Abstract
NODAL and its signaling pathway are known to play a key role in specification and patterning of vertebrate embryos. Mutations in several genes encoding components of the NODAL signaling pathway have previously been implicated in the pathogenesis of human left–right (LR) patterning defects. Therefore, NODAL, a member of TGF-β superfamily of developmental regulators, is a strong candidate to be functionally involved in congenital LR axis patterning defects or heterotaxy. Here we have investigated whether variants in NODAL are present in patients with heterotaxy and/or isolated cardiovascular malformations (CVM) thought to be caused by abnormal heart tube looping. Analysis of a large cohort of cases (n = 269) affected with either classic heterotaxy or looping CVM revealed four different missense variants, one in-frame insertion/deletion and two conserved splice site variants in 14 unrelated subjects (14/269, 5.2%). Although similar with regard to other associated defects, individuals with the NODAL mutations had a significantly higher occurrence of pulmonary valve atresia (P = 0.001) compared with cases without a detectable NODAL mutation. Functional analyses demonstrate that the missense variant forms of NODAL exhibit significant impairment of signaling as measured by decreased Cripto (TDGF-1) co-receptor-mediated activation of artificial reporters. Expression of these NODAL proteins also led to reduced induction of Smad2 phosphorylation and impaired Smad2 nuclear import. Taken together, these results support a role for mutations and rare deleterious variants in NODAL as a cause for sporadic human LR patterning defects.
INTRODUCTION
Heterotaxy, also called situs ambiguus, is a complex congenital disorder characterized by the disruption of the normal left–right (LR) asymmetry of the thoracoabdominal organs. Heterotaxy is thought to arise from disturbances in early developmental processes either directly or indirectly dependent on the establishment of the embryonic LR axis. Heterotaxy includes a broad range of anatomic abnormalities involving segmental discordances and abnormal symmetry of normally asymmetric thoracic and abdominal organs. Patients with segmental discordances in the asymmetric thoracoabdominal organs typically have complex cardiac and vascular abnormalities that are thought to involve partial or complete reversal of heart tube looping, LR patterning of the atria or the failure of asymmetric remodeling of symmetric embryonic structures. Examples of congenital cardiovascular malformations (CVM) that are highly suggestive of defects in embryonic LR patterning include dextrocardia, mesocardia, levo-transposition of great arteries (l-TGA) and atrial isomerisms. Other commonly associated cardiac defects, including pulmonic stenosis, pulmonary atresia (PA), anomalous pulmonary venous return, interrupted inferior vena cava, persistent left superior vena cava, dextro-transposition of the great arteries (d-TGA), double-outlet right ventricle (DORV), ventricular septal defect (VSD), atrial septal defect, single ventricle, hypoplastic left heart and co-arctation of the aorta, apparently can arise by more than one mechanism although they are often observed as part of the complex defects in heterotaxy. For example, d-TGA might result from the misalignment of the atrioventricular canal and conotruncus relative to the ventricle through direct LR patterning abnormalities of the second heart lineage (1), or alternatively may occur as the result of defects in aorticopulmonary septation. Mutations or chromosomal imbalances affecting ZIC3 (2–7), CFC1 (Cryptic) (8–10), ACVR2B (11), LEFTY2 (12), NKX2.5 (13,14), CRELD1 (15), FOXH1, CRIPTO (TDGF1) (16), and GDF1 (17) have been associated with either heterotaxy or related CVM in humans. In addition, a de novo reciprocal translocation t (6,18) (q21;q21) in a subject with heterotaxy was found to disrupt the SESN1 (PA26) locus (18,19).
Situs ambiguus should be distinguished from situs inversus totalis or complete LR transposition of thoracic and abdominal organs. Although situs inversus can occur in otherwise healthy adults, it may also be associated with primary ciliary dyskinesia, infantile nephonophthisis and Bardet–Biedl syndrome (20). All these disorders share a molecular basis in dysfunction of the primary cilia. Further complicating the picture, subjects with these conditions may also exhibit situs ambiguus, although much less commonly than situs inversus.
