Abstract
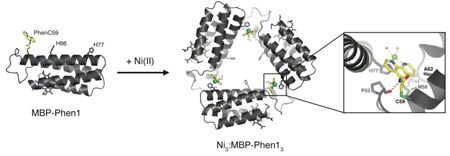
We previously devised a strategy (Metal-Directed Protein Self-Assembly, MDPSA) that utilizes the simultaneous stability, lability and directionality of metal-ligand bonds to drive protein-protein interactions. Here we show that both the structural and the functional scope of MDPSA can be broadened by non-natural metal chelating ligands incorporated onto protein surfaces. A cytochrome cb562 variant, MBP-Phen1, which features a covalently attached phenanthroline (Phen) group on its surface, self-assembles into an unusual triangular architecture (Ni3:MBP-Phen13) upon binding Ni, owing to specific Phenprotein interactions. The crystal structure of Ni3:MBP-Phen13 reveals that the Phen group is buried in a small pocket on the protein surface, which results in an unsaturated Ni coordination environment.
Nature utilizes proteins as building blocks to construct a wide variety of self-assembled nanoscale architectures. Despite advances in protein design and engineering,1 attaining the structural and functional sophistication of such multi-protein architectures remains a distant goal. To bypass the immense challenge of controlling the non-covalent interactions that hold these assemblies together, we devised a strategy, which we now call “Metal-Directed Protein Self-Assembly” (MDPSA), that utilizes the simultaneous stability, lability and directionality of metal-ligand bonds to drive protein-protein interactions.2 The use of metal coordination to control protein self-assembly is attractive from both structural and functional perspectives: whereas the directionality and symmetry inherent in metal coordination can govern overall supramolecular geometry, the resulting interfacial metal centers may potentially offer new reactivities within biological scaffolds. With these advantages in mind, we asked whether the structural and functional scope of MDPSA can be further augmented with non-natural metal ligands. Here we report the Ni-dependent self-assembly properties of a cyt cb562 variant, MBP-Phen1, which features a covalently attached phenanthroline (Phen) group on its surface. MBP-Phen1 not only forms an unusual supramolecular architecture owing to specific Phen-protein interactions, but also presents coordinatively unsaturated Ni centers within this assembly.
To site-selectively nucleate metal binding on protein surfaces, we previously adopted the strategy of employing multidentate motifs (e.g., an i, i+4 di-His arrangement on a helix) to outcompete the mostly monodentate sidechain functionalities for metal coordination.2 We then imagined that non-natural multidentate motifs such as Phen with a single-point attachment would offer more structural flexibility than a di-His motif while also allowing non-proteinaceous functionalities to be incorporated into protein assemblies.3 A cysteine-specific iodoacetamide derivative of Phen (IA-Phen) has previously been used to generate stable metal binding sites on proteins.4 Using IA-Phen, we constructed MBP-Phen1, which contains a single Phen group covalently-bound to Cys59 (PhenC59) near the N-terminus of Helix3, and His77 incorporated at the opposite end to induce head-to-tail arrangements (Fig. S1a). Additionally, MBP-Phen1 contains a His at position 66 (two helix turns away from Cys59) with the idea that this residue could potentially form a i, i +7 tridentate acceptor motif together with PhenC59.
Titrations with late first-row transition metals indicated that Ni(II), in particular, tightly associates with MBP-Phen1, and leads to significant enhancement in overall protein stability, likely through i, i+7 His66-PhenC59 crosslinking (Fig. S2 and Fig. S3). The multitude of possible Ni-mediated protein oligomerization modes (Fig. S1b) and the non-negligible interactions that could be formed between protein surfaces in these states make the a priori assignment of the thermodynamically preferred superstructure challenging. Therefore, we sought to determine the crystal structure of the Ni adduct of MBP-Phen1.
