Abstract
Accumulating data suggest a link between alterations/deficiencies in cytoskeletal proteins and the progression of cardiomyopathy and heart failure, although the molecular basis for this link remains unclear. Cypher/ZASP is a cytoskeletal protein localized in the sarcomeric Z-line. Mutations in its encoding gene have been identified in patients with isolated non-compaction of the left ventricular myocardium, dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy. To explore the role of Cypher in myocardium and to better understand molecular mechanisms by which mutations in cypher cause cardiomyopathy, we utilized a conditional approach to knockout Cypher, specially in either developing or adult myocardium. Cardiac-specific Cypher knockout (CKO) mice developed a severe form of DCM with disrupted cardiomyocyte ultrastructure and decreased cardiac function, which eventually led to death before 23 weeks of age. A similar phenotype was observed in inducible cardiac-specific CKO mice in which Cypher was specifically ablated in adult myocardium. In both cardiac-specific CKO models, ERK and Stat3 signaling pathways were augmented. Finally, we demonstrate the specific binding of Cypher's PDZ domain to the C-terminal region of both calsarcin-1 and myotilin within the Z-line. In conclusion, our studies suggest that (i) Cypher plays a pivotal role in maintaining adult cardiac structure and cardiac function through protein–protein interactions with other Z-line proteins, (ii) myocardial ablation of Cypher results in DCM with premature death and (iii) specific signaling pathways participate in Cypher mutant-mediated dysfunction of the heart, and may in concert facilitate the progression to heart failure.
INTRODUCTION
Dilated cardiomyopathy (DCM), which is characterized primarily by left ventricular dilation and systolic dysfunction, is a leading cause of congestive heart failure in young patients. The recognition that genetic defects may play pivotal roles in the pathogenesis of DCM, especially familial DCM, has received increasing attention during the past decade. Mutations in multiple cytoskeletal and sarcomeric genes, such as dystrophin (1), vinculin (2), desmin (3), titin (4), actin (5), β-myosin heavy chain (MHC) (6) and troponin T (7), have been linked to the pathogenesis of familial DCM in both human and mouse models. The anatomical and pathophysiological phenotypes of these cytoskeletal and sarcomeric gene mutations are associated variably with impaired structural maintenance, deficiencies in intracellular force generation and intercellular force transmission, and alterations in stretch signaling (8–15). However, the molecular mechanisms, linking the gene mutations to a specific DCM phenotype, remain largely unknown.
Cypher is a cytoskeletal protein, which binds to α-actinin in the Z-line of both skeletal and cardiac muscles (16,17). Mutations in Z-line alternatively spliced PDZ-motif protein (ZASP), a human orthologue of Cypher, have been identified in patients with isolated non-compaction of the left ventricular myocardium (INLVM), DCM, hypertrophic cardiomyopathy (HCM) as well as skeletal myopathy (18–23).
Our previous studies of mice with global ablation of Cypher in all cells revealed a pivotal role of Cypher in the structure and functions of striated muscles. These mutant mice developed a severe form of congenital myopathy and died from functional failure in multiple striated muscles within 1 week after birth (17). The perinatal lethality of Cypher knockout (CKO) mice prevented us from further exploring the specific requirement for Cypher in adult myocardium. To overcome this problem and better understand the molecular mechanisms by which mutations in Cypher cause cardiomyopathy, we utilized a conditional approach to knockout Cypher specifically in the heart by crossing floxed Cypher mice with the Mlc2v-Cre mouse line (24) and the MHC-α-MHC mER-Cre-mER tamoxifen-inducible Cre mouse line (25).
In the present study we report that cardiac-specific CKO mice display disrupted cardiomyocyte ultrastructure and decreased cardiac function, which eventually leads to a severe form of DCM with premature death. A similar phenotype is observed in inducible cardiac-specific CKO mice (ICKO) in which Cypher is specifically ablated in adult myocardium, confirming our results showing an essential role of Cypher in adult myocardium. In both cardiac-specific CKO models, cardiac dysfunction is accompanied by augmentation of ERK and Stat3 signaling pathways. Furthermore, we demonstrate specific interaction of Cypher's PDZ domain with the C-terminal region of both calsarcin-1 and myotilin.
Taken together, our studies suggest that (i) Cypher plays a pivotal role in maintaining adult cardiac structure and cardiac function, through protein–protein interactions with other Z-line proteins, (ii) myocardial ablation of Cypher results in DCM with premature death and (iii) specific signaling pathways participate in Cypher mutant-mediated dysfunction of the heart and may, in concert, facilitate the progression to heart failure.
RESULTS
Generation of cardiac-specific Cypher knockout mice
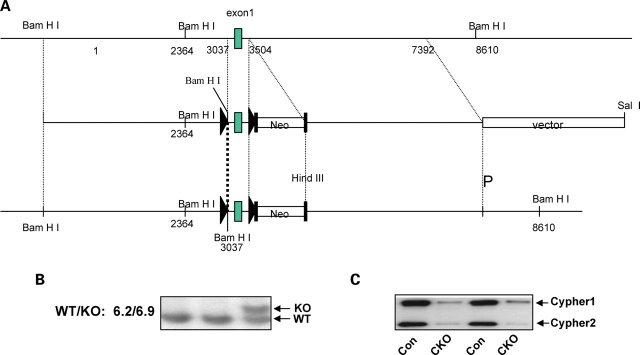
To specifically knockout Cypher in the heart, a targeting construct was generated containing loxP sites (triangles) flanking exon 1 of the Cypher gene, which is conserved in all six Cypher isoforms (Fig. 1A) (26). The neomycin (Neo) selection cassette was flanked by Flpase Recognition Target (FRT) sites, allowing for subsequent excision with FLPase. Targeted embryonic stem (ES) cells were identified by Southern blot analysis, and gave the expected 6.9 kb mutant fragment compared with the 6.2 kb wild-type fragment (Fig. 1B). Cypherf/f mice were crossed with Mlc2v-Cre mice to generate cardiac-specific CKO mice in which Cypher is specifically deleted in ventricular cardiomyocytes. Assessment of Cypher protein expression in CKO hearts, using a cardiac-specific Cypher antibody, demonstrated the specific loss of Cypher in CKO hearts from 1-month-old mice as shown in Figure 1C.
Figure 1.
Generation of gene-targeted mice. (A) Targeting construct for the generation of floxed Cypher mice. In the targeting vector, cypher gene exon 1 is flanked by LoxP sites as indicated with a black triangle. The neomyosin resistant gene is surrounded by FLP cassettes as shown by black rectangles. (B) Southern blot analysis shows a 6.9 kb band in heterozyogous gene-targeted embryonic stem cells when compared with a 6.2 kb band in the wild-type allele. (C) Representative western blot results showing the effective deletion of Cypher in CKO mouse heart at 1 month of age.
To further investigate the specificity and efficiency of Mlc2v-Cre-mediated excision during cardiac development, total protein was extracted from atria and ventricles separately and the Cypher protein level was measured (data not shown). Consistent with previously published data reporting Mlc2v expression only in ventricles (24), decreased expression of Cypher protein was observed only in ventricles, but not in the atria of CKO mouse hearts.
