions were performed in 25 μl of the TNT-coupled reticulocyte lysate system (Promega) containing [35S]methionine or [35S]cysteine, with or without the addition of canine microsomal membranes at 30°C for 90 min. Total translation products were separated by 12% SDS/PAGE and identified by autoradiography.
Analysis of ΔHVR1 Mutant of HCV in Chimpanzees.
The housing, maintenance, and care of the two chimpanzees used in the present study were in compliance with all relevant guidelines and requirements. RNA was transcribed in vitro with T7 RNA polymerase from 10 μg of pH77C(ΔHVR1) linearized with XbaI as described previously (32). RNA transcripts from two transcription mixtures were percutaneously injected into the liver of a chimpanzee (#1590) under ultrasonographic control (36). A second chimpanzee (#96A008) was inoculated intravenously with 90 ml of plasma from chimpanzee 1590 taken 4 weeks after transfection. Serum samples were collected weekly from both chimpanzees and monitored for HCV-RNA [in-house RT-nested PCR (38) and HCV Monitor test version 2.0 (Roche Diagnostics)], HCV antibodies (second generation ELISA, Abbott), and liver enzyme levels (alanine aminotransferase, Anilytics). Liver biopsies were collected weekly and examined for necroinflammatory changes.
The genome (ORF) of virus recovered from infected chimpanzees was amplified by RT-PCR with primers specific for HCV strain H77 and sequenced directly to obtain consensus sequences (32, 38). For sequential analysis, genome regions with mutations were amplified in RT-nested PCR assays with specific primers and sequenced directly (38).
To examine the relative titers of HCV RNA in serum, peripheral blood mononuclear cells (PBMC), and liver tissue in an animal infected with the ΔHVR1 mutant, additional samples were collected from chimpanzee 1590 at weeks 51 and 52 postinoculation (p.i.) PBMC were isolated with Ficoll–Paque Plus (Amersham Pharmacia) from 10 ml of fresh whole blood (collected in ACD tubes). More than 106 cells were isolated and tested in total. Total RNA from PBMC, serum, and liver were extracted in parallel by Trizol (GIBCO/BRL). RNA from PBMC was tested for HCV RNA in RT-nested PCR (38). The genome titer of the RNA extracted from serum or liver was tested in parallel by Monitor 2.0 (Roche) in duplicate experiments.
Cellular Immune Responses in Chimpanzees Infected with ΔHVR1 Mutant of HCV.
PBMC were tested for HCV-specific proliferative capacity with a panel of recombinant HCV proteins [C22 (core), C33-c (NS3), c100 (NS3-NS4), and NS5]. The mean level of thymidine incorporation, after 6 days of culture, in the antigen-stimulated and control cultures was used to calculate the stimulation index. A stimulation index value of >2 was considered positive. The peripheral CD8+ T cell response was tested by stimulating PBMC with a large panel of HCV peptides corresponding to CD8+ T cell (CTL) epitopes located within the viral structural and nonstructural proteins (39). Peptide-specific CTL lines were tested by flow cytometry for peptide-specific interferon–gamma production after 3 weeks of repetitive peptide stimulation. To test for the intrahepatic CD4+ T cell response, liver-infiltrating lymphocytes were isolated from approximately 0.5–1 cm of liver tissue obtained by needle biopsy (16-gauge needle). The tissue was homogenized in 2–3 ml of PBS using a Dounce tissue grinder. Cell suspensions were then incubated with beads coupled to anti-CD4 antibodies (Dynabeads) for 20 min, and bound CD4+ T cells were isolated using a particle magnetic concentrator. Isolated intrahepatic CD4+ T cells were next plated into separate wells in 24-well plates in 1 ml of 10% blood type AB serum, 100 units/ml IL2, and 0.04 μg/ml anti-human CD3 monoclonal antibody as a stimulus to T cell growth and 2 × 106 irradiated autologous PBMC as feeders (40). After 2–3 weeks, the expanded T cells were tested for HCV-specific proliferative responses as described above.
Results
In Vitro Processing of the HVR1 Deletion Mutant.