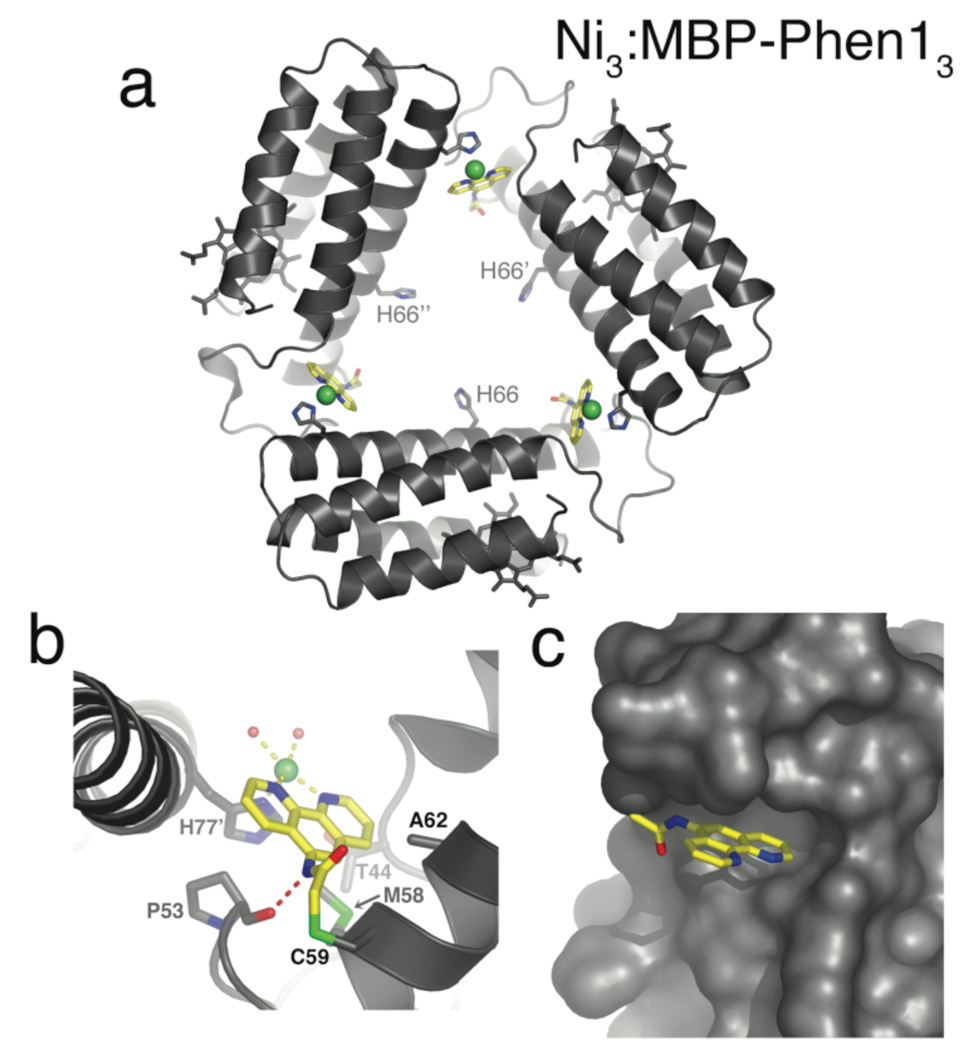
Crystals of MBP-Phen1 were obtained in two spacegroups (P21 and P6322) from two similar but different solution conditions containing equimolar protein and Ni, and their structures were determined at 2.4 and 3.15 Å resolution, respectively. Both structures (PDB IDs: 3FOO, 3FOP) reveal a unique triangular assembly, Ni3:MBP-Phen13, with perfect C3 symmetry (Fig. 1). Each vertex of this triangle is formed by a Ni coordinated to PhenC59 from one protein monomer and His77 (Nδ) from another (Fig. 1b), whereby the three Ni’s lie on the plane of the triangle, approximately 30 Å from one another. Ni-protein coordination appears to be the primary driving force for self-assembly, as the docking interactions between each protein monomer are minimal and non-specific.
Figure 1.
