Abstract
Members of the peroxisome proliferator-activated receptor γ coactivator (PGC) family are potent inducers of mitochondrial biogenesis. We have tested the potential effect of increased mitochondrial biogenesis in cells derived from patients harboring oxidative phosphorylation defects due to either nuclear or mitochondrial DNA mutations. We found that the PGC-1α and/or PGC-1β expression improved mitochondrial respiration in cells harboring a complex III or IV deficiency as well as in transmitochondrial cybrids harboring mitochondrial encephalomyopathy lactic acidosis and stroke A3243G tRNA(Leu)UUR gene mutation. The respiratory function improvement was found to be associated with increased levels of mitochondrial components per cell, although this increase was not homogeneous. These results reinforce the concept that increased mitochondrial biogenesis is a promising venue for the treatment of mitochondrial diseases.
INTRODUCTION
Oxidative phosphorylation (OXPHOS) dysfunctions play a critical pathogenic role in several human diseases (1). To date, more than 200 different mitochondrial DNA (mtDNA) mutations and probably a similar number of nuclear DNA (nDNA) mutations have been identified in patients with mitochondrial diseases. Defects in mitochondrial OXPHOS function affect preferentially high-energy demand tissues such as brain, skeletal muscle, heart, retina, renal tubules and endocrine glands. Almost invariably, the OXPHOS defect in patients is partial, suggesting that a complete defect would be incompatible with life. Therefore, patients either have mutant mtDNA coexisting with the wild-type mtDNA (mtDNA heteroplasmy) or a homoplasmic mtDNA mutation causing a partial impairment of OXPHOS. Likewise, nDNA mutations associated with diseases commonly cause partial defects with OXPHOS residual activity (1). Tissues of many patients with mitochondrial disorders may show massive mitochondrial proliferation, which has been assumed to be a metabolic response to the decreased OXPHOS function (1). The potential mechanisms of mitochondrial proliferation are now being better understood, thanks to the recent discoveries of transcriptional control of metabolic pathways and mitochondrial biogenesis.
Transcriptional coactivators of the PGC-1 gene family are master regulators of mitochondrial biogenesis and oxidative metabolism (2). There are three family members namely PGC-1α, PGC-1β and PGC-1 related coactivator (PRC). PGC-1α and PGC-1β are expressed in tissues with high-energy demand such as brown fat, skeletal muscle, heart, brain and kidney (3,4), whereas PRC is expressed ubiquitously (5). Studies have shown that PGC-1α/β are potent regulators of mitochondrial function and biogenesis (3,6–9). PGC-1α/β co-activates transcription factors that in turn stimulate the expression of a large number of nuclear genes involved in mitochondrial respiration and biogenesis. These transcription factors include the nuclear receptors peroxisome proliferator-activated receptors (PPARs), the NRF-1 and -2 (nuclear respiratory factors) and ERRα (estrogen-related receptor alpha) (10,11).
The present study demonstrated that induced PGC-1α/β upregulation improves mitochondrial respiration and OXPHOS function in cells with partial OXPHOS defects caused by either nDNA or mtDNA mutations.
RESULTS
We have studied three fibroblast cell lines obtained from pediatric patients with mitochondrial disorders (see Patients section). Two of these patients had a defect in complex IV and one in complex III activities. Fibroblasts were isolated and transduced with a retrovirus expressing hTERT (human telomerase). MtDNA sequencing and family history indicated that they had a nDNA defect, which was identified as located in the Surf1 gene in Patient D (Pat.D) (see Patients section). The molecular defects in the other patients remain unknown. In addition, we also studied human osteosarcoma cells harboring the A3243G tRNALeu(UUR) mtDNA mitochondrial encephalomyopathy lactic acidosis and stroke (MELAS) mutation which have been shown to have a respiratory defect due to partial defects in enzyme complexes I and IV (12).
Biochemical characterization of the patient fibroblasts showed OXPHOS complex defects
We performed the biochemical characterization of the cell lines obtained after immortalization of the patients’ fibroblasts and confirmed that the cell lines from Pat.D and Pat.C had isolated complex IV defects, whereas that from Pat.N had an isolated complex III defect (Fig. 1A). Steady-state levels of representative subunits of the involved respiratory complex were shown by western blot analysis (Fig. 1B). The levels of cytochrome oxidase COXII and COXIV subunits of complex IV in Pat.D and Pat.C fibroblasts and those of iron–sulfur protein (ISP) and core-1 subunit of complex III in Pat.N fibroblasts were severely decreased (Fig. 1B).
Figure 1.