In mouse, the TGF-β family member Nodal is known to play a central role in early embryonic development, mesoderm and endoderm formation and LR axis patterning. Nodal is expressed in the epiblast and visceral endoderm during gastrulation, and it induces its co-receptor Cripto, which is also necessary for antero-posterior patterning. Nodal-deficient mice lack the primitive streak and do not form mesoderm (21–24). These embryos produce excessive embryonic and extra-embryonic ectoderm, probably due to the failure of mesoderm differentiation. The pattern of Nodal expression, as well as its developmental function, appears to be largely conserved among vertebrates. In zebrafish, double mutants for Nodal-related genes, cyclops and squint, are deficient in head and trunk mesoderm (25). In Xenopus, five different related factors (Xnr1, Xnr2, Xnr4, Xnr5, Xnr6) have been shown to have similar functions (26,27). In all species examined, Nodal orthologs are expressed asymmetrically in the left side of the node as well as in left lateral plate mesoderm (LPM). The Nodal signaling cascade within the LPM mesoderm is required for LR patterning in vertebrates (28–33), and ectopic expression of Nodal leads to reversal of polarity of visceral organs and heart looping (34).
Nodal signaling uses an Activin/TGF-β-like pathway mediated by several Activin-like receptors (ALKs). Nodal is believed to signal via ALK4 and AKL7, type I receptors, in association with either ActRIIA or ActRIIB, type II receptors (32,35). The activated receptor complex phosphorylates intracellular receptor-regulated Smads (R-Smads; Smad2 and Smad3). Phospho-Smad2/3 binds to Smad4, resulting in translocation to the nucleus (36), where the Smad complex either binds DNA directly (e.g. Smad3) or can interact with other DNA-binding proteins (e.g. FoxH1, Mixer, Jun/Fos). These complexes interact with promoters of various target genes. FoxH1 also regulates Nodal signaling by binding to the Nodal and Lefty2 asymmetric enhancer element (37). Extracellular EGF-CFC proteins are also important components of the Nodal signaling pathway. These include Cripto (TDGF1) and Cryptic (CFC1) in mouse and human, CFC in chick, FRL-1 in frog and oep (one-eyed pinhead) in zebrafish. Biochemical studies indicate that Cripto and Cryptic form complexes with Nodal, ALK4/ALK7 and ActRIIB (38,39) and are, thus, co-receptors for Nodal at the cell surface. Cripto and oep mutants resemble Nodal mutants in mice and cylops:squint double mutants in zebrafish (40,41).
Mutation in several components of the Nodal signaling pathway such as ActRIIB, Cryptic/Cfc1, Cripto/Tdgf1, Smad2 and FoxH1 in mouse and oep and schmalspur in zebrafish exhibit defects in LR axis development (31,39–46). Although null mutation in Nodal arrests mouse embryonic development at gastrulation stage, a conditional hypomorphic Nodal mutant (47) has demonstrated the requirement for Nodal in the establishment of the embryonic LR axis. In order to assess the involvement of NODAL in human heterotaxy, we screened a large cohort of heterotaxy patients for mutations in NODAL. We identified four missense, one in-frame insertion/deletion and two conserved splice site variants in patients with severe laterality disorders. Functional analysis of the missense variants demonstrated impairment of Activin/Nodal-Smad signaling. These results support a model in which imbalance in dosage-sensitive NODAL signaling is a final common pathway for heterotaxy and related CVM.