Cypher knockout mice develop dilated cardiomyopathy and die within 5 months
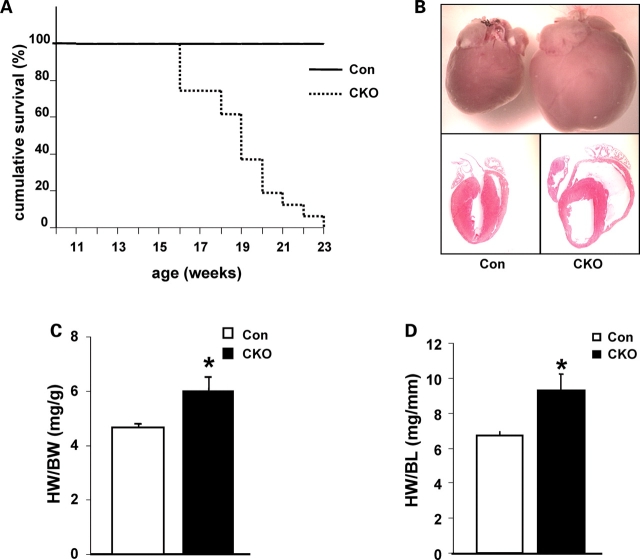
Cardiac-specific deletion of Cypher does not affect the Mendelian frequency of viable CKO male and female mice produced at birth. CKO mutants began to die at 16 weeks of age and all died before 23 weeks of age (Fig. 2A) when compared with 100% survival of control littermates, indicating the essential functional role of Cypher in the heart. Histological analysis of CKO hearts with hematoxylin and eosin staining revealed enlarged right and left ventricular chambers with thinner walls (Fig. 2B). Both ratios of heart weight to body weight and heart weight to tibia bone length were significantly increased from 4.65 (mg/g) in control to 6.01 (mg/g) in CKO mice and 6.72 (mg/mm) in control to 9.3 (mg/mm) in CKO mice, respectively (Fig. 2C and D).
Figure 2.
Survival curve and enlarged Cypher knockout (CKO) mouse hearts. (A) Cumulative survival curve of CKO and control littermate mice. CKO mice die from 16 weeks of age and are all dead before 23 weeks of age (n = 29 for CKO mice and n = 33 for control littermates). (B) Hematoxylin and eosin staining showing dilated ventricular chambers in 3-month-old CKO mouse heart compared with control littermate. (C) Heart weight to body weight ratio of CKO mice versus control littermates (n = 15 for CKO mice and n = 28 for control littermates). (D) Heart weight to tibia bone length ratio of CKO mice versus control littermates (n = 12 for CKO mice and n = 12 for control littermates). HW/BW, heart weight to body weight ratio; HW/BL, heart weight to tibia bone length ratio; *P < 0.05.
We next evaluated heart performance by echocardiography on CKO and control mice from 1 to 4 months of age. Whereas no significant differences were found between 1-month-old CKO and control littermate mice, 2–4 months old CKO mice showed enlarged left ventricular chamber size as evidenced by a significant increase in left ventricular internal dimension values at end-diastole (LVIDd) and end-systole (LVIDs) (Fig. 3A–C) in agreement with the histological observations (above). In addition, significant wall thinning was seen in CKO hearts as indicated by a significant decrease in interventricular septal wall thickness (IVSd) at end-diastole compared with controls (Table 1). More importantly, the changes in cardiac dimensions in CKO mice were accompanied by a dramatic decrease in LV systolic function, showed by a significant decrease in fraction shortening (%FS) and the velocity of circumferential fiber shortening (VCF) when compared with littermate controls (Fig. 3D, Table 1). Despite wall thinning, the significant increase in heart size in CKO mice led to an increase in calculated left ventricular mass (LVMd) as well as left ventricular mass normalized to body weight (LV/BW) at 4 months of age (Table 1). Electrocardiography (ECG) analyses also revealed AV block in CKO mice, which was further confirmed by conscious telemetric monitoring of electrocardiograms (data not shown).
Figure 3.

Echocardiography. (A) Typical example of echocardiography data showing LVIDd, LVIDs and FS in Cypher knockout (CKO) and control littermate hearts. (B) LVIDd, (C) LVIDs, (D) FS and (E) EDD/PWD results in CKO mice and control littermates at various ages as indicated (n = 4) for each group. LVIDd, end-diastolic left ventricular internal dimension at end-diastole; LVIDs, end-systolic left ventricular internal dimension; %FS, percentage of fractional shortening; EDD/PWD, end-diastolic dimension/diastolic posterior wall thickness; #P < 0.01.
Table 1.
Echocardiographic assay of cardiac function in Cypher knockout and control mice
| 1 month |
2 month |
3 month |
4 month |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CKO (n = 4) | Con (n = 4) | P | CKO (n = 4) | Con (n = 4) | P | CKO (n = 4) | Con (n = 4) | P | CKO (n = 4) | Con (n = 4) | P | |
| IVSd (mm) | 0.57 ± 0.02 | 0.58 ± 0.01 | 0.65 | 0.62 ± 0.04 | 0.61 ± 0.02 | 0.95 | 0.54 ± 0.02 | 0.63 ± 0.01 | 0.0040 | 0.55 ± 0.03 | 0.61 ± 0.01 | 0.05795 |
| LVIDd (mm) | 2.99 ± 0.14 | 3.11 ± 0.37 | 0.76 | 4.20 ± 0.18 | 3.43 ± 0.12 | 0.006 | 4.75 ± 0.17 | 3.61 ± 0.31 | 0.0093 | 5.33 ± 0.15 | 3.60 ± 0.12 | 0.0001 |
| LVIDs (mm) | 1.87 ± 0.11 | 1.75 ± 0.21 | 0.56 | 3.27 ± 0.06 | 2.03 ± 0.08 | 0.00001 | 4.03 ± 0.12 | 1.93 ± 0.24 | 0.0001 | 4.34 ± 0.09 | 1.90 ± 0.12 | 0.000004 |
| LVPWs (mm) | 1.11 ± 0.08 | 1.10 ± 0.10 | 0.92 | 1.03 ± 0.08 | 1.20 ± 0.10 | 0.18 | 0.88 ± 0.13 | 1.25 ± 0.05 | 0.0197 | 0.95 ± 0.11 | 1.20 ± 0.05 | 0.008182 |
| %FS | 37.40 ± 3.74 | 43.70 ± 1.56 | 0.12 | 21.92 ± 3.04 | 40.62 ± 249 | 0.0015 | 15.02 ± 3.43 | 46.30 ± 3.04 | 0.0018 | 18.44 ± 2.81 | 47.33 ± 2.72 | 0.000215 |
| EDD/PWD | 5.43 ± 0.33 | 5.46 ± 0.64 | 0.96 | 7.04 ± 0.24 | 5.61 ± 0.25 | 0.0032 | 8.97 ± 0.21 | 5.60 ± 0.33 | 0.0001 | 9.18 ± 0.53 | 5.77 ± 0.21 | 0.000148 |
| VCF (circ/s) | 8.02 ± 1.19 | 9.16 ± 0.56 | 0.36 | 4.92 ± 0.73 | 8.30 ± 0.85 | 0.0130 | 3.39 ± 0.76 | 10.32 ± 1.7 | 0.0051 | 4.04 ± 0.82 | 9.39 ± 0.65 | 0.001116 |
| LVDd/BW | 0.15 ± 0.02 | 0.15 ± 0.01 | 0.92 | 0.16 ± 0.02 | 0.14 ± 0.01 | 0.3267 | 0.16 ± 0.02 | 0.13 ± 0.01 | 0.1252 | 0.16 ± 0.02 | 0.13 ± 0.01 | 0.068602 |
| LVM(d) (mg) | 45.36 ± 3.58 | 50.97 ± 9.68 | 0.55 | 90.83 ± 15.16 | 63.70 ± 4.85 | 0.0967 | 94.87 ± 10.36 | 74.01 ± 11.5 | 0.1707 | 125.42 ± 13.14 | 70.14 ± 5.31 | 0.001530 |
| LV/BW (index%) | 0.23 ± 0.02 | 0.24 ± 0.02 | 0.66 | 0.34 ± 0.04 | 0.26 ± 0.03 | 0.0976 | 0.32 ± 0.02 | 0.27 ± 0.04 | 0.2384 | 0.38 ± 0.06 | 0.25 ± 0.03 | 0.031141 |
Echocardiographic data were obtained both from CKO and control mice at the indicated ages. Results are expressed as means ± standard errors. IVSd, interventricular septal wall thickness at end-diastole; LVIDd, left ventricular internal dimension values at end-diastole; LVIDs, left ventricular internal dimension values at end-systole; LVPWs, systolic left ventricular posterior wall thickness; % FS, percentage of fractional shortening; EDD/PWD, end-diastolic dimension/diastolic posterior wall thickness; VCF, velocity of circumferential fiber shortening; LV/BW, left ventricular mass normalized to body weight.