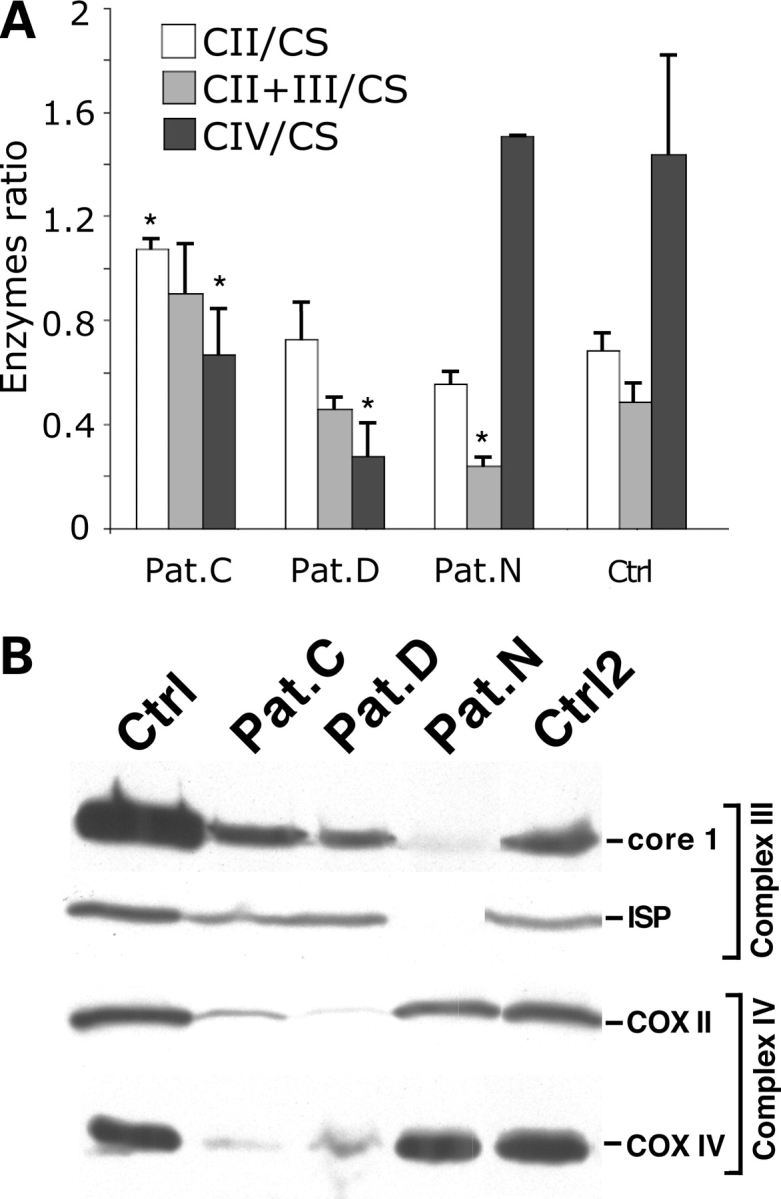
Characterization of fibroblast lines from patients with mitochondrial disorders. Three fibroblast lines from patients previously diagnosed with mitochondrial disorders associated with complex IV (Pat.C and Pat.D) and complex III (Pat.N) deficiencies were analyzed for their respiratory complexes function as a ratio to citrate synthase (A) (n = 3 independent measurements, bars = SD) and steady-state levels of OXPHOS components by western blot (B). The western blot on (B) was probed with antibodies against two complex III subunits (ISP and core-1) and two complex IV subunits (COXII and COXIV). Error bars represent SD of at least three independent measurements. *P < 0.05 in comparisons with the enzyme activities observed in the control.
PGC-1α/β expression improved mitochondrial respiration in OXPHOS deficient patient fibroblasts and MELAS cybrids
To investigate if an increase in mitochondrial biogenesis could compensate for partial OXPHOS defects, we transduced the patients’ fibroblasts and the MELAS A3243G cybrids with recombinant adenovirus (rAd) expressing PGC-1α and/or PGC-1β. Successful expression of the PGC-1α vector was monitored by green fluorescent protein (GFP) expression (from its own cytomegalovirus promoter). Transductions at titers conferring >70% GFP positive cells were used for analysis. Similar titers were used for rAd expressing PGC-1β. At these concentrations of rAd, we could not observe toxicity. The increased expression of PGC-1α after rAd–PGC-1α transduction could be documented by western blot (Supplementary Material, Fig. S1).
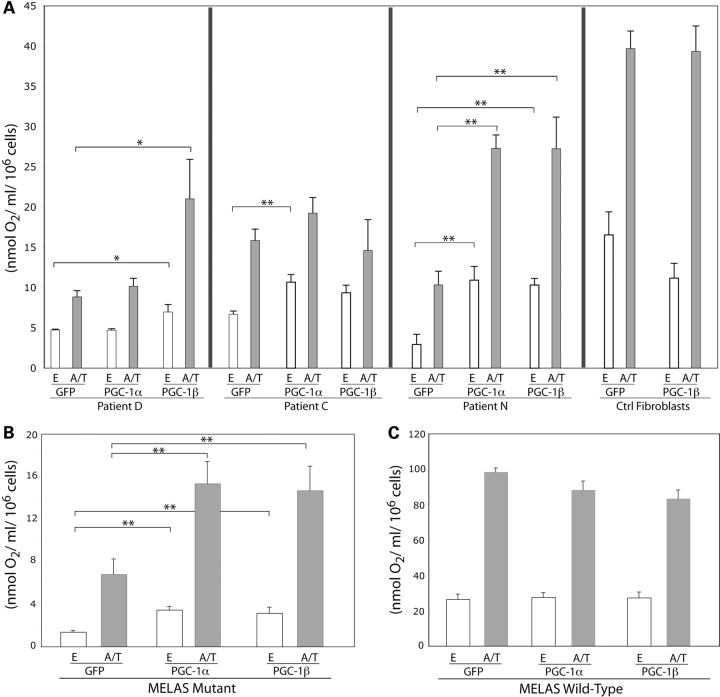
Induced expression of PGC-1α/β in patient fibroblasts significantly increased respiration both from the endogenous substrates (electrons entering at complexes I and II) and from the ascorbate/TMPD where electrons are donated to cytochrome c and subsequently to cytochrome c oxidase and oxygen. Interestingly, the two fibroblasts with defective complex IV, caused by distinct nuclear gene defects, showed different response to PGC-1 isoforms overexpression. Pat.D fibroblasts showed ∼50% increase both in endogenous and ascorbate/TMPD driven respiration after PGC-1β expression but not after PGC-1α expression. In contrast, Pat.C fibroblasts showed ∼50% increase after PGC-1α expression (Fig. 2A). On the other hand, Pat.N fibroblasts showed >2-fold increase in endogenous as well as ascorbate/TMPD driven respiration with expression of either PGC-1α or PGC-1β isoforms (Fig. 2A). Interestingly, the wild-type fibroblasts did not show increase in respiration with PGC-1α nor PGC-1β expression (Fig. 2A), suggesting that in wild-type cells there may be limiting factors that were not increased by the treatment.