RESULTS
Mutation analysis
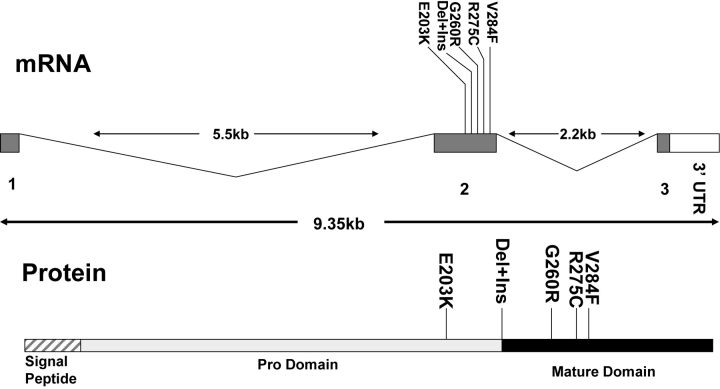
Two hundred and sixty-nine unrelated probands (either sporadic or familial cases) (Supplementary Material, Table 1S) meeting inclusion criteria for classic heterotaxy or related isolated CVM were screened for variants in NODAL (NM_018055) by denaturing high-pressure liquid chromatography (DHPLC). Amplicons bearing heteroduplexes were then analyzed by DNA sequencing. This revealed four missense variants (E203K, G260R, R275C and V284F) and a 24 base deletion with a 9 base insertion in exon 2 (Fig. 1 and Table 1). We also observed two variants in conserved non-coding sequence which are predicted to alter the IVS1-exon 2 splice acceptor site (c.194-1G>T) and the exon 2-IVS2 splice donor (c.891+1G>A). A previously characterized common non-synonymous polymorphic variant (rs1904589, H165R) was also observed but was not investigated further. In addition, three other rare synonymous variants (L196L, Q204Q and V308V) were detected (Supplementary Material, Table 2S) but were not further characterized, although it should be noted that non-synonymous SNPs can sometimes affect splicing via the regulation of splice enhancers or inhibitors (48,49).
Figure 1.
Schematic diagram of NODAL structure with location of the approximate position of amino acid variants observed in this study.
Table 1.
Non-synonymous or conserved splice site nucleotide changes identified in probands
| Nucleotide | Protein | Affected | Controls |
|---|---|---|---|
| c.607A>G | E203K | 1/269 | 0/395 |
| c.700_723delinsTTGACTTCC | p.R234_P241delinsLTS | 1/269 | 0/298 |
| c.778G>A | G260R | 8/269 | 1/298 |
| c.823C>T | R275C | 1/269 | 0/298 |
| c.850G>T | V284F | 1/269 | 0/298 |
| c194-1G>T | IVS1 splice as | 1/269 | ND |
| c.891+1G>A | IVS2 splice ds | 1/269 | 0/298 |
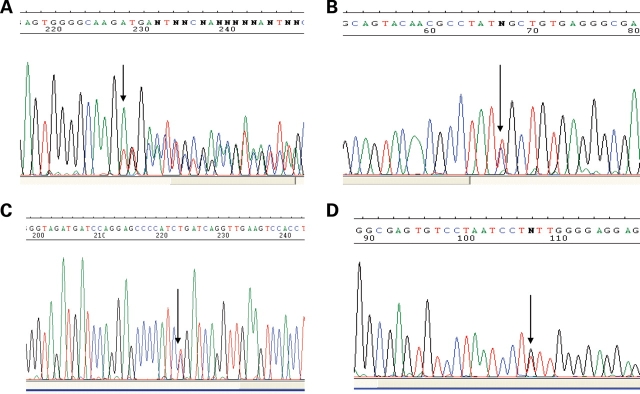
A heterozygous nucleotide change (607A>G) in exon 2 of the gene leading to the E203K variant was found in a sporadic patient with abdominal situs inversus and CVM, consisting of single ventricle with d-TGA. The variant was absent in 395 control individuals including 98 ethnic-matched Southeast Asian controls. However, this variant has been observed in the HapMap CHB sample (rs10900334) with an allele frequency of 0.022. E203K was not observed in the other HapMap population samples including the other East Asian sample, JPT (www.hapmap.org). The proband harboring the R275C rare variant (corresponding to the nucleotide 823C>T transition, Fig. 2B) exhibited abdominal situs inversus, asplenia, midline liver and complex CVM (d-TGA and complete atrioventricular canal-type septal defects). Although not found in 298 controls, analysis of DNA samples of family members revealed that this rare variant was inherited from the phenotypically normal mother. Another heterozygous nucleotide change, 850G>T (V284F), was observed in a sporadic case with dextrocardia, levo-transposition of the great arteries (l-TGA), atrioventricular block and interruption of the inferior vena cava (parents unavailable to assess inheritance). In another sporadic case with d-TGA, PA and double-inlet left ventricle (DILV), a 24 base deletion (700–723) and a 9 base in-frame insertion (Fig. 2A) were observed. Parental DNA samples were not available in this case. None of these variants was detected in 298 controls (95 Caucasian, 95 African-American and 108 Hispanic) individuals.