Inducible Cypher knockout mice exhibit similar dilated cardiomyopathy
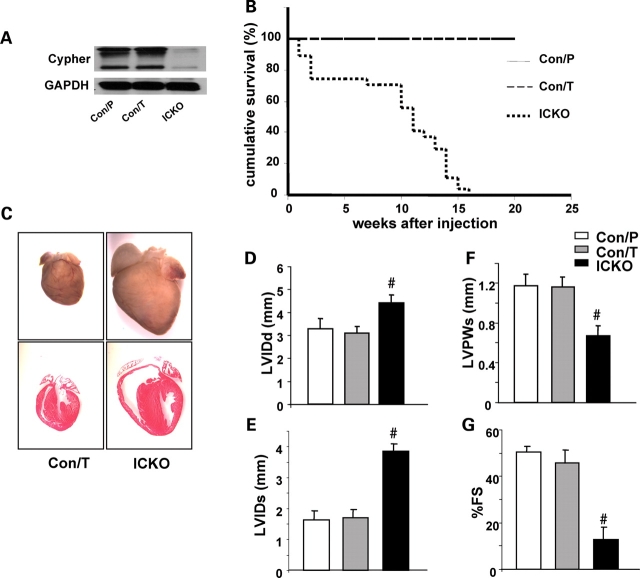
In CKO mouse heart, Cypher protein was knocked-out by Cre recombinase, mediated by the endogenous Mlc2v promoter, which drives Cre expression from embryonic day 8.75 (24). To determine whether adult cardiac dysfunction in CKO mice was due to the effect of decreased Cypher in the developing myocardium, we further generated an ICKO mouse strain, α-MHC-mER-Cre-mER/Cypherf/f to specifically explore the role of Cypher in adult myocardium. The ICKO mice were generated by crossing Cypherf/f mice with α-MHC-mER-Cre-mER mice. One month after tamoxifen injection for 5 consecutive days, expression of Cypher protein in α-MHC-mER-Cre-mER/Cypherf/f mouse heart was almost totally abolished when compared with controls of both α-MHC-mER-Cre-mER/Cypherf/f mice injected with peanut oil and Cypherf/f mice injected with tamoxifen (Fig. 4A).
Figure 4.
Inducible cardiac-specific deletion of Cypher. (A) Western blot results showing inducible deletion of Cypher in the heart of Cypherf/f/Cre mice 1 month after injection with tamoxifen [ICKO (inducible cardiac-specific CKO)] or Cypherf/f/Cre and Cypherf/f littermate mouse injected with peanut oil (Con/P) and tamoxifen (Con/T), respectively, as controls. (B) Accumulative survival curve showing that ICKO mice die from 1 week after tamoxifen injection, and all die before 15 weeks after injection, compared with 100% survival of both the control groups (n = 27 for ICKO group, n = 20 for tamoxifen control group and n = 22 for peanut oil control group). (C) Hematoxylin and eosin staining showing severe dilated ventricules in ICKO mouse heart 1 month after injection, compared with tamoxifen control mouse heart (Con/T). Echocardiography data showing (D) LVIDd, (E) LVIDs, (F) LVPWs and (G) FS results in ICKO and control littermate mice 1 month after tamoxifen or peanut oil injection (n = 4 for each group). LVIDd, end-diastolic left ventricular internal dimension; LVIDs, end-systolic left ventricular internal dimension; LVPWs, systolic left ventricular posterior wall thickness; % FS, percentage of fractional shortening; #P < 0.01.
ICKO mice began to die from 1 week after tamoxifen injection and all died before 16 weeks after injection. In contrast, 100% were alive in both control groups at least to the time indicated (Fig. 4B). DCM could be readily determined by the manifestation of a huge heart and severe dilated chambers of both left and right ventricles in ICKO mouse heart (Fig. 4C). Consistent with CKO mouse, echocardiography performed one month after treatment showed decreased cardiac function and increased chambers in ICKO mice compared with both control groups (Fig. 4D–G). Moreover, ECG analysis revealed a similar pattern in ICKO mouse as in CKO mouse (data not shown). Thus, our data obtained from the inducible Cypher mutant mice are in agreement with our observations from the CKO mouse model, showing that Cypher is essential for adult cardiac function.
Cypher knockout mice develop disruption of Z-line structure
Since we have previously shown the localization of Cypher at the Z-line, we performed ultrastructural transmission electron microscopy (TEM) to examine Z-line organization in left ventricular tissues from CKO and control littermate mice (Fig. 5). Whereas some Z-lines were still intact in 1-month-old CKO heart (Fig. 5B), cardiomyocytes from 3-month-old CKO mice had severely disorganized and disrupted Z-lines with relatively normal M-lines (Fig. 5D and F). Abnormal mitochondria were also observed in cardiac myocytes from both 1- and 3-month-old CKO mice (Fig. 5B, D and F). These observations are consistent with previous studies in conventional CKO mice, which showed ultrastructural disorganization of both skeletal and cardiac muscles at embryonic day 17.5 and postnatal day 1 (17). These data confirm the important role of Cypher in maintaining cardiac structure and function.
Figure 5.

Disrupted ultrastructure in Cypher knockout (CKO) mouse heart. Transmission electron microscopy showing some disorganized Z-lines in cardiomyocytes from CKO mice at 1 month of age while most Z-line structures remain intact (B) compared with the well-organized and condensed Z-line structures in control littermates (A). At 3 months of age, Z-lines are completely disrupted in cardiomyocytes from CKO mice (D) when compared with normal Z-lines in control littermates (C). Magnified figures to show the dispersed and punctured Z-lines better in a 3-month-old CKO mouse (inducible cardiac-specific CKO mice) compared with intact Z-lines in the control littermate (E).
Cypher interacts with calsarcin and myotilin
To understand the molecular basis for the disrupted Z-line structure in CKO mouse hearts, we next studied the effect of Cypher ablation on the protein level and localization of α-actinin, Cypher's interaction partner in the Z-line (16). As demonstrated by Western blot analysis and immunostainings, neither the protein level, nor the localization of α-actinin was altered in CKO mouse hearts (data not shown). To search for other Cypher-interacting proteins, full-length Cypher1C and Cypher2C were fused to the GAL4 DNA-binding domain and tested in the yeast two-hybrid system. Unfortunately, cypher1C caused autoactivation of the GAL4-dependent reporter genes, precluding its use as a bait. Using Cypher2C as a bait for screening of a pretransformed human cardiac muscle cDNA library, six clones were identified, all of which were confirmed by the α-galactosidase assay. Two clones encoded α-actinin-2, confirming the previously identified interaction between Cypher and α-actinin (16), while two other clones corresponded to calsarcin-1/FATZ-2/myozenin-2. The remaining two clones were leaky and appeared to be false-positive clones.