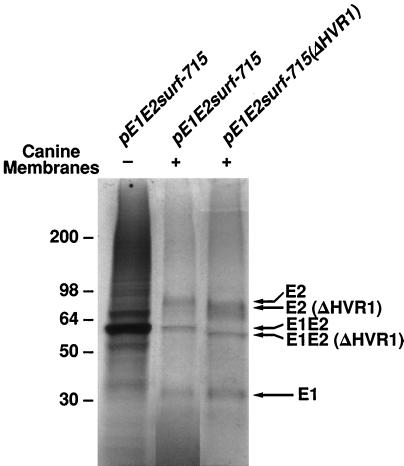
We previously reported that E2 protein lacking HVR1 appeared to be properly folded since it could be targeted to the cell surface and could bind to the large extracellular loop of CD81 (37), which was recently proposed as the receptor for HCV (41). However, these studies were done with E2 protein expressed by itself, whereas the E2 protein is normally synthesized as part of a polyprotein. Because HVR1 is located immediately downstream from the cleavage site between E1 and E2, it was important to determine whether deletion of HVR1 would affect the host-mediated processing of E1 and E2. Two expression vectors encoding E1 and E2, which were identical except that one lacked HVR1, were transcribed and translated in the TNT-coupled reticulolysate system. When microsomal membranes were added to the mixture, cleavage between E1 and E2 was observed for both constructs; as expected for correct processing, the E1 protein of each was the same size, but the E2 protein derived from the expression vector lacking HVR1 was slightly smaller than its counterpart (Fig. 1). These results suggested that deletion of HVR1 did not have major effects on the processing of the HCV E2 protein.
Figure 1.
Proteolytic cleavage of an E1–E2 precursor lacking HVR1. Expression vectors pE1E2surf-715 and pE1E2surf-715(ΔHVR1) were assayed for in vitro protein synthesis and processing. Reactions were performed in the TNT -coupled reticulocyte lysate system (Promega) containing [35S]cysteine, with or without the addition of canine microsomal membranes. Total translation products were separated by 12% SDS/PAGE and identified by autoradiography. The reaction of pE1E2surf-715(ΔHVR1) without addition of canine membranes is not shown. In both cases, cleavage between E1–E2 occurred in the presence of microsomal membranes, yielding E1 glycoproteins of comparable size; for pE1E2surf-715(ΔHVR1), an E2 protein of slightly smaller size was generated. The presence of uncleaved, nonglycosylated E1E2 proteins is indicated.
Intrahepatic Transfection of a Chimpanzee with HCV RNA Lacking HVR1.
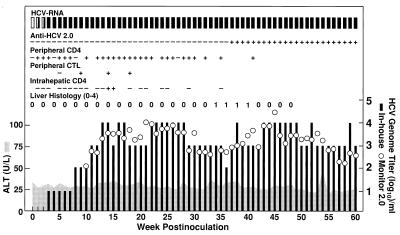
To determine whether an infectious clone of HCV (pCV-H77C) (32) retained its viability after HVR1 was deleted [pH77C(ΔHVR1)], we transcribed RNA from linearized pH77C(ΔHVR1) in vitro and inoculated it into the liver of a naive chimpanzee (#1590). The virological and immunological correlates of HCV infection following the transfection are shown in Fig. 2. Serum HCV-RNA was detected by RT-nested PCR (38) at week one p.i., and the animal remained HCV-RNA positive throughout the entire follow-up period of 72 weeks. Thus, this chimpanzee developed a chronic infection. The genome titer during the first 7 weeks was ≤10 genome equivalents (GE)/ml, which was very low, compared with titers of 103 to 105 GE/ml reported for the complete virus following transfection (32). However, during weeks 8–14, the titer increased progressively to ≈104 GE/ml and remained at 103 to 104 GE/ml throughout follow-up. The transfected chimpanzee became positive for antibodies to HCV (second generation ELISA) at week 37. Third generation RIBA (Chiron) confirmed the presence of antibodies to C22 (core), C33-c (NS3), and c100 (NS3-NS4). An HCV-specific proliferative CD4+ T cell response to C22 (core), C33-c (NS3), c100 (NS3-NS4), and/or NS5 antigens of HCV was detected in the PBMC beginning at week one posttransfection and throughout follow-up; at most weeks, however, we detected only a monospecific response to core. Peripheral CTL responses were tested at weeks 5, 9, 14, and 18 by an in vitro peptide stimulation assay. CTL were detected with two NS4 peptides at weeks 9 and 14 and with one of these at week 18. A transient and monospecific (NS5) proliferative T cell response was detected in the liver at weeks 14 and 15. Although serum liver enzyme values remained normal during the entire follow-up, necroinflammatory changes indicative of hepatitis were detected in liver biopsies during weeks 34–40.
Figure 2.