Figure 2.
DNA sequence chromatograms. (A) Complex re-arrangement with 24 bp deletion (700_723) with an in-frame 9 bp insertion (c.700_723delinsTTGACTTCC). (B) C823T nucleotide substitution causing R275C. (C) G778A nucleotide transition causing G260R (reverse sequence shown). (D) G850T nucleotide substitution V284F.
In eight of the 82 unrelated Hispanic cases, a heterozygous 778G>A nucleotide change predicting the G260R variant was observed (Hispanic case allele frequency 0.049). Parental DNA sample was not available for seven of these cases, but analysis of parental DNA in one case demonstrated this sequence variant in the father of the affected child. One of 108 Hispanic controls also carried with the same variant (Hispanic control allele frequency 0.0046, two-tailed Fisher exact P = 0.005 comparing control versus affected frequency). None of the Caucasian or African-American controls was found to have this sequence variant. All the patients detected with this variant presented with severe cardiac and multi-organ abnormalities (Table 2). Two other variants were observed in conserved splice donor and acceptor sites of exon 2. The case harboring c.194-1G>T exhibited dextrocardia, single ventricle, DILV and PA. The subject bearing the c891+1G>A mutation presented with dextrocardia, l-TGA, DORV, pulmonary valve stenosis and a VSD. These two variants occurred in residues highly conserved across species. In silico analyses indicate that these variants are likely to alter splicing activity (http://www.fruitfly.org/seq_tools/splice.html). Overall, PA was observed more commonly than expected (8/14 or 57%) in the cases bearing the NODAL variants compared with complete cohort of affected cases without a NODAL variant (45/255 or 18%; Supplementary Material, Table 1S, Fisher exact test two-sided, P = 0.015).
Table 2.
Genotype–phenotype correlations of heterotaxy cases carrying NODAL mutations
| Patient No. | Gender and ethnicity | Mutation | Cardiovascular malformation | Other abnormalities |
|---|---|---|---|---|
| LAT0022 | Female Caucasian | c.194-1 G>T | Dextrocardia, single ventricle, DILV, PA, hypoplastic tricuspid valve annulus | Asplenia, bronchectasis |
| LAT0123 | Male, Vietnamese | E203K | d-TGA, single ventricle, | Abdominal situs inversus, cerebellar and cerebral atrophy |
| LAT0191 | Male Hispanic | c.700indel | d-TGA, single ventricle, DILV, PA, ASD, intra-atrial re-entrant tachycardia | |
| LAT0040 | Female Hispanic | G260R | d-TGA, right atrial isomerism, PA, PAPVR with pulmonary vein confluence, ASD, VSD, interrupted IVC with large hemiazygous vein draining to a left SVC | Abdominal situs inversus, normal spleen |
| LAT0201 | Female Hispanic | G260R | d-TGA, PA, ASD, single ventricle (LV morphology), PDA, intra-atrial re-entrant tachycardia | |