(a) Topview of Ni3:MBP-Phen13. Ni ions are shown as green spheres, and PhenC59 is highlighted in yellow. (b) Coordination environment of Ni-PhenC59. The H-bond between the P53 carbonyl and the PhenC59 amide nitrogen is indicated with a red dashed line. Aquo/chloride ligands are shown as red spheres. (c) Burial of PhenC59 under the 50’s loop.
The Phen group, instead of extending into the solvent, is positioned in a small hydrophobic crevice underneath the 50’s loop, further stabilized by a H-bond between the PhenC59 amide nitrogen and the Pro53 backbone carbonyl (Figs. 1b, c). In accord with these favorable interactions, MBP-Phen1 is found to be ~1.5 kcal/mol more stable than its unlabeled counterpart through unfolding titrations (Fig. S2). The placement of Phen is the key to the open Ni3:MBP-Phen13 architecture: it protects the Ni ion from coordination by a second Phen group (or His66) and allows only one other protein monomer to coordinate through His77 in the cis position, which ultimately results in an unsaturated, roughly square-pyramidal Ni coordination geometry. While His77 (dNi-N = 2.1 ± 0.1 Å) and PhenC59 (dNi-N1/N2 = 2.1 ± 0.1 Å) are clearly defined in the electron density maps (Fig. S.11), the two other coordination sites cannot be unambiguously assigned due to the resolution limits. We tentatively ascribe the diffuse electron density near Ni to two aquo/chloride ligands coordinated trans to PhenC59 at distances of 2.8 and 2.6 (± 0.3) Å from Ni and 2.7 (± 0.2) Å from each other, averaged over the twelve independent metal sites in the asymmetric unit of P21 crystals (see Supp. Info. for a detailed discussion).
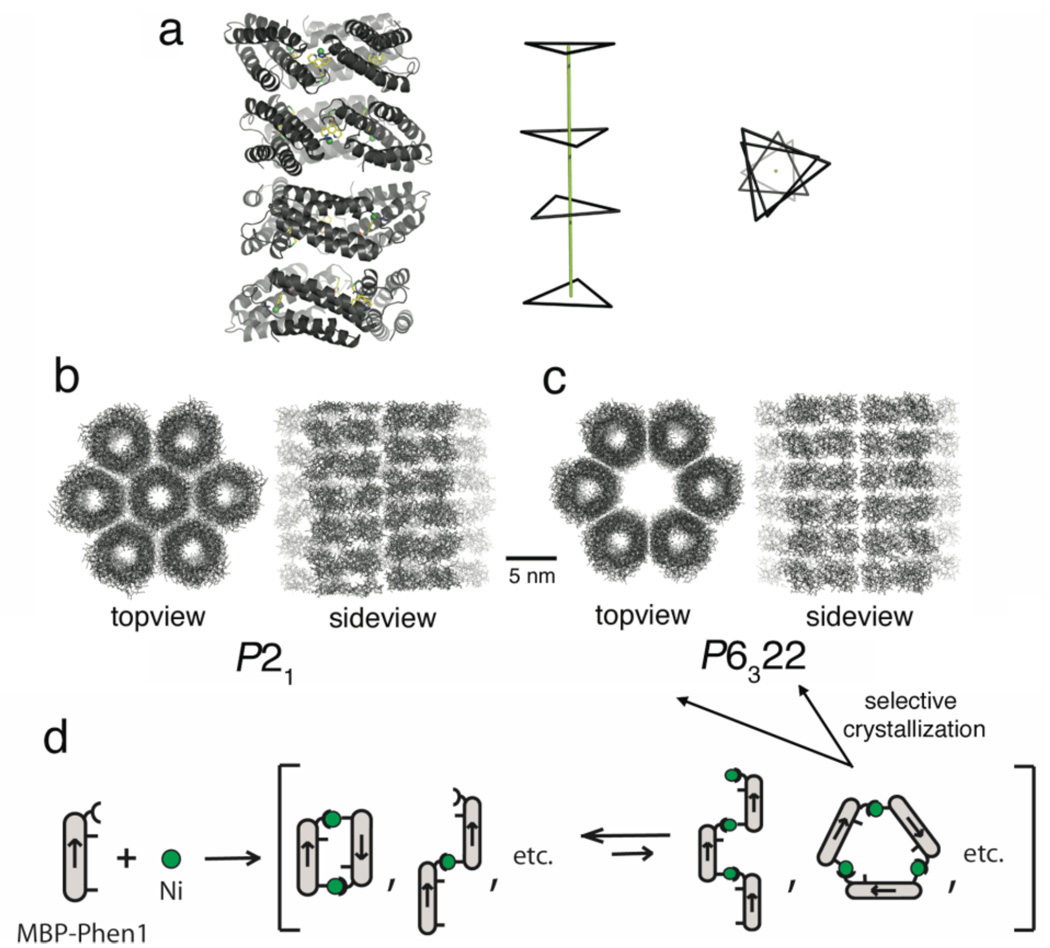
Lattice packing interactions in both Ni3:MBP-Phen13 crystals are particularly noteworthy. In the P21 form, there are four crystallographically distinct, but identical (overall rmsd-Cα= 0.3 Å) copies of the trimer in the asymmetric unit, which stack up along their threefold symmetry axes to form a tubular architecture (Fig. 2a). Each of the four Ni3:MBP-Phen13 trimers adopts a different orientation around the long axis of the tube, giving rise to three distinct trimer-trimer interactions. In the lattice, the tubular units are further stacked end-on-end infinitely, which, due to the superposition of the four different trimer orientations, adopt an apparent hexagonal geometry. The resulting hexagonal tubes form a tightly-packed 2-D array (50% solvent content) (Fig. 2b). In the P6322 crystals, the Ni3:MBP-Phen13 trimers are similarly arranged to form hexagonal tubular