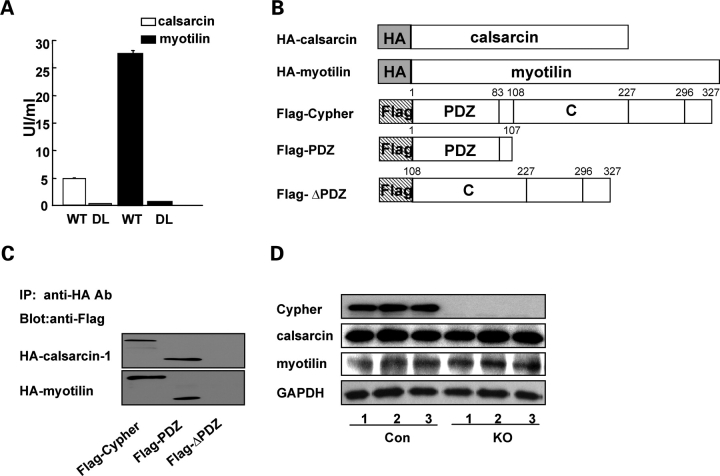
The interaction between Cypher with calsarcin-1 has previously been described (27), but the specific domains essential for the interaction have not been identified. To determine the exact interaction site for calsarcin-1 in Cypher2C, truncated constructs of Cypher2C were cotransformed with calsarcin-1. This demonstrated that the PDZ domain of Cypher is sufficient for interaction with calsarcin-1 (data not shown). It has been shown that PDZ can bind to the C-terminal sequences of proteins. One C-terminal consensus sequences for PDZ domains have been shown to be D/E-X-ψ where ψ is a hydrophobic amino acid and X is an unspecified amino acid (28). Consistent with a consensus PDZ motif, the last three amino acids of calsarcin-1 are DDL. Correspondingly, cotransformations of truncated constructs of calsarcin-1 with Cypher2C demonstrated that the C-terminal 64 amino acids of calsarcin-1 are sufficient for its binding to Cypher2C. Furthermore, deletion of the last amino acid of calsarcin-1 nearly completely ablated the binding (Fig. 6A).
Figure 6.
Cypher's PDZ domain interacts with calsarcin-1 and myotilin. (A) Deletion of the last residue leucine in calsarcin-1 (DL) and myotilin (DL) results in nearly complete ablation of their interaction with Cypher's PDZ domain in the yeast two-hybrid system. The strength of the interactions was quantified in a liquid α-galactosidase assay using PNP-α-gal as a substrate. (B) Schematic presentation of HA-tagged calsarcin-1 (HA-calsarcin) and myotilin (HA-myotilin) and Flag-tagged fragments encoding Cypher2C (Flag-Cypher), truncated Cypher2C containing the PDZ domain (Flag-PDZ), and PDZ-less Cypher2C (Flag-ΔPDZ). (C) In vivo immunoprecipitation of Cypher with calsarcin-1 and myotilin showing the interaction of Cypher with calsarcin-1 and myotilin through its PDZ domain. (D) Western blot analysis showing protein levels of Cypher, calsarcin-1 and myotilin in hearts from wild-type and conventional CKO mice at embryonic day 17.5.
To search for other ligands of Cypher's PDZ domain, we searched for consensus sequences within known Z-line proteins. This revealed the presence of a PDZ binding consensus motif (the last three amino acids are EEL) within myotilin, a Z-line protein known to interact with α-actinin, calsarcin-1, calsarcin-2/FATZ-1/myozenin-1 and filamins (29–31). The potential binding of myotilin to Cypher2C was tested in the yeast two-hybrid system, which revealed that Cypher's PDZ domain does indeed bind to myotilin. Consistently, deletion of the last amino acid in myotilin nearly completely abolished its binding to Cypher (Fig. 6A).
To further confirm the interaction of Cypher's PDZ domain with myotilin and calsarcin, HA-tagged myotilin or calsarcin were cotransfected with Flag-tagged Cypher2C (Flag-Cypher), Cypher's PDZ domain (Flag-PDZ) or PDZ-deleted Cypher (Flag-ΔPDZ) in HEK293 cells, Following immunoprecipitation, western blot analyses using anti-Flag antibody showed coprecipitation of myotilin and calsarcin both with full-length Cypher2C and the PDZ domain alone, but not with PDZ-deleted Cypher. Thus, Cypher's 107 amino acid N-terminal PDZ domain is sufficient and required for Cypher's binding to both myotilin and calsarcin (Fig. 6B and C).
To test the effect of Cypher deficiency on the expression and localization of calsarcin and myotilin, western blot analyses (Fig. 6D) and immunostainings (data not shown) were performed on hearts from conventional CKO mice and control littermates at embryonic day 17.5. This revealed no differences in the expression or localization of either calsarcin or myotilin.
Altered signals in the hearts of Cypher mutant mice
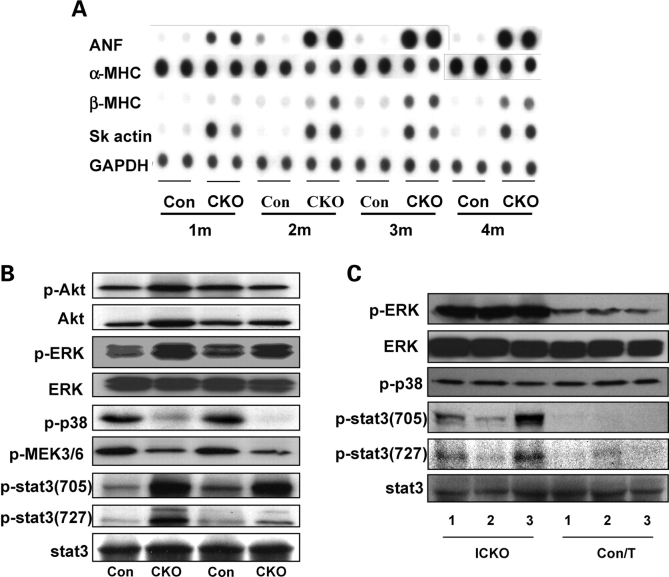
To examine cardiac hypertrophic biomarkers, dot-blot analysis was performed using mRNA isolated from hearts of CKO mice and control littermates at different ages (1, 2, 3 and 4 months of age). As expected, levels of atrial natriuretic factor, β-MHC to α-MHC ratio and skeletal muscle actin were increased at all four ages (Fig. 7A).
Figure 7.
Alterations of signals in Cypher-deficient heart. (A) Dot-blot assay showing altered expression of fetal genes in Cypher knockout (CKO) mouse compared with control littermate mouse hearts at the indicated ages. (B) Western blot analyses for activation of proteins involved in intracellular signaling in mouse hearts from CKO and control littermates at 2 months of age as well as in (C) Inducible cardiac-specific CKO mice (ICKO) and control littermates 1 month after tamoxifen injection.
We next investigated several growth-related signaling pathways. Although previous studies have suggested that the phosphatidylinositol 3-kinase–Akt signaling pathway plays a pivotal role in cardiac hypertrophy, we found no activation of Akt in CKO mouse hearts, showing that Akt is not involved in cardiac hypertrophy due to ablation of Cypher. In contrast, phosphorylation levels of two other growth-related proteins in the heart, extracellular signal-regulated kinase 1/2 (ERK1/2) and signal transducer and activator of transcription 3 (Stat3) were significantly increased (Fig. 7B), whereas phosphorylation levels of MEK3/6 and p38 MAPK were decreased in 2-month-old CKO mouse hearts (Fig. 7B). Surprisingly, in ICKO hearts 1 month after tamoxifen injection, the phosphorylation level of p38 MAPK was unaltered compared with control littermates, while phosphorylation of ERK1/2 and Stat3 were increased to levels similar to those found in CKO mouse hearts (Fig. 7C).
DISCUSSION
Cypher/ZASP is a striated muscle-specific protein localized in the sarcomeric Z-line (16,32). Our previous studies have demonstrated that conventional ablation of Cypher results in neonatal lethality of mice within 1 week after birth, caused by multiple striated muscle failure with symptoms that include decreased milk intake, limb muscle weakness and cyanosis (17). To further investigate the functional role of Cypher in the heart, we generated a conditional cardiac-specific CKO model using the Cre-loxP system. Homozygous CKO/Cre mice developed a severe form of DCM with disrupted cardiac ultrastructure, increased heart weight-to-body weight and heart weight-to-tibia bone length ratios, and decreased cardiac function, resulting in the death of all CKO mice before 5 months of age. Similar observations were made in an inducible cardiac-specific CKO mouse model. This indicates an important role of Cypher in maintaining cardiac function.