Figure 2.
PGC-1α/β overexpression improves respiration of patients’ fibroblasts. Fibroblasts were transduced with Ad-PGC-1α or Ad-PGC-1β. After 72 h, cells were collected and oxygen consumption measured both with endogenous substrates (E) and with ascorbate/TMPD (A/T), which donates electrons to cytochrome c. Infections with adGFP were used as controls. (A) Shows the results with the fibroblasts lines. (B) Shows the results for the MELAS mutant cybrids, whereas (C) shows the wild-type cybrids. Error bars represent SD of at least three independent measurements. *P < 0.01; **P < 0.001.
Induced expression of PGC-1α/β in MELAS A3243G transmitochondrial cybrids showed similar results. The endogenous and ascorbate/TMPD driven respiration were significantly increased (∼2-fold) after PGC-1α or PGC-1β expression in MELAS A3243G mutant cybrids compared with the GFP expressing wild-type controls (Fig. 2B). The wild-type control cybrids did not show an increase in respiration after PGC-1α/β expression (Fig. 2B). MELAS cells were also transduced with PGC-1α+PGC-1β, but a synergistic effect was not observed (data not shown).
PGC-1α/β expression enhanced mitochondrial enzyme complexes activity in patient fibroblasts and MELAS cybrids
To search for the mechanisms of increased mitochondrial respiration in patient fibroblasts and MELAS A3243G cybrids overexpressing PGC-1α/β, we determined whether it was associated with increase in the previously defective mitochondrial activity. There were modest increases in the OXPHOS enzymes. Pat.D showed the greater increases, followed by Pat.C and finally by Pat.N who essentially showed no significant increases in complexes I+III or IV. Complex I+III was increased in MELAS mutant cells (Fig. 3). These results showed that the improvement in respiration associated with PGC-1α/β expression was not necessarily associated with an increase of respiratory chain levels, including the defective enzyme complexes. The activity of the TCA cycle enzyme, citrate synthase (CS), was significantly increased in all the cells: both the patient fibroblasts and the MELAS A3243G cybrids, after PGC-1α/β expression (Fig. 3C). An increase in the TCA cycle enzyme activity would generate an increased number of reducing equivalents that in turn might stimulate the electron transport chain. We also observed that the activity of complex II (succinate dehydrogenase) was increased after PGC-1α/β expression both in patient fibroblasts and MELAS A3243G cybrids (data not shown).