| LAT0248 | Male Hispanic | G260R | d-TGA, single ventricle (RV morphology), single atrium, CAVC, PA, supracardiac TAPVR, primum ASD, LPA hypoplasia, IVC to LA, PDA, Wolf–Parkinson–White | Asplenia, abdominal situs inversus |
| LAT0658 | Male Hispanic | G260R | Dextrocardia, single ventricle (LV morphology), absent pulmonary trunk, TAPVR (to liver), ASD, persistent left SVC, bilateral IVC, each SVC and IVC drained into left and right atrium, respectively | Abdominal situs inversus, bilateral trilobed lungs, asplenia, mild hydronephrosis |
| LAT0858 | Male Hispanic | G260R | d-TGA, single ventricle (RV morphology), PS, DORV, ASD, VSD | |
| LAT0909 | Male Caucasian | G260R | d-TGA, ASD, PDA | |
| LAT1016 | Male Hispanic | G260R | d-TGA, Aortic arch hypoplasia, DORV, ASD, VSD, severe CoA, PDA | |
| LAT1028 | Female Hispanic | G260R | Dextrocardia, d-TGA, dilated ascending aorta, PA, DILV, VSD, hypoplastic RV, criss-cross AV connections | |
| LAT0165 | Male Hispanic | R275C | d-TGA, CAVC, PA, AVVR, TAPVR, ASD | Abdominal situs inversus, asplenia, intestinal malrotation |
| LAT0368 | Male Caucasian | V284F | Dextrocardia, l-TGA, ventricular inversion, interrupted IVC, ventricular inversion, AV block, dilated cardiomyopathy | S/P orthotopic cardiac transplant |
| LAT0457 | Male Hispanic | c.891+1 G>A | Dextrocardia, hypoplastic subpulmonary conus with PS, l-TGA, DORV with posterior main PA, VSD |
ASD, atrial septal defect; AV, atrioventricular; c.700indel, c.700_723delinsTTGACTTCC; CAVC, complete atrioventricular canal defect; CoA, co-arctation of the aorta; DILV, double-inlet left ventricle; DORV, double-outlet right ventricle; d-TGA, dextro-transposition of the great arteries; IVC, inferior vena cava; LA, left atrium; LPA, left pulmonary artery; l-TGA, levo-transposition of the great arteries; LV, left ventricle; PA, pulmonary atresia; PS, pulmonic stenosis; PAPVR, partial anomalous pulmonary venous return; RV, right ventricle; SVC, superior vena cava; TAPVR, total anomalous pulmonary venous return; VSD, ventricular septal defect.
Functional deficits in signaling caused by non-synonymous amino acid substitutions
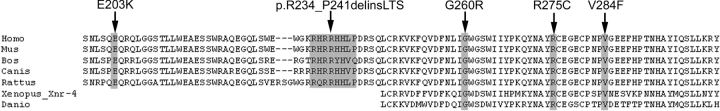
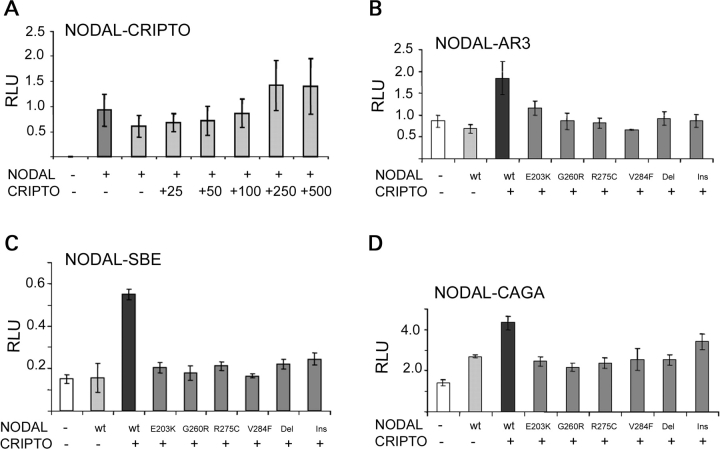
The amino acid substitutions identified occur in positions that are highly conserved across species (Fig. 3) as well as across family members (data not shown). Analysis of the sequence variants performed using PolyPhen (http://www.bork.embl-heidelberg.de/PolyPhen/) suggested possible damaging effects of the identified missense changes. In order to further evaluate the effect of these NODAL variants on its signaling pathway, reporter expression from three different NODAL-responsive promoters was examined. Luciferase