Intrahepatic inoculation of chimpanzee 1590 with RNA transcripts of pH77C (ΔHVR1). Serum samples were collected weekly from the chimpanzee and monitored for HCV-RNA (In-house RT-nested PCR and HCV Monitor test version 2.0; Roche), HCV antibodies (second generation ELISA, Abbott), and liver enzyme levels (alanine aminotransferase, Anilytics). PBMC were collected weekly and tested for HCV-specific proliferative capacity (Peripheral CD4) with a panel of recombinant HCV proteins [C22 (core), C33-c (NS3), C100 (NS3-NS4), and NS5]. The peripheral CTL was tested by stimulating PBMC with a large panel of HCV peptides corresponding to known CTL epitopes. Expanded T cells isolated from liver biopsy samples were tested for HCV-specific proliferative responses (Intrahepatic CD4). +, positive; −, negative. Liver biopsies were examined also for necroinflammatory changes (0, normal; 1+, 2+, 3+, 4+).
We sequenced the genome of viruses recovered from chimpanzee 1590 following transfection with clone H77C(ΔHVR1). The consensus sequence of the entire ORF (8952 nucleotides encoding 2984 amino acids) was determined at weeks 13 and 24 p.i. We confirmed that the recovered virus lacked the HVR1 region. Compared with the cDNA clone, six nucleotide substitutions were identified in the virus, and five of these were nonsynonymous (Table 1). The temporal occurrence of these six mutations was determined by sequential analysis of the relevant regions (Table 1). During weeks 4–7, mutations were not detected. However, during weeks 8–14, and coinciding with the increase in viral titer, we detected amino acid substitutions at four positions (two in E2, one in NS3, and one in NS5B). The NS3 mutation reverted to the wild-type sequence at week 18, but, at the three other positions, the wild-type sequence was replaced completely with the mutant sequence. An additional amino acid substitution in NS3 appeared at week 18 p.i.
Table 1.
Evolution of HCV lacking HVR1 in chimpanzee 1590
| Week | Titer in-house* | Titer monitor† | E2
|
NS3
|
NS5B
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nt 1881 G | aa 514 V | nt 2185 T | aa 615 L | nt 3769 G | aa 1143 R | nt 4708 C | aa 1456 T | nt 8234 G | aa 2631 K | nt 8966 A | aa 2875 E | |||
| 1 | <1 | Neg | ||||||||||||
| 2 | <1 | Neg | ||||||||||||
| 3 | 1 | Neg | ||||||||||||
| 4 | 1 | Neg | T | L | G | R | ||||||||
| 5 | 1 | Neg | G | V | T | L | G | R | A | E | ||||
| 6 | 1 | Neg | G | V | T | L | G | R | A | E | ||||
| 7 | 1 | Neg | G | V | T | L | G | R | A | E | ||||
| 8 | 2 | Neg | G | V | T/A | L/H | G | R | A/t | E/d | ||||
| 9 | 2 | Neg | G | V | T/A | L/H | G/a | R/h | T/a | D/e | ||||
| 10 | 2 | 2.08 | G | V | A/t | H/l | G/a | R/h | T/a | D/e | ||||
| 11 | 3 | 2.88 | G | V | A | H | G/a | R/h | T | D | ||||
| 12 | 3 | 2.83 | G/a | V/m | A | H | G/A | R/H | T | D | ||||
| 13 | 4 | 3.30 | G/A | V/M | A | H | G/A | R/H | C | T | G/a | K | T | D |
| 14 | 4 | 3.50 | G/A | V/M | G/A | R/H | C | T | G | K | ||||
| 15 | 3 | 3.48 | ||||||||||||
| 16 | 4 | 3.52 | A/g | M/v | G/a | R/h | C | T | G/a | K | ||||
| 17 | 4 | 3.29 | ||||||||||||
| 18 | 3 | 3.72 | A | M | G | R | C/t | T/m | G/A | K | ||||
| 19 | 3 | 3.20 | ||||||||||||
| 20 | 3 | 3.37 | A | M | G | R | C/T | T/M | A/g | K | ||||
| 21 | 3 | 4.04 | ||||||||||||
| 22 | 4 | 3.94 | A | M | G | R | C/T | T/M | A/g | K | ||||
| 23 | 4 | 3.48 | ||||||||||||
| 24 | 4 | 3.74 | A | M | A | H | G | R | T/c | M/t | A | K | T | D |
We performed sequence analysis of the entire open reading frame of the virus recovered from chimpanzee 1590 at weeks 13 and 24 posttransfection. We confirmed that the HVR1 region was not present and identified differences from the wild-type cDNA clone as indicated. Nucleotide (nt) and amino acid (aa) positions of pCV-H77C (32). Sequence of H77C(ΔHVR1) cDNA is shown on top. Dominant sequences recovered from the chimpanzee are shown in capital letters. Minor sequences recovered from the chimpanzee are shown in lower case letters.