structures (Fig. 2c). In contrast to the P21 lattice, the trimers of all adjacent tubes are coplanar, which is required for generating the two-, three- and six-fold symmetry components of the P6322 spacegroup. Moreover, a central hexagonal tube is not accommodated in this lattice, leading to a large cavity of 6-nm diameter and an increased crystal solvent content of 64%. Though only observed in crystals, such arrangements suggest that open, symmetrical protein superstructures such as Ni3:MBP-Phen13 could be in principle be utilized as building blocks for porous protein frameworks.
Figure 2.
(a) Four Ni3:MBP-Phen13 trimers in the asymmetric unit of the P21 crystals, and their triangular representation (Ni’s as vertices) viewed from the side and the top. (b,c) Lattice packing arrangement of Ni3:MBP-Phen13 in P21 and P6322 crystal forms. (d) Suggested Ni-induced oligomerization behavior of MBP-Phen1 in solution.
The fact that the same Ni3:MBP-Phen13 structure is found in several different lattice packing environments in two crystal forms provides strong evidence for its existence in solution and its rigidity. Sedimentation velocity measurements, on the other hand, indicate that the predominant species at low protein/Ni concentrations (<1 mM) is dimeric (Fig. S10). We suggest that the trimeric forms (including Ni3:MBP-Phen13), which should be entropically disfavored compared to any dimeric species, are only significantly populated at high protein concentrations required for crystallization.5 The high internal symmetry of Ni3:MBP-Phen13 and its rigidity likely promote its selective crystallization from among other species that coexist in solution (Fig. 2d). Studies are currently underway to stabilize Ni3:MBP-Phen13 and destabilize other possible conformations through surface engineering, such that it can be isolated in solution and the reactivity of the interfacial Ni centers may be assessed.6
Synthetic metal coordinating functionalities have previously been employed for stabilizing coiled-coil assemblies,7 constructing reactive metal binding sites in protein interiors,4a and tuning the potentials of redox centers,8 among others.3 We have demonstrated here that such non-natural ligands incorporated onto protein surfaces can lead to novel biological architectures as well as coordinatively unsaturated metal sites within these scaffolds. The wide array of functionalities available in the synthetic inorganic chemist’s toolbox thus could provide a powerful means to generate structural and functional diversity in protein self-assembly.