Mutations in Cypher have been found in human patients with DCM, INLVM, HCM and skeletal muscle myopathy. Consistent with the genetic and clinical observations in humans, our mouse models in which Cypher has been specifically ablated in the heart developed DCM, eventually leading to heart failure and premature death. The cardiac abnormalities induced by Cypher deficiency were not only observed in Mlc2v-cre-mediated cardiac-specific CKO mouse but also in mice in which Cypher was specifically ablated in adult myocardium, suggesting that Cypher is essential for the maintenance of cardiac function and plays a critical role in the pathogenesis of cardiac diseases, especially DCM.
A growing number of deficiencies in Z-line and Z-line-associated proteins have been found to be related to cardiomyopathy as well as skeletal muscle myopathies. In addition to Cypher/ZASP, these proteins include desmin, α-actinin, myotilin, nebulin and others (3,18,19,33–35). Studies in various model systems have revealed that a wide variety of deficiencies in distinct Z-line and Z-line-associated proteins lead to similar phenotypes, suggesting a converging mechanism shared by all of these proteins, although the exact mechanisms are still largely unknown. The Z-line not only serves as a mechanical stretch sensor that transmits structural or contractile changes to downstream signaling pathways and initiates consequent dysfunctions (36), but is essential for coordinating contraction and maintaining the cytoskeleton in muscle cells.
Previous studies have demonstrated that Cypher is localized at the Z-line where it binds to α-actinin through its PDZ domain. In the current study we find that Cypher's PDZ domain also interacts with other two Z-line-associated proteins, myotilin and calsarcin. However, although ablation of Cypher results in severe DCM, it has no effect on the expression or localization of myotilin, calsarcin and α-actinin. It should be pointed out that α-actinin is the major component of the Z-line that crosslinks and interacts with many other Z-line proteins, including myotinin and calsarcin, which also bind to each other (30,37). This suggests that α-actinin, Cypher, myotilin and calsarcin are components of a Z-line macromolecular complex, which is essential for the maintenance of Z-line structure during constant muscle contraction. Interestingly, ablation of myotilin and calsarcin also has no effect on the expression or localization of other Z-line proteins (38,39), suggesting that the Z-line macromolecular complex is different from the dystrophin–dystroglycan complex in which ablation of one component downregulates protein levels of many other components of the complex (40). Nevertheless, we speculate that although ablation of Cypher, calsarcin or myotilin has no effect on the protein levels of other components of the Z-line complex, it may weaken the protein connections within the complex, thereby disrupting the protein structure and lead to consequent cardiac dysfunction, maladaptive remodeling, signaling changes and heart failure.
DCM and heart failure are usually accompanied by activation of intracellular signaling mechanisms and cardiac remodeling. In our study, cardiac deletion of Cypher resulted in increased expression of molecular markers for cardiomyocyte hypertrophy (ANP, β-MHC/α-MHC and SK actin) and consequently mild cardiac fibrosis (data not shown). Increasing evidence shows that multiple intracellular signals are involved in the sarcomeric protein deficiency-induced cardiomyopathy (41). Among them, MAPKs are implicated to play an important role in the pathogenesis of heart failure (42). In the current study, constantly increased ERK activation was observed in both CKO and ICKO mouse hearts. Surprisingly, p38 MAPK, another MAPK family member which is usually found to be related with cardiac injury and recently with cardiac development (43–45), shows distinct activation patterns with decreased activation in CKO hearts and unaltered activation in ICKO hearts. The difference may reflect the timing of the cardiac Cypher deletion, with Mlc2v-Cre driving Cypher deletion from an early embryonic stage and α-MHC-mER-Cre-mER-mediating Cypher depletion in adulthood; however, the precise mechanism needs further elucidation.
A growing body of evidence also indicates a role of another important cardiac signaling protein, Stat 3, in cardiomyopathy. Altered Stat3 activation has been observed in patients with end-stage DCM (46). Consistent with the observations in human, transgenic mouse with cardiac-Stat3 overexpression induces a protective signal against cardiomyopathy (47), whereas cardiac-specific knockout of Stat3 results in increased susceptibility to cardiac injury, reduced cardiac function and increased mortality (48). It has also recently been shown that the specific activation of Stat3 is a potential signaling mechanism underlying the development of heart failure in the TnI-203/MHC-403 double-mutant murine model of familial hypertrophic cardiomyopathy (49). Consistent with this observation, we found increased Stat3 activation, as indicated by increased phosphorylation of Stat3, in both CKO and ICKO mouse hearts.
From the data presented here, intracellular signaling pathways associated with cardiac protection appear to be increased in the cardiac Cypher-ablated mouse heart, suggesting that these pathways may play a common role in mediating cardiac compensation and subsequently the progression to heart failure. Furthermore, the signal transduction pathways that participate in the pathogenesis of cardiac dysfunction may provide potential targets for therapeutic interventions.
In summary, cardiac-specific knockout of Cypher in mouse leads to severe cardiac dysfunction, including DCM and premature lethality from heart failure, with disrupted cellular architecture and altered activities of multiple cellular signaling pathways. Together, the cardiac dysfunction caused by Cypher deficiency likely reflects the integrated effects of impairment of sarcomeric structure and alterations in intracellular signaling cascades.
MATERIALS AND METHODS
Generation of gene-targeted mice
Floxed Cypher mice were generated by standard techniques using a targeting vector containing a neomycin selection cassette flanked by FRT sites (50). Briefly, exon 1 of Cypher was inserted into 2 flanking LoxP sites (Fig. 1A). After electroporation of the targeting vector into R1 embryonic stem (ES) cells, G418-resistant ES cells were screened for homologous recombination by Southern blot analysis as described below. One heterozyogous recombinant ES clone was identified and microinjected into blastocysts from C57BL/6J mice to generate male chimeras. Male chimeras were bred with female Black Swiss mice to generate germline transmitted floxed heterozygous mice (Cypherf/+). Cypherf/+ mice were then crossed with FLPase deleter mice to remove the neomycin gene, and subsequently intercrosses of Cypherf/+ were used to generate homozygous Cypher-floxed alleles (Cypherf/f).
To generate cardiac-specific CKO mice, Cypherf/f mice were bred with Mlc2v-Cre mice in which Cre recombinase expression was controlled by the myosin light chain 2v promoter (24) to generate double heterozygous Mlc2v-Cre and Cypher-floxed mice (Mlc2v-Cre/Cypherf/+). The mice were then backcrossed with homozygous Cypherf/f mice to generate Cypherf/f Cre+ as cardiac-specific CKO mice and Cypherf/f Cre− mice as control littermates. In a subset experiment to obtain inducible cardiac-specific CKO mice, Cypherf/f mice were crossed with α-MHC-mER-Cre-mER mice. α-MHC-mER-Cre-mER/Cypherf/f and Cypherf/f mice were treated with tamoxifen (Sigma) dissolved in peanut oil by intraperitoneal injection once a day for 5 consecutive days at a dosage of 1 mg/mouse per day, and used as inducible cardiac-specific CKO mice and control littermates, respectively. An additional control group was obtained by intraperitoneal injection of peanut oil in α-MHC-mER-Cre-mER/Cypherf/f mice.