Log10 titer determined by RT-nested PCR on 10-fold serially diluted extracted RNA (38).
Log10 titer determined by second generation Monitor test (Roche).
The relative genome titer of HCV lacking HVR1 was assessed in serum, PBMC, and liver obtained from the transfected chimpanzee at weeks 51 and 52 p.i. We did not detect HCV RNA in PBMC. However, the HCV titers determined from liver tissue (104.8 GE/ml and 104.3 GE/ml at weeks 51 and 52 p.i., respectively) were higher than the titers determined from the serum (103.4 GE/ml and 103.9 GE/ml at weeks 51 and 52 p.i., respectively).
Transmission of HCV Lacking HVR1 to a Second Chimpanzee.
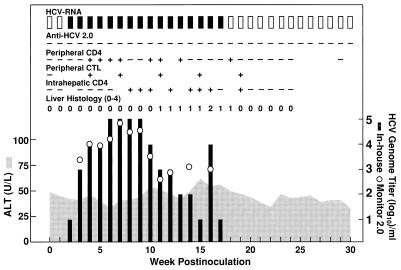
To investigate whether the HCV mutant lacking HVR1 was transmissible, we inoculated a naive chimpanzee (#96A008) intravenously with plasma from the transfected chimpanzee (#1590). As inoculum, we used 90 ml of plasma taken at week 4 from chimpanzee 1590, before appearance of consensus mutations in that chimpanzee (Table 1). The virological and immunological correlates of HCV infection in chimpanzee 96A008 are shown in Fig. 3. Serum HCV-RNA was first detected at week 2 p.i., with a genome titer of 101 GE/ml, and the titer increased to peak levels of 104 to 105 GE/ml during weeks 3–9. The infection was resolved at week 18 p.i. Antibodies to HCV were not detected. A peripheral monospecific proliferative CD4+ T cell response against core (C22) was detected during weeks 4–13 p.i.; a multispecific response against NS3 (C33-c), NS3-NS4 (c100), and NS5 was detected at week 18. The chimpanzee mounted an early and multispecific peripheral CD8+ T cell response to a total of seven different epitopes (representing core, NS3, NS4, and NS5) at the weeks tested (weeks 4, 7, 11, 15, and 19). Finally, a multispecific (C33-c, c100, and NS5) and sustained proliferative CD4+ T cell response was detected in the liver from week 8 p.i. Liver enzyme values were marginally elevated during weeks 10–18 p.i., and necroinflammatory changes were detected in liver biopsies during weeks 11–18 p.i.
Figure 3.
Infection of chimpanzee 96A008 with HCV lacking HVR1. At week 0, the chimpanzee was inoculated intravenously with 90 ml of plasma from chimpanzee 1590 (week 4 after transfection). At a titer of 10 GE/ml this represented an inoculum of approximately 900 genome equivalents of HCV. See also legend to Fig. 2.
Sequence analysis of the entire ORF of HCV recovered from chimpanzee 96A008 at weeks 4 and 9 p.i. showed that the transmitted virus lacked HVR1. Compared with the cDNA clone of the mutant, four nucleotide substitutions were identified in the virus, and all four mutations were nonsynonymous (Table 2). Sequence analysis of regions encompassing the described mutations showed that a mutation in E2 and NS3 each had replaced the wild-type nucleotide by the time the virus was first detected (week 2). An identical change in E2 had occurred in the transfected chimpanzee (Table 1). A change in E1 appeared as a quasispecies at week 2 but had replaced the wild-type nucleotide completely by week 3. The other mutation, in NS3, appeared at week 3 and persisted as a quasispecies during weeks 3–10 and then reverted to wild-type. This latter mutation had been seen also in the transfected chimpanzee (Table 1).
Table 2.