Figure 3.
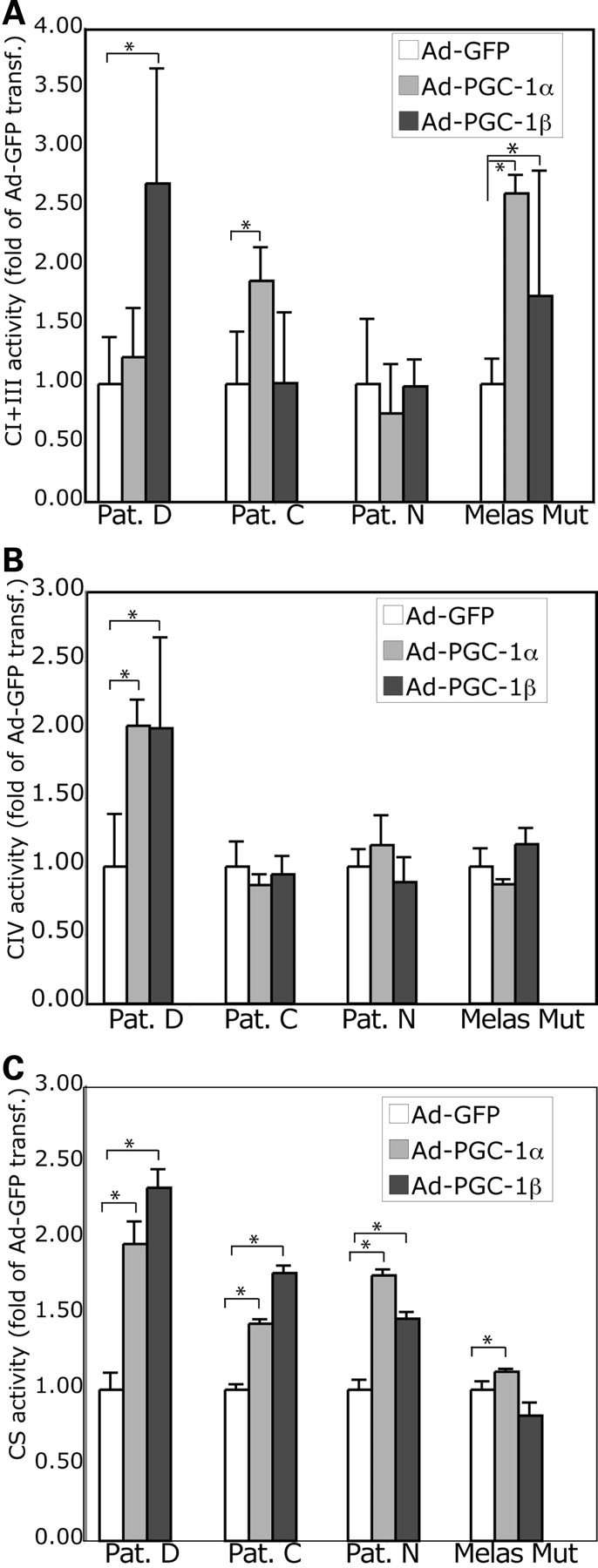
PGC-1α/β-related increase in respiration is associated with increased OXPHOS enzyme activities. Complexes I + III (A), IV (B) and citrate synthase (C) activities were measured spectrophotometrically in total cell homogenates (per mg/protein) obtained 72 h after recombinant adenovirus transduction. Although there was an overall increase in citrate synthase activity, OXPHOS enzyme increases were more variable.
PGC-1α/β upregulation increased expression of various OXPHOS subunits in the patients’ fibroblasts and the MELAS A3243G cybrids
We further investigated whether the expression of PGC-1α/β isoforms altered the steady-state levels of OXPHOS proteins. The endogenous levels of several (but not all) OXPHOS subunits were significantly increased after PGC-1α/β expression both in homogenates from patient fibroblasts and MELAS A3243G cybrids. The steady-state levels of ND39, SDH, Cyt c, Core-1, ISP, COXIV, COXVb and ATPase α subunits were significantly increased after PGC-1α/β induced expression both in patient fibroblasts and the MELAS A3243G cybrids (Fig. 4A–C), suggesting that an increase in the steady-state levels of various OXPHOS subunits could help enhance the electron transport function via the enzyme complexes that in turn may lead to increase in respiration.
Figure 4.
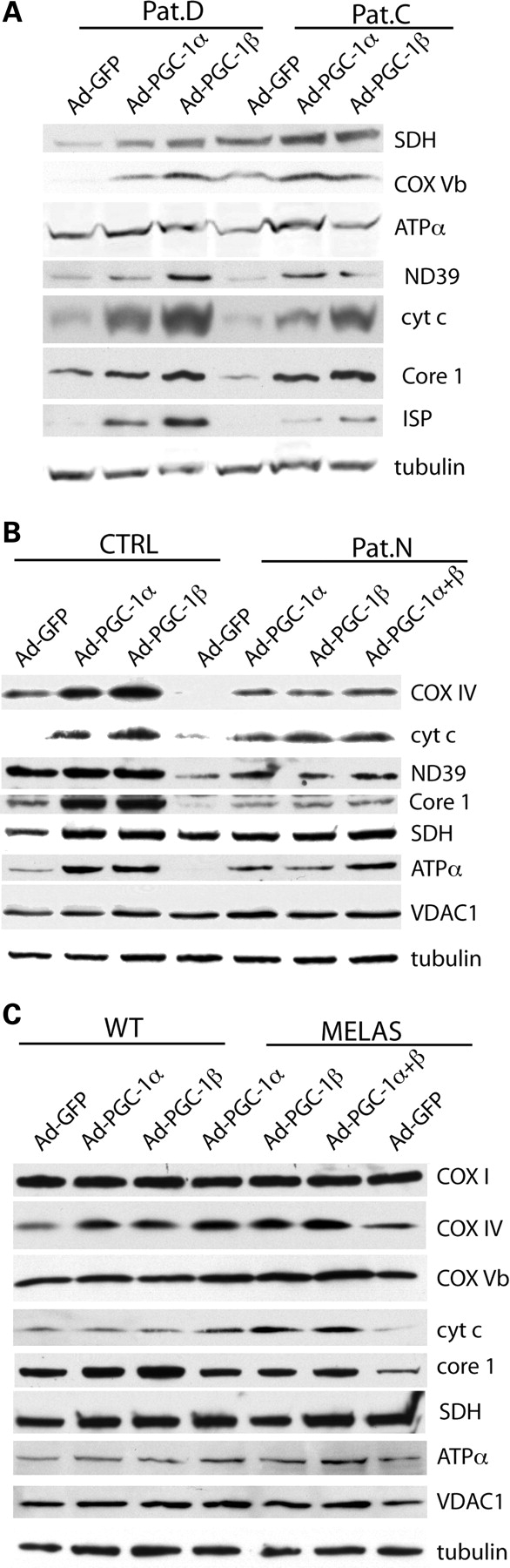
PGC-1α/β-related increase in respiration is associated with increased levels of OXPHOS components. Western blot analyses showed that PGC-1α/β overexpression-related increase in respiration in cells from patients with OXPHOS defects was associated with increased expression of OXPHOS components. Lanes were loaded with 100 μg of cell lysates. (A) Shows protein levels in Pat.C and Pat.D. (B) Shows the analyses for Pat.N and CTRL. (C) Describes similar analyses for MELAS and WT control. The antibodies used were the ones against: SDH (flavoprotein of complex II), ND39 (complex I), COXI, IV, Vb (complex IV), core-1 and Rieske iron–sulfur protein (complex III), cytochrome c, ATPase subunit α (complex V), VDAC1 (outer membrane protein) and tubulin.