reporter plasmids included: pAR3-lux, responsive to FoxH1-mediated TGF-β, Activin and Nodal signaling (containing the Xenopus Mix.2 promoter element); p(SBE)4 reporter, responsive to general TGF-β and Bmp2 signaling; and p(CAGA)12 reporter, specifically responsive to Smad3 signaling, i.e. FoxH1-independent. Co-transfection of CRIPTO (250 ng), a co-receptor for NODAL, was an essential co-factor for ligand-induced reporter activity in the mouse P19 embryonic carcinoma cell model (Fig. 4A) and was used for subsequent transactivation studies. Using the pAR3-lux reporter, NODAL variants E203K, G260R, R275C, V284F, c.700_723del and c.700_723delinsTTGACTTCC exhibited significant reductions (37, 53, 56, 66, 50 and 47%, respectively) in pAR3 luciferase expression. Similarly, the activity of the p(SBE)4 reporter construct (Fig. 4C) was found to be reduced (63, 67, 61, 70, 60 and 56%, respectively) with these same mutant constructs. Finally, the p(CAGA)12 activity (Fig. 4D) was impaired with reductions in the activity of NODAL variant constructs of 43, 48, 45, 40, 41 and 21%, respectively.
Figure 3.
Cross-species sequence alignment of NODAL showing conservation of amino acid positions in which variant amino acids were identified (highlighted at arrows).
Figure 4.
Functional analysis of the transactivation of NODAL variants. (A) Optimization of CRIPTO co-transfection for efficient NODAL activation of AR3 luciferase reporter. (B) Variant NODAL constructs have decreased transactivation of the AR3 luciferase reporter compared with wild type (wt). (C) Transactivation results of wt and variant NODAL constructs using the SBE4 luciferase reporter. (D) Transactivation results of wt and variant NODAL constructs using the CAGA12 luciferase reporter. Del, c.700_723del; Ins., c.700_723delinsTTGACTTCC. RLU, relative luciferase units.
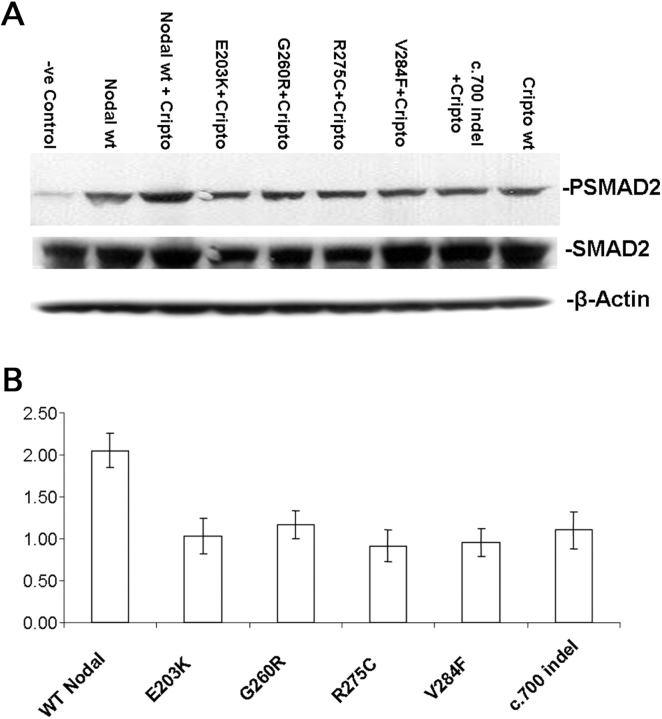
In order to elucidate the effects of mutations in the NODAL gene on the Smad2/3 signaling pathway, we studied the Smad2 phosphorylation in P19 cells after transfection either with wild-type or variant NODAL constructs. We used antisera that specifically detect endogenous Smad2 only when dually phosphorylated at its C-terminal serine 465 and serine 467 residues. Western blot analysis demonstrated a 2-fold (45–55%) reduction in Smad2 phosphorylation in the extracts of P19 cells transfected with the constructs carrying NODAL variants (E203K, G260R, R275C, V284F and c.700_723delinsTTGACTTCC) compared with the wild type (Fig. 5). A high level of endogenous NODAL and Smad2/3 was observed in both the transfected and non-transfected P19 cells as determined by western blot (data not shown). The level of phosphorylated Smad2/3 was normalized on the basis of the level of total Smad2.