Evolution of HCV lacking HVR1 in chimpanzee 96A008
| Week | Titer in-house* | Titer monitor† | E1
|
E2
|
NS3
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| nt 1097 A | aa 252 K | nt 1881 G | aa 514 V | nt 3769 G | aa 1143 R | nt 4269 T | aa 1310 Y | |||
| 1 | Neg | Neg | ||||||||
| 2 | 1 | Neg | A/T | K/N | A | M | G | R | C | H |
| 3 | 3 | 3.33 | T | N | A | M | G/a | R/h | C | H |
| 4 | 4 | 4.00 | T | N | A | M | G/a | R/h | C | H |
| 5 | 4 | 3.98 | G/A | R/H | ||||||
| 6 | 5 | 4.25 | G/A | R/H | ||||||
| 7 | 5 | 4.81 | G/A | R/H | ||||||
| 8 | 5 | 4.51 | G/A | R/H | ||||||
| 9 | 5 | 4.62 | T | N | A | M | G/A | R/H | C | H |
| 10 | 4 | 3.30 | G/a | R/h | ||||||
| 11 | 3 | 2.63 | ||||||||
| 12 | 3 | 2.92 | G | R | ||||||
| 13 | 2 | Neg | ||||||||
| 14 | 2 | 3.14 | G | R | ||||||
| 15 | 1 | Neg | ||||||||
| 16 | 4 | 3.06 | G | R | ||||||
| 17 | 1 | Neg | ||||||||
| 18 | Neg | Neg | ||||||||
| 19 | Neg | Neg | ||||||||
| 20 | Neg | Neg | ||||||||
We performed sequence analysis of the entire open reading frame of the virus recovered from chimpanzee 96A008 at weeks 4 and 9 post-inoculation. We confirmed that the HVR1 region was not present and identified differences from the wild-type cDNA clone as indicated. Nucleotide (nt) and amino acid (aa) positions of pCV-H77C (32). Sequence of H77C(ΔHVR1) cDNA is shown on top. Dominant sequences recovered from the chimpanzee are shown in capital letters. Minor sequences recovered from the chimpanzee are shown in lower case letters.
Log10 titer determined by RT-nested PCR on 10-fold serially diluted extracted RNA (38).
Log10 titer determined by second generation Monitor test (Roche).
Discussion
This study provides definitive evidence that the HVR1 region of the E2 protein is not essential for the life cycle of HCV. A transfected chimpanzee remained viremic with HCV lacking HVR1 for more than 1 year, and the ΔHVR1 virus was transmitted to a second chimpanzee. However, the virus kinetics were different in chimpanzees infected with the ΔHVR1 virus when compared with chimpanzees transfected with the full-length wild-type genome (31, 32) or inoculated intravenously with the wild-type monoclonal virus (42). In particular, during the first weeks of the infection, ΔHVR1 virus replicated at unusually low levels, suggesting that the deletion mutant was attenuated. However, some mutations apparently compensated for the removal of HVR1 because their appearance coincided with the initial increase in virus titers. All such mutations were coding, suggesting that the virus was under strong selective pressure. Two amino acid substitutions were located within the ectodomain of E2. Both E2 mutations are at positions that are conserved among HCV genotype reference strains (38), and one of these appeared in both chimpanzees (Tables 1 and 2). These mutations are not within the regions believed to be involved in binding to the putative HCV receptor, CD81 (43). A single amino acid substitution was found in E1 in one chimpanzee; this mutation is at a position that is variable among HCV genotype reference strains (38). The rapid appearance of mutations in the glycoproteins in the present study contrasted with the previously reported absence of mutations within the structural region during the first year following transfection with transcripts of the wild-type pCV-H77C (44). Thus, it is reasonable to assume that mutations within E1 and/or E2 proteins compensated for the absence of the HVR1 and improved virus fitness in the chimpanzees. Once mutations were selected, infection with the ΔHVR1 virus resembled wild-type HCV infection, including development of hepatitis. Thus, whereas our study clearly demonstrates that the HVR1 is nonessential, it does also suggest a functional role of HVR1 since the deletion mutant without compensating mutations was impaired. Further studies are required to define the putative function of HVR1 in the HCV infection cycle.
The primary replication site of HCV is the liver, but low-level replication has also been reported in lymphocytes (1). If HVR1 played an important role in cell attachment or entry, it is possible that deletion of this region would change its cell tropism. However, this apparently was not the case. We did not detect the ΔHVR1 virus in lymphocytes but did detect it at higher levels in the liver than in the serum of the chronically infected chimpanzee. More importantly, chimpanzees infected with the ΔHVR1 virus had histological evidence of viral hepatitis. Additionally, in these chimpanzees, T cell responses were detected in the liver, as well as peripherally. These data indicate that lack of HVR1 did not prevent HCV infectivity for hepatocytes.