PGC-1α/β upregulation increased the steady-state levels of OXPHOS complexes in the patients’ fibroblasts and the MELAS A3243G cybrids, but did not alter the fraction included in supercomplexes
The levels of assembled OXPHOS complexes were also increased upon the expression of PGC-1α/β (Fig. 5A). MELAS cells showed a higher increase in complex III than in complexes I and IV, a pattern that was also observed with the patients’ fibroblasts. However, complexes I and IV were also increased in most samples. Complex IV in Pat.D and complex III in Pat.N were very defective and essentially absent in these samples. There was a very small increase in their levels after treatment, observed only in overexposed films (data not shown). We also investigated whether the effect on respiration was due to a preferential increase in assembled supercomplexes (Fig. 5B). Samples solubilized in digitonin, to preserve the supercomplex structure were analyzed by blue native PAGE (BN-PAGE). Western blots of these gels showed that supercomplexes were not preferentially increased relative to the levels of the individual complexes. In these cell lines, a very small fraction of complex IV was observed in supercomplexes (data not shown).
Figure 5.
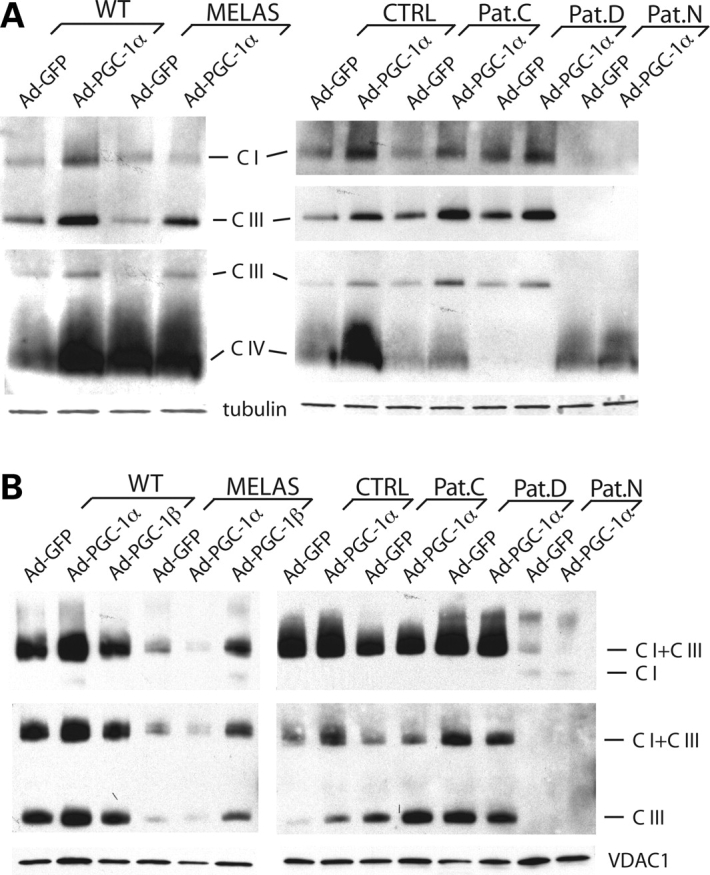
PGC-1α/β expression increases the levels of assembled OXPHOS complexes but does not change the relative fraction incorporated into supercomplexes. Cell samples were solubilized in either lauryl maltoside (A) or digitonin (B) to preserve the complex or supercomplex structure, respectively. Samples were separated in a 4–16% PAGE as described in Materials and Methods, transferred to a PVDF membrane and probed with antibodies representative of OXPHOS complexes. COXI (complex IV), core-2 (complex III), ND39 (complex I), VDAC1 and tubulin.
DISCUSSION
We have previously described the peculiar features of a colorectal tumor cell line with severe mtDNA mutations (null mutations) affecting complexes I and IV activity. Surprisingly, these cells respired at close to normal levels and had extremely high levels of PGC-1α and PGC-1β (13). When this mtDNA was transferred to a different nuclear background, the cells were unable to respire. PGC-1 family members are master regulators of mitochondrial biogenesis and there is strong evidence that mitochondrial proliferation is a natural compensatory phenomenon in mitochondrial diseases. Unfortunately, in diseases associated with heteroplasmic mtDNA mutations, mitochondria proliferation is usually observed in small domains of extreme OXPHOS deficiency where mtDNA mutation focal levels reach close to 100% (14,15). Increasing PGC-1 family members’ expression in all cells could allow healthy mitochondria to also proliferate, thus increasing the ATP generating capacity of the cell. This initial observation led us to investigate whether the overexpression of PGC-1 isoforms could have a beneficial effect for cells with an OXPHOS defect caused by either mtDNA or nDNA abnormalities. This concept was supported by our recent observations showing that increased expression of PGC-1α improved a mitochondrial myopathy associated with a COX10 conditional gene deletion in mice (16). Moreover, Bastin et al. (17) found that bezafibrate, a PPAR pan-agonist improved respiration of OXPHOS defective cells in culture. Their data suggested that not only PPAR was being activated but also PGC-1α. The use of more specific PPAR agonists showed that the activation of PPARδ was mostly responsible for the effect (17).