Supplementary Material
Acknowledgment
This work was supported by UCSD, a Beckman Young Investigator Award (F.A.T.), and an NIH Heme and Blood Program Training Grant (R.J.R). Portions of this research were carried out at SSRL, operated by Stanford University on behalf of DOE.
Footnotes
Supporting Information Available: Additional experimental details and discussions. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Ghirlanda G, Lear JD, Ogihara NL, Eisenberg D, DeGrado WF. J. Mol. Biol. 2002;319:243–253. doi: 10.1016/S0022-2836(02)00233-4. [DOI] [PubMed] [Google Scholar]; (b) Andre I, Bradley P, Wang C, Baker D. Proc. Natl. Acad. Sci. USA. 2007;104:17656–17661. doi: 10.1073/pnas.0702626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Salgado EN, Faraone-Mennella J, Tezcan FA. J. Am. Chem. Soc. 2007;129:13374–13375. doi: 10.1021/ja075261o. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Salgado EN, Lewis RA, Faraone-Mennella J, Tezcan FA. J. Am. Chem. Soc. 2008;130:6082–6084. doi: 10.1021/ja8012177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu Y. Curr. Opin. Chem. Biol. 2005;9:118–126. doi: 10.1016/j.cbpa.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 4.(a) Qi DF, Tann CM, Haring D, DiStefano MD. Chem. Rev. 2001;101:3081–3111. doi: 10.1021/cr000059o. [DOI] [PubMed] [Google Scholar]; (b) Chen CHB, Milne L, Landgraf R, Perrin DM, Sigman DS. ChemBioChem. 2001;2:735–740. doi: 10.1002/1439-7633(20011001)2:10<735::aid-cbic735>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 5.Since the dimeric and trimeric species are in equilbrium as dictated by the exchange lability of Ni(II), we carried out kinetic trapping experiments to isolate any metal-induced oligomeric forms. To this end, we alternately utilized a) homobifunctional imidoester crosslinkers (DMS, DMP, DMA) aimed at linking Lys pairs across protein interfaces, and b) Ru(II) as a substitution- inert surrogate for Ni(II) (Supp. Info). In accord with the scenario in Fig. 2d, electrophoresis results indicate that all three crosslinkers and Ru(II) lead to the capture of predominantly dimeric and some trimeric products. In the case of the covalent crosslinkers, Ni(II) is required for trapping both dimeric and trimeric species. We subsequently isolated the trimeric species from Ru- as well as DMS-treated MBP-Phen1 samples by chromatography, and subjected them to metal analysis and hydrodynamic measurements. ICP-OES measurements on thoroughly dialyzed samples yield Ni:protein and Ru:protein ratios of 1.1 and 1.4 for the DMS- and Ru-crosslinked species. Sedimentation coefficient distributions for both species are centered at 3.3 S, in good agreement with hydrodynamic calculations based on the Ni3:MBP-Phen13 crystal structure. While these studies by themselves do not prove that the crosslinked trimeric species include Ni3:MBP-Phen13, they are consistent with its composition and shape.
- 6.Preliminary IR studies (Fig. S13) using Ni3:MBP-Phen13 crystals suggest that the Ni sites can coordinate isocyanate ligands within the crystal lattice.
- 7.(a) Ghadiri MR, Soares C, Choi C. J. Am. Chem. Soc. 1992;114:4000–4002. [Google Scholar]; (b) Lieberman M, Sasaki T. J. Am. Chem. Soc. 1992;113:1470–1471. [Google Scholar]; (c) Mutz MW, McLendon GL, Wishart JF, Gaillard ER, Corin AF. Proc. Natl. Acad. Sci. U. S. A. 1996;93:9521–9526. doi: 10.1073/pnas.93.18.9521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Privett HK, Reedy CJ, Kennedy ML, Gibney BR. J. Am. Chem. Soc. 2002;124:6828–6829. doi: 10.1021/ja025534+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.