Genetic variation within HVR1 is thought to play an essential role in determining whether patients have acute resolving or chronic infection (22). At least 15% of acutely infected individuals have spontaneous viral clearance, but the mechanism for this is unknown. A role for HVR1 in viral persistence through an immune escape mechanism was suggested in numerous studies (7–10). Our study proves that a host immune response to HVR1 is not essential for viral clearance since one chimpanzee infected with the ΔHVR1 virus cleared the virus at week 18 of the infection. Antibodies to HCV, including antibodies to E2 (data not shown), were not detected in this chimpanzee. However, the clearance in the chimpanzee was preceded by a strong, sustained, and multispecific intrahepatic CD4+ T cell response to HCV nonstructural proteins, as well as by a peripheral multispecific CD8+ T cell response to multiple epitopes representing core and nonstructural proteins. In contrast, HCV persisted for more than 1 year in the other chimpanzee infected with ΔHVR1. This animal did develop antibodies to structural and nonstructural proteins. In this chimpanzee, the ΔHVR1 virus persisted in the face of an early peripheral, multispecific CD4+ T cell response and a transient monospecific intrahepatic CD4+ T cell response as well as a peripheral monospecific CD8+ T cell response. Thus, although HVR1 contains a neutralization epitope, this region is required neither for clearance nor persistence of HCV.
The outcome of infection in the chimpanzees studied might have correlated with the quality of the cellular immune response to HCV. Recent studies suggested an association of peripheral CD4 or CD8+ T cell responses and viral clearance in acutely infected patients (45–48). In acutely infected chimpanzees, an association of an intrahepatic CD8+ T cell response and viral clearance was reported (49). We did not observe an association between the vigor of the peripheral CD4+ T cell response and the outcome of infection. However, our study suggested an important role of the intrahepatic CD4+ T cell response as well as the peripheral CTL response. In the chimpanzee with acute resolving infection, the proliferative CD4+ response occurred in the liver earlier and was much stronger, more broadly directed and durable compared with the intrahepatic CD4+ response in the chimpanzee that developed a chronic infection. In addition, in the chimpanzee with acute resolving infection, the peripheral CTL response appeared earlier and was directed against several additional epitopes compared with the CTL response in the chimpanzee that developed a chronic infection. These data suggest that the homing efficiency of the virus-specific T cells to the liver and the breadth of the CTL response may play an important role in the outcome of acute HCV infection. It was recently reported that clearance of hepatitis B virus from liver cells of experimentally infected chimpanzees occurs primarily without destruction of the infected cells (50). In the present study, clearance of HCV in chimpanzee 96A008 was associated with only marginally elevated transaminase levels even though high titers of virus were eliminated. This observation implies that noncytolytic mechanisms might play a role also for HCV clearance. Detailed analysis of the T cell response to HCV in these and other chimpanzees will be reported separately to address further the role of the cellular immune response for viral clearance during the early acute phase of HCV infection.
Our study challenges the concept that HVR1, located in the envelope protein thought to promote attachment and entry into host cells, has a pivotal role in the natural history of HCV. We have shown that this region was not essential for the infection cycle of HCV. However, our study did suggest a functional role of HVR1 since wild-type HCV without HVR1 was attenuated. Removal of HVR1 did not restrict the outcome of the infection. Although this does not necessarily mean that the HVR1 has no influence on whether the disease is resolved or progresses following infections with wild-type virus, these data suggest that other factors do play a role and need to be identified.
Acknowledgments
We thank Dr. Marisa St. Claire and Mr. Max Shapiro (Bioqual, Inc.) for animal care, Ms. Lynn Rasmussen and Mr. John Elser (Science Applications International Corporation, Frederick) for assistance in sequence analysis, and Mr. Carl L. Apgar and Mr. Ronald Engle (Hepatitis Viruses Section, Laboratory of Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health) and Ms. Janell Pemberton (Division of Experimental Pathology, Department of Molecular and Experimental Medicine, The Scripps Research Institute) for technical assistance. We are grateful to Dr. Masayuki Yanagi (Hepatitis Viruses Section, Laboratory of Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health) for helpful discussions. This study was supported in part by National Institutes of Health Contracts N01-AI-52705 and N01-CO-56000 and National Institutes of Health Grant R01-AI20001. R.T. was also supported by Grant TH 719/1-1 from the Deutsche Forschungsgemeinschaft, Bonn, Germany.