DNA analysis
Genomic DNA was extracted from G418-resistant ES cell clones and mouse tails, as previously described (50). ES cell DNA was digested by BamHI, electrophoresed on a 1% (w/v) agarose gel and subsequently blotted onto a nitrocellulose membrane. A 400 bp fragment was generated by polymerase chain reaction (PCR) using mouse genomic DNA and specific Cypher primers (forward, GAAGGCCCTGAAGAAAGGAGAG; reverse, GACCCTCCTGTTTCTCCAGGTC). The PCR product was subsequently radiolabeled using α-[32P]dATP by random priming (Invitrogen). DNA blots were hybridized with the radiolabeled probe and visualized by autoradiography. Mice were genotyped by PCR analysis using mouse tail DNA, Cypher primers (forward, ATCAGTGTCGATGGCATTCA; reverse, TGCAGGCAAACAGTACGAAG) and Cre primers (forward, GTTCGCAAGAACCTGATGGACA; reverse, CTAGAGCCTGTTTTGCACGTTC) were used to detect floxed-Cypher and Cre, respectively.
Plasmid construction for yeast two-hybrid analysis
For yeast two-hybrid analysis, full-length cypher1C (Genbank: AF114378) and full-length cypher2C (Genbank: AY206011) were amplified from a mouse heart cDNA library by PCR using the primers: P1 (5′-cggcgaattcATGTCTTACAGTGTGACTCTGAC-3′) and P2 (5′-ctcagtcgacgaGCTCTTAGCTTTTAACATTAAGG-3′) and cloned into yeast-two hybrid vector pGBKT7. Full-length calsarcin-1 (Genbank: BC005195) was amplified from a positive clone obtained from a yeast two-hybrid screen using the primers: P3 (5′-aggccgaattcATGCTATCACATAATACTATGATGAAGC-3′) and P4 (5′-aatgtcgacgagctcGCACATACAACTTTCTT TTCATAGG-3′) and full-length mouse myotilin (Genbank: BC016214) was amplified from ATCC clone MCG-28747 using primers P5 (5′-ccaagaattcATGTTTAACTACGAACGTCC-3′) and P6 (5′-gtaaaggatccAAGTTCTTCACTTTCATAGAG-3′). All the fragments were cloned into the yeast-two hybrid vectors pGADT7 and/or pGADT7. Truncated constructs of calsarcin-1 and myotilin were generated by PCR from full-length constructs and cloned into pGADT7.
For co-immunoprecipitations, full-length calsarcin and myotilin were cloned into the mammalian expression vector pXJ40-HA, while full-length and truncated Cypher2C was inserted into the pXJ40-Flag vector (both kindly provided by Dr Tong Zhang). All constructs were checked by sequencing.
Yeast two-hybrid studies
Full-length human cypher2C cDNA, fused to the GAL4 DNA-binding domain (pGBKT7-2C), was used as a bait in a yeast two-hybrid screen of approximately 4 × 106 clones from a pretransformed human heart cDNA library (Matchmaker 638833, Clontech) according to the instructions provided by the manufacturer. Briefly, positive clones growing on selective plates, lacking histidine, leucine, adenine and tryptophan, were picked and screened for α-galactosidase activity by plating onto selective plates containing X-α-galactosidase. Plasmid DNA from α-galactosidase-positive clones were isolated as described by the manufacturer and sequenced. To confirm interactions, obtained clones were retransformed into yeast with cypher2C or empty pGBKT7 vector. In addition, inserts of pGBKT7 and pGADT7 were swapped and cotransformed into yeast. To pinpoint the interaction sites, truncated or mutant constructs of cypher2C, calsarcin-1 and myotilin were cotransformed into yeast and positive clones were analyzed for α-galactosidase activity in liquid culture using PNP-α-gal as a substrate (described in the yeast protocols handbook, PT3024-1, Clontech).
Western blot analysis
To assess protein expression of cardiac isoforms of Cypher, western blots analyses were performed on heart lysates from CKO or ICKO mice and control mice using affinity-purified Cypher antibodies as described previously (17). In subset experiments, antibodies against calsarcin-1 (Alpha diagnostic international), myotilin (kindly provided by Olli Carpen), α-actinin (Sigma), Flag(Sigma), HA (Santa Cruz), phospho-p44/42 MAPK (Thr202/Tyr204), phospho-p38 and phospho-stat3 (Cell Signaling Technology) were used as described (17,51). GAPDH antibody was used for normalization.
In vivo co-immunoprecipitation
Two hundred and ninety-three cells were harvested 36 h after co-transfection with constructs generated in the pXJ40-flag and pXJ40-HA vectors at a concentration of 16 µg/100 mm plate, using the Hyfect reagent according to the manufacturer's instruction. Cell lysate (600 µg total protein) was immunoprecipitated with 2 µg HA polyclonal antibody and 20 µl Protein G Sepharose 4 Fast Flow beads (Amersham Biosciences), and subjected to western blot analysis using anti-flag M2 monoclonal antibody (1:20,000).
Hematoxylin & eosin staining
Hearts were harvested, relaxed in 50 mm KCl in phosphate-buffered saline, fixed in 4% paraformaldehyde, embedded in paraffin and sectioned into 7-µm thick sections. Hematoxylin and eosin (H&E) staining was performed according to standard procedures.
Transmission electron microscopy
Hearts were perfused with 50 mm KCl in tyrode solution followed by 2% PFA and 2% glutaraldehyde in 0.15 m Na cacodylate buffer. The tissues were processed for TEM as described previously (17).
RNA dot-blot
Total RNA was isolated from ventricles using Trizol reagent (Invitrogen) following the manufacturer's protocol. RNA dot-blot was performed as described (52). Briefly, 2 mg denatured RNA of each sample was blotted on a nitrocellulose membrane and cross-linked by UV. The membrane was then probed with α-[32P]dATP-labeled probes and signals were visualized by autoradiography. GAPDH mRNA levels were detected as RNA loading control.
Echocardiography analysis
Mice were anaesthetized with isoflurane and subjected to echocardiography as previously described (53).
Electrocardiography analysis
Surface electrocardiogram recordings were performed on mice anaesthetized with 1–1.5% isoflurane. Needle electrodes were used and placed in the conventional lead II position. A differential amplifier (DP-304, Warner Instruments Corp, Hamden, CT, USA) amplified the signals in the bandwidth of 0.1–1000 Hz, and signals were filtered using an adaptive 60 Hz filter (Humbug, Quest Scientific, Vancouver, Canada). The signals were digitized at 3000 Hz, and analyzed using ECG Analysis Software (QRS phenotyping, Calgary, Canada).
Statistical analysis
Statistical analyses were done using Student's unpaired t-test. P < 0.05 was considered statistically significant. Data are represented as means ± standard errors of the means.
FUNDING
Grants from NIH, MDA and Children's Cardiomyopathy Foundation to J.C., partially by National Science Foundation of China (30770870 and 30500182) and National Key Basic Research Program of China (2007CB512100) to M.Z.