Figure 5.
Western blot analysis of P19 cells transfected with wild-type and mutant NODAL constructs. (A) Expression of phosphorylated Smad2 and Smad2 in P19 cells transfected with either wild-type or mutant NODAL. β-Actin serves as a loading control. (B) Quantitation of western blots for phosphorylated Smad2 versus Smad2 in cells treated with either wild-type or mutant NODAL expression vectors. c700indel, c.700_723delinsTTGACTTCC.
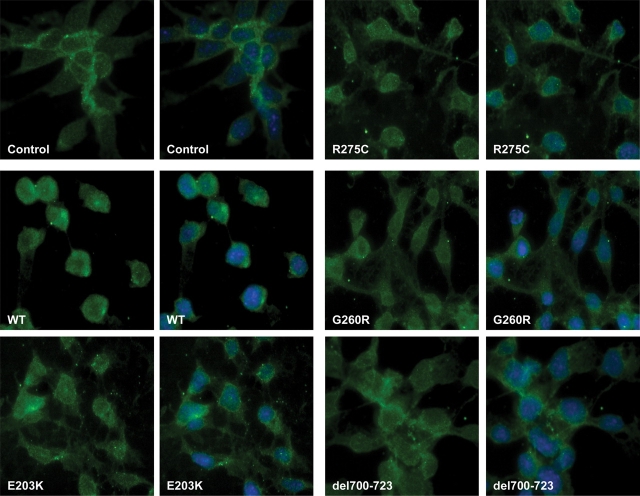
In order to investigate the subcellular distribution of Smad2 in the P19 cells transfected with either wild-type or variant NODAL constructs, we carried out immunostaining of non-transfected and transfected cells using anti-phospho-Smad2 antibodies. Non-transfected P19 cells revealed a low level of phospho-Smad2 mostly localized to the cytoplasm. However, in cells transfected with wild-type NODAL plus CRIPTO, phospho-Smad2 accumulated in the nucleus revealing nearly 100% nuclear translocation (Fig. 6). In contrast, the distribution of phospho-Smad2 in the cells transfected with NODAL variants plus CRIPTO was predominantly cytoplasmic and the overall level was decreased. Decreased phospho-Smad2 (Figs 5 and 6) and increased cytoplasmic retention of phospho-Smad2 clearly suggest reduced Smad2/3-mediated signaling by variant NODAL proteins compared with wild type.
Figure 6.
Immunostaining of P19 cells showing inhibition of nuclear translocation of phospho-Smad2 in cells transfected with NODAL variant constructs (green: anti-phospho-Smad2, blue: DAPI). Control, non-transfected cells showing endogenous distribution of phopho-Smad2.