In the present study, we found that increasing expression of PGC-1α or PGC-1β leads to an improvement in the respiratory capacity of cell lines with OXPHOS defects due to a mtDNA or a nDNA gene alteration. Interestingly, control cells showed increased components of mitochondria without a significant increase in their respiration rate. This observation suggests the presence of factors regulating the respiration rate that are independent from the amount of OXPHOS components, not necessarily induced by PGC-1 family members. Likely, candidates are the ratio ADP/ATP and the flux of production of substrates for the respiratory chain. In contrast, defective cells consistently showed improvement in respiration, but with different features. Overexpression of PGC-1β was efficient in improving Pat.D’s respiration defect due to a Surf1 mutation, whereas PGC-1α was more efficient for Pat.C’s defect. The increase in both these patients was not as strong as in Pat.N (complex III deficiency) or the MELAS mutant cybrids. Therefore, respiration can be improved by PGC-1α and/or PGC-1β overexpression, but at different levels depending on the host cell, the defect and the PGC-1 isoform.
The increase in the steady-state levels of OXPHOS components observed with both PGC-1 isoforms involved numerous proteins but it was heterogeneous. Increase in mitochondrial biogenesis was not accompanied by a stoicheometric increase in all mitochondrial components. Curiously, the increases did not correlate well with the specific defect. In other words, patients with a COX deficiency did not have COX subunits preferentially increased when compared with other mitochondrial proteins. The reason for this heterogeneity is not well understood, but not surprising, as target genes respond differently to PGC-1 activation (18). This heterogeneity could also be observed in the analyses of assembled complexes and supercomplexes. Complex III appears to be very responsive to induction by PGC-1 isoforms, and that may explain the increased respiration observed in Pat.N. The reason for the increase in respiration in Pat.N and in complex III levels in most samples is unknown, but as mentioned above, it may be due to the responsiveness of complex III genes to PGC-1 activation. Cytochrome c, for example, is highly induced by PGC-1α, whereas other genes coding for mitochondrial proteins are less sensitive (18). Because the levels of complex III were very low and increased very little upon expression of PGC-1 isoforms in Pat.N, it is likely that other adaptations have a role in the increased respiration. A similar argument can be made for complex IV in Pat.D. In fact, the samples with the greatest increase in respiration were the ones with the most severe defects (Pat.D, Pat.N and MELAS). These observations suggest that the respiratory rescue is due to an increase of not only specific subunits, but also electron donors, transporters, assembly factors, translational activators, etc.
The improved respiration of OXPHOS defective cells when mitochondrial biogenesis is induced has important implications for the treatment of mitochondrial disorders. Although overexpression of PGC-1 isoforms may be difficult to achieve, the work of Bastin et al. (17). and ours (16) suggest that similar effects can be achieved pharmacologically (17). In summary, we now showed that PGC-1α/β overexpression can lead to a marked improvement in OXPHOS defects caused by mutations in nDNA or mtDNA. The mechanisms appear to be related to an increase in both the levels of OXPHOS limiting factors and the overall mitochondrial environment, thereby increasing the electron transfer and ATP production per cell.
MATERIALS AND METHODS
Patients
Pat.C was a boy, second child of consanguineous parents (first cousins). One older sibling was subjected to therapeutic abortion for renal and cerebral malformations. One younger brother had a similar disease with hypotonia, encephalopathy, hypertrophic cardiomyopathy and COX deficiency. He also had one healthy sister. He presented with hypotonia and repeated apneic episodes since birth necessitating mechanic ventilation. Hypertrophic myocardiopathy was detected at 1 month of age. Hyporeactivity, pyramidal syndrome, monotonous EEG, altered evoked visual potentials and abnormal brain imaging with leukodystrophy, basal ganglia lucencies and progressive cortical atrophy were observed at 2 months of age. Lactate levels were consistently increased in blood and CSF since birth (3–7 mm in blood, 6 mm in CSF). He also had increased intermediates of Krebs cycle in the urine. Muscle biopsy at 1 month of age showed mild accumulation of lipids and isolated respiratory COX deficiency (25% residual activity). Cultured skin fibroblasts showed 35% residual COX activity. MtDNA had normal amount and sequence. The patient died at 3 months of age; post-mortem muscle and liver samples showing isolated respiratory complex IV defect (20 and 2% residual activity in muscle and liver, respectively). Transfection of the child’s skin fibroblasts with the wild-type cDNA of Surf1, COX10, Sco1 or Sco2 genes did not restore complex IV histochemical activity thus demonstrating that none of these genes was responsible for the complex IV defect.