ACKNOWLEDGEMENTS
We thank Dr F Sheikh for critical reading of the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Ortiz-Lopez R., Li H., Su J., Goytia V., Towbin J.A. Evidence for a dystrophin missense mutation as a cause of X-linked dilated cardiomyopathy. Circulation. 1997;5:2434–2440. doi: 10.1161/01.cir.95.10.2434. [DOI] [PubMed] [Google Scholar]
- 2.Olson T.M., Illenberger S., Kishimoto N.Y., Huttelmaier S., Keating M.T., Jockusch B.M. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation. 2002;105:431–437. doi: 10.1161/hc0402.102930. [DOI] [PubMed] [Google Scholar]
- 3.Li D., Tapscoft T., Gonzalez O., Burch P.E., Quiñones M.A., Zoghbi W.A., Hill R., Bachinski L.L., Mann D.L., Roberts R. Desmin mutation responsible for idiopathic dilated cardiomyopathy. Circulation. 1999;100:461–464. doi: 10.1161/01.cir.100.5.461. [DOI] [PubMed] [Google Scholar]
- 4.Gerull B., Gramlich M., Atherton J., McNabb M., Trombitás K., Sasse-Klaassen S., Seidman J.G., Seidman C., Granzier H., Labeit S., et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat. Genet. 2002;30:201–204. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- 5.Olson T.M., Michels V.V., Thibodeau S.N., Tai Y.S., Keating M.T. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science. 1998;280:750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- 6.Villard E., Duboscq-Bidot L., Charron P., Benaiche A., Conraads V., Sylvius N., Komajda M. Mutation screening in dilated cardiomyopathy: prominent role of the beta myosin heavy chain gene. Eur. Heart J. 2005;26:794–803. doi: 10.1093/eurheartj/ehi193. [DOI] [PubMed] [Google Scholar]
- 7.Morimoto S., Lu Q.W., Harada K., Takahashi-Yanaga F., Minakami R., Ohta M., Sasaguri T., Ohtsuki I. Ca(2+)-desensitizing effect of a deletion mutation Delta K210 in cardiac troponin T that causes familial dilated cardiomyopathy. Proc. Natl Acad. Sci. USA. 2002;99:913–918. doi: 10.1073/pnas.022628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knöll R., Hoshijima M., Hoffman H.M., Person V., Lorenzen-Schmidt I., Bang M.L., Hayashi T., Shiga N., Yasukawa H., Schaper W., et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 9.Kamisago M., Sharma S.D., DePalma S.R., Solomon S., Sharma P., McDonough B., Smoot L., Mullen M.P., Woolf P.K., Wigle E.D., et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N. Engl. J. Med. 2000;343:1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- 10.Robinson P., Mirza M., Knott A., Abdulrazzak H., Willott R., Marston S., Watkins H., Redwood C. Alterations in thin filament regulation induced by a human cardiac troponin T mutant that causes dilated cardiomyopathy are distinct from those induced by troponin T mutants that cause hypertrophic cardiomyopathy. J. Biol. Chem. 2002;277:40710–40716. doi: 10.1074/jbc.M203446200. [DOI] [PubMed] [Google Scholar]
- 11.Chen J., Chien K.R. Complexity in simplicity: monogenic disorders and complex cardiomyopathies. J. Clin. Invest. 1999;103:1483–1485. doi: 10.1172/JCI7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seidman J.G., Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 13.Towbin J.A., Bowles N.E. Dilated cardiomyopathy: a tale of cytoskeletal proteins and beyond. J. Cardiovasc. Electrophysiol. 2006;17:919–926. doi: 10.1111/j.1540-8167.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- 14.Lapidos K.A., Kakkar R., McNally E.M. The dystrophin glycoprotein complex: signaling strength and integrity for the sarcolemma. Circ. Res. 2004;94:1023–1031. doi: 10.1161/01.RES.0000126574.61061.25. [DOI] [PubMed] [Google Scholar]
- 15.Dellefave L., McNally E.M. Sarcomere mutations in cardiomyopathy, noncompaction, and the developing heart. Circulation. 2008;117:2847–2849. doi: 10.1161/CIRCULATIONAHA.108.781518. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Q., Ruiz-Lozano P., Martone M.E., Chen J. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J. Biol. Chem. 1999;274:19807–19813. doi: 10.1074/jbc.274.28.19807. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Q., Chu P.H., Huang C., Cheng C.F., Martone M.E., Knoll G., Shelton G.D., Evans S., Chen J. Ablation of Cypher, a PDZ-LIM domain Z-line protein, causes a severe form of congenital myopathy. J. Cell. Biol. 2001;155:605–612. doi: 10.1083/jcb.200107092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vatta M., Mohapatra B., Jimenez S., Sanchez X., Faulkner G., Perles Z., Sinagra G., Lin J.H., Vu T.M., Zhou Q., et al. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J. Am. Coll. Cardiol. 2003;42:2014–2027. doi: 10.1016/j.jacc.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 19.Xing Y., Ichida F., Matsuoka T., Isobe T., Ikemoto Y., Higaki T., Tsuji T., Haneda N., Kuwabara A., Chen R., et al. Genetic analysis in patients with left ventricular noncompaction and evidence for genetic heterogeneity. Mol. Genet. Metab. 2006;88:71–77. doi: 10.1016/j.ymgme.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Arimura T., Hayashi T., Terada H., Lee S.Y., Zhou Q., Takahashi M., Ueda K., Nouchi T., Hohda S., Shibutani M., et al. A Cypher/ZASP mutation associated with dilated cardiomyopathy alters the binding affinity to protein kinase C. J. Biol. Chem. 2004;279:6746–6752. doi: 10.1074/jbc.M311849200. [DOI] [PubMed] [Google Scholar]
- 21.Theis J.L., Bos J.M., Bartleson V.B., Will M.L., Binder J., Vatta M., Towbin J.A., Gersh B.J., Ommen S.R., Ackerman M.J. Echocardiographic-determined septal morphology in Z-disc hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 2006;351:896–902. doi: 10.1016/j.bbrc.2006.10.119. [DOI] [PubMed] [Google Scholar]
- 22.Sheikh F., Bang M.L., Lange S., Chen J. ‘Z’eroing in on the role of Cypher in striated muscle function, signaling, and human disease. Trends Cardiovasc. Med. 2007;17:258–262. doi: 10.1016/j.tcm.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selcen D., Engel A.G. Mutations in ZASP define a novel form of muscular dystrophy in humans. Ann. Neurol. 2005;57:269–276. doi: 10.1002/ana.20376. [DOI] [PubMed] [Google Scholar]
- 24.Chen J., Kubalak S.W., Minamisawa S., Price R.L., Becker K.D., Hickey R., Ross J.J., Chien K.R. Selective requirement of myosin light chain 2v in embryonic heart function. J. Biol. Chem. 1998;273:1252–1256. doi: 10.1074/jbc.273.2.1252. [DOI] [PubMed] [Google Scholar]
- 25.Sohal D.S., Nghiem M., Crackower M.A., Witt S.A., Kimball T.R., Tymitz K.M., Penninger J.M., Molkentin J.D. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ. Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 26.Huang C., Zhou Q., Liang P., Hollander M.S., Sheikh F., Li X., Greaser M., Shelton G.D., Evans S., Chen J. Characterization and in vivo functional analysis of splice variants of Cypher. J. Biol. Chem. 2003;278:7360–7365. doi: 10.1074/jbc.M211875200. [DOI] [PubMed] [Google Scholar]
- 27.Frey N., Olson E.N. Calsarcin-3, a novel skeletal muscle-specific member of the calsarcin family, interacts with multiple Z-disc proteins. J. Biol. Chem. 2002;277:13998–14004. doi: 10.1074/jbc.M200712200. [DOI] [PubMed] [Google Scholar]