Abbreviations
- HCV
hepatitis C virus
- p.i.
postinoculation
- HVR1
hypervariable region 1
- E
envelope
- GE
genome equivalents
- CTL
CD8+ T cell
- PBMC
peripheral blood mononuclear cells
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230453597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230453597
References
- 1.Houghton M. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 1035–1058. [Google Scholar]
- 2.National Institutes of Health Consensus Development Conference Panel. Hepatology. 1997;26, Suppl. 1:2S–10S. doi: 10.1002/hep.510260701. [DOI] [PubMed] [Google Scholar]
- 3.Rice C M. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 931–959. [Google Scholar]
- 4.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Biochem Biophys Res Commun. 1991;175:220–228. doi: 10.1016/s0006-291x(05)81223-9. [DOI] [PubMed] [Google Scholar]
- 5.Ogata N, Alter H J, Miller R H, Purcell R H. Proc Natl Acad Sci USA. 1991;88:3392–3396. doi: 10.1073/pnas.88.8.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiner A J, Brauer M J, Rosenblatt J, Richman K H, Tung J, Crawford K, Bonino F, Saracco G, Choo Q-L, Houghton M, et al. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 7.Bukh J, Miller R H, Purcell R H. Semin Liver Dis. 1995;15:41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 8.Farci P, Bukh J, Purcell R H. Springer Semin Immunopathol. 1997;19:5–26. doi: 10.1007/BF00945022. [DOI] [PubMed] [Google Scholar]
- 9.Forns X, Purcell R H, Bukh J. Trends Microbiol. 1999;7:402–410. doi: 10.1016/s0966-842x(99)01590-5. [DOI] [PubMed] [Google Scholar]
- 10.Smith D B. J Viral Hepatitis. 1999;6, Suppl. 1:41–46. doi: 10.1046/j.1365-2893.1999.00010.x. [DOI] [PubMed] [Google Scholar]
- 11.Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. J Virol. 1993;67:3923–3930. doi: 10.1128/jvi.67.7.3923-3930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taniguchi S, Okamoto H, Sakamoto M, Kojima M, Tsuda F, Tanaka T, Munekata E, Muchmore E E, Peterson D A, Mishiro S. Virology. 1993;195:297–301. doi: 10.1006/viro.1993.1378. [DOI] [PubMed] [Google Scholar]
- 13.Tsai S-L, Chen Y-M, Chen M-H, Huang C-Y, Sheen I-S, Yeh C-T, Huang J-H, Kuo G C, Liaw Y-F. Gastroenterology. 1998;115:954–965. doi: 10.1016/s0016-5085(98)70268-9. [DOI] [PubMed] [Google Scholar]
- 14.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, et al. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter H J, Purcell R H. Proc Natl Acad Sci USA. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu Y K, Igarashi H, Kiyohara T, Cabezon T, Farci P, Purcell R H, Yoshikura H. Virology. 1996;223:409–412. doi: 10.1006/viro.1996.0497. [DOI] [PubMed] [Google Scholar]
- 17.Rosa D, Campagnoli S, Moretto C, Guenzi E, Cousens L, Chin M, Dong C, Weiner A J, Lau J Y N, Choo Q-L, et al. Proc Natl Acad Sci USA. 1996;93:1759–1763. doi: 10.1073/pnas.93.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zibert A, Schreier E, Roggendorf M. Virology. 1995;208:653–661. doi: 10.1006/viro.1995.1196. [DOI] [PubMed] [Google Scholar]
- 19.Allander T, Beyene A, Jacobson S H, Grillner L, Persson M A A. J Infect Dis. 1997;175:26–31. doi: 10.1093/infdis/175.1.26. [DOI] [PubMed] [Google Scholar]
- 20.Zibert A, Meisel H, Kraas W, Schulz A, Jung G, Roggendorf M. Hepatology. 1997;25:1245–1249. doi: 10.1002/hep.510250530. [DOI] [PubMed] [Google Scholar]
- 21.Zibert A, Kraas W, Meisel H, Jung G, Roggendorf M. J Virol. 1997;71:4123–4127. doi: 10.1128/jvi.71.5.4123-4127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder J C, Strazzera A, Chien D Y, Munoz S J, Balestrieri A, et al. Science. 2000;288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 23.Ray S C, Wang Y-M, Laeyendecker O, Ticehurst J R, Villano S A, Thomas D L. J Virol. 1999;73:2938–2946. doi: 10.1128/jvi.73.4.2938-2946.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang K-M, Rehermann B, McHutchison J G, Pasquinelli C, Southwood S, Sette A, Chisari F V. J Clin Invest. 1997;100:2376–2385. doi: 10.1172/JCI119778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frasca L, Del Porto P, Tuosto L, Marinari B, Scotta C, Carbonari M, Nicosia A, Piccolella E. J Immunol. 1999;163:650–658. [PubMed] [Google Scholar]