Pat.D was a girl, second child of consanguineous parents (second cousins). She presented with normal development up to 15 months of age, followed by progressive onset of anorexia, recurrent vomiting, motor difficulties with ataxia and pyramidal syndrome. At 3 years of age, brain imaging showed typical lesions of Leigh syndrome with basal ganglia, brainstem and cerebellum alterations. At 5 years of age, she presented with major failure to thrive (height at −8 SD), severe hypotonia, ataxia, pyramidal syndrome and muscle weakness. Lactate was constantly increased in blood and CSF. Muscle biopsy showed mild accumulation of lipids and severe defect of respiratory complex IV by histochemistry and biochemistry (5% residual activity). Cultured skin fibroblasts were also deficient in complex IV activity (20% residual activity). mtDNA had normal amount and sequence. Transfection of the child’s skin fibroblasts with the wild-type Surf1 cDNA restored complex IV histochemical activity thus demonstrating that Surf1 gene alterations were responsible for the complex IV defect. Sequencing of the Surf1 gene showed several alterations including an homozygous 7 bp deletion in the 85 bp long intron between exons 1 and 2 (+30deltgcgggg), an heterozygous G > A mutation in exon 5 (gly124arg), an heterozygous T > C mutation in exon 9 (leu281pro) and an heterozygous C > T mutation in exon 6 creating a synonymous change (phe181phe).
Pat.N was a girl, third child of consanguineous parents (first cousins). She had three episodes of coma with hyperlactatemia and ketoacidosis between 2 and 3.5 years of age. Each episode was triggered by infection (urinary infection, chicken pox). During the third episode, recurrent vomiting preceded the onset of coma and polypnea. Biological investigations disclosed severe metabolic acidosis (pH 6.94, bicarbonates 3 mm), hyperlactatemia (11 mm, n < 2 mm) with high lactate/pyruvate ratio (31, n < 15), hyperammonemia (300 µm, n < 50 μm), a 2-fold increase of blood transaminase levels and moderate hepatic insufficiency (factor V 49%). Muscle biopsy at 3 years of age showed normal morphology and isolated respiratory complex III defect (50% residual activity). Cultured skin fibroblasts also disclosed complex III defect (50% residual activity). MtDNA had normal levels and the cytochrome b gene had a normal sequence.
Fibroblasts obtained from a patient with typical MELAS syndrome were enucleated and the mtDNA transferred to a osteosarcoma rho zero cell as described (19). Clones with different levels of A3243G mutated mtDNA were obtained and a clone containing only mutated mtDNA and a clone containing exclusively the wild-type mtDNA were selected and used in this study.
Cell culture
Fibroblasts derived from the above described patients (C, D and N) were immortalized under cell culture conditions by infecting the cells with a recombinant retrovirus encoding hTERT (human telomerase) (20). Immortalized patient fibroblasts and the MELAS A3243G tRNALeu(UUR) transmitochondrial cybrids were grown in Dulbecco’s modified Eagle’s medium supplemented with 1 mm pyruvate, 50 µg/ml uridine and 10% fetal bovine serum.
Adenoviral infection
The patient fibroblasts and MELAS transmitochondrial cybrids were transduced with adenovirus expressing GFP, PGC-1α or PGC-1β constructs. Ad-GFP was obtained from the viral vector core facility at Colorado State University (Fort Collins, CO, USA). Ad-PGC-1α and Ad-PGC-1β were a gift from Dr Bruce M. Spiegelman (Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA, USA) and also contained a GFP marker expressed from an independent promoter. Serial dilutions of the virus were made for Ad-GFP (titer 9.2 × 1012 virus particles/ml), Ad-PGC-1 α (titer 6.9 × 1012 virus particles/ml) and Ad-PGC-1 β (titer 8.5 × 1012 virus particles/ml) to determine the amount of virus needed for ∼100% infection. Approximately 5–7 × 1010 virus particles/ml were used to infect patient fibroblasts or MELAS transmitochondrial cybrid cell lines. Twenty-four hours after infection, the cell culture medium was replaced with fresh medium. Cells were analyzed 72 h after infection.
Polarographic studies
Cell respiration (endogenous and ascorbate/TMPD) was measured in Ad-GFP, Ad-PGC-1α or Ad-PGC-1β transduced patient fibroblast or MELAS cybrids. Respiration was measured in buffer containing 25 mm Tris–HCl, 10 mm K2HPO4 and 150 mm sucrose (pH 7.4) using a Clark oxygen electrode in a water-jacketed cell, magnetically stirred at 37°C (Hansatech Instruments, Norfolk, UK) as described (21).
Western blot analysis
Total cell extracts (100 μg) prepared from GFP or PGC-1α/β transduced patient fibroblasts and MELAS A3243G transmitochondrial cybrids were run on SDS–PAGE and transferred to polyvinylidene difluoride membranes (Bio-Rad). Blots were blocked overnight in 5% milk and incubated with the primary antibody for 2 h followed by a secondary anti-mouse IgG conjugated to horseradish peroxidase (HRP). The chemiluminescent signal was detected using Phototope-HRP western blot detection kit (New England Biolabs, Beverly, MA). Monoclonal antibodies against ND39, SDH(Fp) (succinate dehydrogenase flavoprotein subunit), core-1 of complex III, ISP subunit of complex III, cytochrome c, COXI, COXIV, COXVb, ATPase α subunit, VDAC1 and tubulin were used for western detection. The ND39, SDH(Fp), core-1, ISP, COXIV, COXVb and VDAC1 antibodies were obtained from Molecular Probes (Eugene, OR, USA). The COXI and cytochrome c antibodies were obtained from Mitosciences (Eugene, OR, USA). Separation and western blot of BN-PAGE were performed as described (22).