- 28.Nourry C., Grant S.G., Borg J.P. PDZ domain proteins: plug and play. Sci. STKE. 2003;179:RE7. doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- 29.Salmikangas P., Mykkänen O.M., Grönholm M., Heiska L., Kere J., Carpén O. Myotilin, a novel sarcomeric protein with two Ig-like domains, is encoded by a candidate gene for limb-girdle muscular dystrophy. Hum. Mol. Genet. 1999;8:1329–1336. doi: 10.1093/hmg/8.7.1329. [DOI] [PubMed] [Google Scholar]
- 30.Gontier Y., Taivainen A., Fontao L., Sonnenberg A., van der Flier A., Carpen O., Faulkner G., Borradori L. The Z-disc proteins myotilin and FATZ-1 interact with each other and are connected to the sarcolemma via muscle-specific filamins. J. Cell. Sci. 2005;118:3739–3749. doi: 10.1242/jcs.02484. [DOI] [PubMed] [Google Scholar]
- 31.van der Ven P.F., Wiesner S., Salmikangas P., Auerbach D., Himmel M., Kempa S., Hayess K., Pacholsky D., Taivainen A., Schröder R., et al. Indications for a novel muscular dystrophy pathway: gamma-filamin, the muscle-specific filamin isoform, interacts with myotilin. J. Cell. Biol. 2000;151:235–248. doi: 10.1083/jcb.151.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faulkner G., Pallavicini A., Formentin E., Comelli A., Ievolella C., Trevisan S., Bortoletto G., Scannapieco P., Salamon M., Mouly V., et al. ZASP: a new Z-band alternatively spliced PDZ-motif protein. J. Cell. Biol. 1999;146:465–475. doi: 10.1083/jcb.146.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hauser M.A., Horrigan S.K., Salmikangas P., Torian U.M., Viles K.D., Dancel R., Tim R.W., Taivainen A., Bartoloni L., Gilchrist J.M., et al. Myotilin is mutated in limb girdle muscular dystrophy 1A. Hum. Mol. Genet. 2000;9:2141–2147. doi: 10.1093/hmg/9.14.2141. [DOI] [PubMed] [Google Scholar]
- 34.Selcen D., Engel A.G. Mutations in myotilin cause myofibrillar myopathy. Neurology. 2004;62:1363–1371. doi: 10.1212/01.wnl.0000123576.74801.75. [DOI] [PubMed] [Google Scholar]
- 35.Lehtokari V.L., Pelin K., Sandbacka M., Ranta S., Donner K., Muntoni F., Sewry C., Angelini C., Bushby K., Van den Bergh P., et al. Identification of 45 novel mutations in the nebulin gene associated with autosomal recessive nemaline myopathy. Hum. Mutat. 2006;27:946–956. doi: 10.1002/humu.20370. [DOI] [PubMed] [Google Scholar]
- 36.Chang A.N., Potter J.D. Sarcomeric protein mutations in dilated cardiomyopathy. Heart Fail Rev. 2005;10:225–235. doi: 10.1007/s10741-005-5252-6. [DOI] [PubMed] [Google Scholar]
- 37.Sanger J.M., Sanger J.W. The dynamic Z bands of striated muscle cells. Sci. Signal. 2008;1:e37. doi: 10.1126/scisignal.132pe37. [DOI] [PubMed] [Google Scholar]
- 38.Moza M., Mologni L., Trokovic R., Faulkner G., Partanen J., Carpén O. Targeted deletion of the muscular dystrophy gene myotilin does not perturb muscle structure or function in mice. Mol. Cell. Biol. 2007;27:244–252. doi: 10.1128/MCB.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frey N., Barrientos T., Shelton J.M., Frank D., Rütten H., Gehring D., Kuhn C., Lutz M., Rothermel B., Bassel-Duby R., et al. Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat. Med. 2004;10:1336–1343. doi: 10.1038/nm1132. [DOI] [PubMed] [Google Scholar]
- 40.Michele D.E., Campbell K.P. Dystrophin-glycoprotein complex: post-translational processing and dystroglycan function. J. Biol. Chem. 2003;278:15457–15460. doi: 10.1074/jbc.R200031200. [DOI] [PubMed] [Google Scholar]
- 41.Solaro R.J. Multiplex kinase signaling modifies cardiac function at the level of sarcomeric proteins. J. Biol. Chem. 2008;283:26829–26833. doi: 10.1074/jbc.R800037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ravingerová T., Barancík M., Strnisková M. Mitogen-activated protein kinases: a new therapeutic target in cardiac pathology. Mol. Cell. Biochem. 2003;247:127–138. doi: 10.1023/a:1024119224033. [DOI] [PubMed] [Google Scholar]
- 43.Hernández-Torres F., Martínez-Fernández S., Zuluaga S., Nebreda A., Porras A., Aránega A.E., Navarro F. A role for p38alpha mitogen-activated protein kinase in embryonic cardiac differentiation. FEBS Lett. 2008;582:1025–1031. doi: 10.1016/j.febslet.2008.02.050. [DOI] [PubMed] [Google Scholar]
- 44.Aouadi M., Bost F., Caron L., Laurent K., Le Marchand Brustel Y., Binétruy B. p38 mitogen-activated protein kinase activity commits embryonic stem cells to either neurogenesis or cardiomyogenesis. Stem Cells. 2006;24:1399–1406. doi: 10.1634/stemcells.2005-0398. [DOI] [PubMed] [Google Scholar]
- 45.Petrich B.G., Wang Y. Stress-activated MAP kinases in cardiac remodeling and heart failure; new insights from transgenic studies. Trends Cardiovasc. Med. 2004;14:50–55. doi: 10.1016/j.tcm.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 46.Podewski E.K., Hilfiker-Kleiner D., Hilfiker A., Morawietz H., Lichtenberg A., Wollert K.C., Drexler H. Alterations in Janus kinase (JAK)-signal transducers and activators of transcription (STAT) signaling in patients with end-stage dilated cardiomyopathy. Circulation. 2003;107:798–802. doi: 10.1161/01.cir.0000057545.82749.ff. [DOI] [PubMed] [Google Scholar]
- 47.Kunisada K., Negoro S., Tone E., Funamoto M., Osugi T., Yamada S., Okabe M., Kishimoto T., Yamauchi-Takihara K. Signal transductor and activator of transcription 3 in the heart transduces not only a hypertrophic signal but a protective signal against doxorubicin induced cardiomyopathy. Proc. Natl Acad. Sci. USA. 2000;97:315–319. doi: 10.1073/pnas.97.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacoby J.J., Kalinowski A., Liu M.G., Zhang S.S., Gao Q., Chai G.X., Ji L., Iwamoto Y., Li E., Schneider M., et al. Cardiomyocyte-restricted knockout of STAT3 results in higher sensitivity to inflammation, cardiac fibrosis, and heart failure with advanced age. Proc. Natl Acad. Sci. USA. 2003;100:12929–12934. doi: 10.1073/pnas.2134694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsoutsman T., Kelly M., Ng D.C., Tan J.E., Tu E., Lam L., Bogoyevitch M.A., Seidman C.E., Seidman J.G., Semsarian C. Severe heart failure and early mortality in a double-mutation mouse model of familial hypertrophic cardiomyopathy. Circulation. 2008;117:1820–1831. doi: 10.1161/CIRCULATIONAHA.107.755777. [DOI] [PubMed] [Google Scholar]
- 50.Li X., Zima A.V., Sheikh F., Blatter L.A., Chen J. Endothelin-1-induced arrhythmogenic Ca2+ signaling is abolished in atrial myocytes of inositol-1,4,5-trisphosphate(IP3)-receptor type 2-deficient mice. Circ. Res. 2005;96:1274–1281. doi: 10.1161/01.RES.0000172556.05576.4c. [DOI] [PubMed] [Google Scholar]
- 51.Zheng M., Reynolds C., Jo S.H., Wersto R., Han Q., Xiao R.P. Intracellular acidosis-activated p38 MAPK signaling and its essential role in cardiomyocyte hypoxic injury. FASEB J. 2005;19:109–111. doi: 10.1096/fj.04-2607fje. [DOI] [PubMed] [Google Scholar]
- 52.Jones W.K., Grupp I.L., Doetschman T., Grupp G., Osinska H., Hewett T.E., Boivin G., Gulick J., Ng W.A., Robbins J. Ablation of the murine alpha myosin heavy chain gene leads to dosage effects and functional deficits in the heart. J. Clin. Invest. 1996;98:1906–1917. doi: 10.1172/JCI118992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka N., Dalton N., Mao L., Rockman H.A., Peterson K.L., Gottshall K.R., Hunter J.J., Chien K.R., Ross J., Jr Transthoracic echocardiography in models of cardiac disease in the mouse. Circulation. 1996;94:1109–1117. doi: 10.1161/01.cir.94.5.1109. [DOI] [PubMed] [Google Scholar]