- 26.Kaneko T, Moriyama T, Udaka K, Hiroishi K, Kita H, Okamoto H, Yagita H, Okumura K, Imawari M. Eur J Immunol. 1997;27:1782–1787. doi: 10.1002/eji.1830270728. [DOI] [PubMed] [Google Scholar]
- 27.McAllister J, Casino C, Davidson F, Power J, Lawlor E, Yap P L, Simmonds P, Smith D B. J Virol. 1998;72:4893–4905. doi: 10.1128/jvi.72.6.4893-4905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puntoriero G, Meola A, Lahm A, Zucchelli S, Ercole B B, Tafi R, Pezzanera M, Mondelli M U, Cortese R, Tramontano A, et al. EMBO J. 1998;17:3521–3533. doi: 10.1093/emboj/17.13.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger E A. AIDS. 1997;11, Suppl. A:S3–S16. [PubMed] [Google Scholar]
- 30.Wyatt R, Sullivan N, Thali M, Repke H, Ho D, Robinson J, Posner M, Sodroski J. J Virol. 1993;67:4557–4565. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 32.Yanagi M, Purcell R H, Emerson S U, Bukh J. Proc Natl Acad Sci USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolykhalov A A, Mihalik K, Feinstone S M, Rice C M. J Virol. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yanagi M, Purcell R H, Emerson S U, Bukh J. Virology. 1999;262:250–263. doi: 10.1006/viro.1999.9889. [DOI] [PubMed] [Google Scholar]
- 35.Yanagi M, St. Claire M, Emerson S U, Purcell R H, Bukh J. Proc Natl Acad Sci USA. 1999;96:2291–2295. doi: 10.1073/pnas.96.5.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanagi M, St. Claire M, Shapiro M, Emerson S U, Purcell R H, Bukh J. Virology. 1998;244:161–172. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]
- 37.Forns X, Allander T, Rohwer-Nutter P, Bukh J. Virology. 2000;274:75–85. doi: 10.1006/viro.2000.0419. [DOI] [PubMed] [Google Scholar]
- 38.Bukh J, Apgar C L, Engle R, Govindarajan S, Hegerich P A, Tellier R, Wong D C, Elkins R, Kew M C. J Infect Dis. 1998;178:1193–1197. doi: 10.1086/515683. [DOI] [PubMed] [Google Scholar]
- 39.Cerny A, McHutchison J G, Pasquinelli C, Brown M E, Brothers M A, Grabscheid B, Fowler P, Houghton M, Chisari F V. J Clin Invest. 1995;95:521–530. doi: 10.1172/JCI117694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erickson A L, Houghton M, Choo Q L, Weiner A J, Ralston R, Muchmore E, Walker C M. J Immunol. 1993;151:4189–4199. [PubMed] [Google Scholar]
- 41.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner A J, Houghton M, Rosa D, Grandi G, et al. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 42.Forns X, Payette P J, Ma X, Satterfield W, Eder G, Mushahwar I K, Govindarajan S, Davis H L, Emerson S U, Purcell R H, et al. Hepatology. 2000;32:618–625. doi: 10.1053/jhep.2000.9877. [DOI] [PubMed] [Google Scholar]
- 43.Flint M, Maidens C, Loomis-Price L D, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating J A. J Virol. 1999;73:6235–6244. doi: 10.1128/jvi.73.8.6235-6244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bukh J, Yanagi Y, Emerson S U, Purcell R H. Hepatology. 1998;28:319A. [Google Scholar]
- 45.Diepolder H M, Zachoval R, Hoffmann R M, Wierenga E A, Santantonio T, Jung M C, Eichenlaub D, Pape G R. Lancet. 1995;346:1006–1007. doi: 10.1016/s0140-6736(95)91691-1. [DOI] [PubMed] [Google Scholar]
- 46.Gruner N H, Gerlach T J, Jung M C, Diepolder H M, Schirren C A, Schraut W W, Hoffmann R, Zachoval R, Santantonio T, Cucchiarini M, et al. J Infect Dis. 2000;181:1528–1536. doi: 10.1086/315450. [DOI] [PubMed] [Google Scholar]
- 47.Lechner F, Wong D K, Dunbar P R, Chapman R, Chung R T, Dohrenwend P, Robbins G, Phillips R, Klenerman P, Walker B D. J Exp Med. 2000;191:1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Missale G, Bertoni R, Lamonaca V, Valli A, Massari M, Mori C, Rumi M G, Houghton M, Fiaccadori F, Ferrari C. J Clin Invest. 1996;98:706–714. doi: 10.1172/JCI118842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooper S, Erickson A L, Adams E J, Kansopon J, Weiner A J, Chien D Y, Houghton M, Parham P, Walker C M. Immunity. 1999;10:439–449. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 50.Guidotti L G, Rochford R, Chung J, Shapiro M, Purcell R, Chisari F V. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]