Spectrophotometric analysis
Mitochondrial enzyme complex activities were measured spectrophotometrically in cells using a DU-640 spectrophotometer (Beckman Instruments Inc., Fullerton, CA, USA) as described elsewhere (23). The activities of NADH–cytochrome c oxidoreductase (complex I + III), cytochrome c oxidase (complex IV) and CS were determined by following the cytochrome c reduction (complexes I + III) or oxidation (complex IV) at 550 nm (23). All assays were performed at 37°C (except the CS at 30°C). Total cell lysates were normalized by protein content and aliquots used for the different assays.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by Muscular Dystrophy Association and PHS grants NS041777, CA085700 and EY10804 (to C.T.M.). F.D. work was supported by the James & Esther King Biomedical Research Program.
ACKNOWLEDGEMENTS
We are grateful to Dr Bruce M. Spiegelman (Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA) for the recombinant adenoviruses.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Dimauro S., Schon E.A. Mitochondrial disorders in the nervous system. Annu. Rev. Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- 2.Handschin C., Spiegelman B.M. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 3.Lin J., Puigserver P., Donovan J., Tarr P., Spiegelman B.M. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 4.Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 5.Andersson U., Scarpulla R.C. PGC-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol. Cell. Biol. 2001;21:3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 7.Lehman J.J., Barger P.M., Kovacs A., Saffitz J.E., Medeiros D.M., Kelly D.P. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J., Wu H., Tarr P.T., Zhang C.Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N., et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;a 418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 9.St-Pierre J., Lin J., Krauss S., Tarr P.T., Yang R., Newgard C.B., Spiegelman B.M. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J. Biol. Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 10.Scarpulla R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 11.Spiegelman B.M. Transcriptional control of energy homeostasis through the PGC-1 coactivators. Novartis Found. Symp. 2007;286:3–6. discusssion 6–12, 162–163, 196–203. [PubMed] [Google Scholar]
- 12.King M.P., Koga Y., Davidson M., Schon E.A. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNA(Leu(UUR)) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Mol. Cell. Biol. 1992;12:480–490. doi: 10.1128/mcb.12.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srivastava S., Barrett J.N., Moraes C.T. PGC-1alpha/beta upregulation is associated with improved oxidative phosphorylation in cells harboring nonsense mtDNA mutations. Hum. Mol. Genet. 2007;16:993–1005. doi: 10.1093/hmg/ddm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sciacco M., Bonilla E., Schon E.A., DiMauro S., Moraes C.T. Distribution of wild-type and common deletion forms of mtDNA in normal and respiration-deficient muscle fibers from patients with mitochondrial myopathy. Hum. Mol. Genet. 1994;3:13–19. doi: 10.1093/hmg/3.1.13. [DOI] [PubMed] [Google Scholar]
- 15.Moraes C.T., Ricci E., Petruzzella V., Shanske S., DiMauro S., Schon E.A., Bonilla E. Molecular analysis of the muscle pathology associated with mitochondrial DNA deletions. Nat. Genet. 1992;1:359–367. doi: 10.1038/ng0892-359. [DOI] [PubMed] [Google Scholar]
- 16.Wenz T., Diaz F., Spiegelman B.M., Moraes C.T. Activation of the PPAR/PGC-1alpha pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab. 2008;8:249–256. doi: 10.1016/j.cmet.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Bastin J., Aubey F., Rotig A., Munnich A., Djouadi F. Activation of peroxisome proliferator-activated receptor pathway stimulates the mitochondrial respiratory chain and can correct deficiencies in patients’ cells lacking its components. J. Clin. Endocrinol. Metab. 2008;93:1433–1441. doi: 10.1210/jc.2007-1701. [DOI] [PubMed] [Google Scholar]
- 18.Scarpulla R.C. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann. N. Y. Acad. Sci. 2008;1147:321–334. doi: 10.1196/annals.1427.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bacman S.R., Moraes C.T. Transmitochondrial technology in animal cells. Methods Cell. Biol. 2007;80:503–524. doi: 10.1016/S0091-679X(06)80025-7. [DOI] [PubMed] [Google Scholar]
- 20.Krupp G., Bonatz G., Parwaresch R. Telomerase, immortality and cancer. Biotechnol. Annu. Rev. 2000;6:103–140. doi: 10.1016/s1387-2656(00)06020-8. [DOI] [PubMed] [Google Scholar]
- 21.Villani G., Attardi G. Polarographic assays of respiratory chain complex activity. Methods Cell. Biol. 2007;80:121–133. doi: 10.1016/S0091-679X(06)80005-1. [DOI] [PubMed] [Google Scholar]
- 22.Vempati U.D., Han X., Moraes C.T. Lack of cytochrome c in mouse fibroblasts disrupts assembly/stability of respiratory complexes I and IV. J. Biol. Chem. 2009;284:4383–4391. doi: 10.1074/jbc.M805972200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrientos A., Kenyon L., Moraes C.T. Human xenomitochondrial cybrids. Cellular models of mitochondrial complex I deficiency. J. Biol. Chem. 1998;273:14210–14217. doi: 10.1074/jbc.273.23.14210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.