DISCUSSION
NODAL is a secreted signaling molecule known to play a key role in vertebrate embryonic development. Genetic studies in mouse and zebrafish, as well as misexpression studies in chick and Xenopus, suggest that Nodal and its family members are important signaling molecules for LR axis determination and left-specific organogenesis. Although several other members of the NODAL signaling pathway have been reported to be involved in human heterotaxy, NODAL mutations have not been investigated in large studies of patients with CVM. Here we report four novel missense variants (E203K, R275C, G260R and V284F), a deletion/insertion mutation (24 base deletion with a 9 base insertion) and two conserved splice site variants in NODAL in individuals affected with heterotaxy.
There have been three previous preliminary reports of NODAL associated with human heterotaxy. Gebbia et al. (4) reported a mutation in NODAL (R183Q) in an affected female and her unaffected mother. This individual was part of an extended pedigree in which a mutation in ZIC3 (T325M) segregated with heterotaxy among affected males. This female did not carry the ZIC3 T325M mutation, however. In addition, Bamford et al. (8) noted a single affected case bearing both a CFC1 mutation (G174fs) and a NODAL missense variant. Bassi et al. (50) described an individual with heterotaxy and variants in both NODAL and FOXA2. Taking these observations together with the present biochemical studies which demonstrate reduced activity of the missense variants, we conclude that these changes most likely play a role in the LR axis malformations.
Functional analyses of rare variants in NODAL indicate a decrease in signal transduction. In this study, we observed a significant reduction in p(AR3) and p(SBE)4 as well as in p(CAGA)12 reporter activity. The p(AR3) and p(SBE)4 reporter constructs contain FoxH1-dependent Xenopus Mix.2 promoter elements, whereas the p(CAGA)12 reporter is Smad3-specific and its activation is independent of FoxH1. Our results demonstrate reduced NODAL signaling through both FoxH1-dependent and -independent pathways. Phenotypic differences observed among mouse mutants in Nodal, Smad2, ActRIIB and FoxH1 suggest overlapping signaling pathways during embryonic development. Unlike the Nodal, ALK4 and ActRIIB mutants, which fail to gastrulate, FoxH1-/- embryos form a primitive streak and develop relatively normally through gastrula stages (45). This suggests that there are other key transcription factors acting downstream of early Nodal signaling.
Additional functional investigation demonstrated abnormal subcellular localization of phospho-Smad, an important mediator of NODAL signal transduction upon transfection of NODAL variants. Activation of Smad2 phosphorylation is required for the specification of both endoderm and mesoderm, embryonic differentiation and organogenesis (51–53). Our observation of reduced Smad2 phosphorylation comparing wild-type NODAL with the patient-derived variants in P19 cells implies reduced efficiency of signaling. Impaired Smad2/3 signaling in individuals bearing those mutations could therefore be an important pathological mechanism.
Interestingly, the complex mutation consisting of a 24 base deletion with a 9 base insertion is exactly at the cleavage site of the NODAL mature domain. A highly arginine- and histidine-rich domain (Arg–His–Arg–Arg–His–His–Leu–Pro) is apparently replaced by three amino acids (Leu–Thr–Ser). This alteration would eliminate the proprotein convertase (SPC) recognition motif (R-Q/H-R-R) and is, thus, predicted to cause an impairment of NODAL maturation. The proprotein convertases (SPCs), FURIN and PACE4, stimulate NODAL maturation after its secretion and are required in vivo for both NODAL and BMP4 signaling (54). Both the activity and the signaling range of NODAL are believed to be dependent on the rate of proteolytic maturation (55–57). A long-range signaling effect has also been suggested with a relatively stable NODAL precursor (56). However, ablation of SPC-recognition site or inhibition of SPCs caused NODAL inactivation. Impairment of human NODAL protein precursor maturation is likely to decrease its activity.
Occurrence of the G260R variant in eight Hispanic cases suggests an ethnicity-specific predilection. However, the detection of this variant in one unaffected Hispanic control and the unaffected father points to incomplete penetrance. This result is fully consistent with the low recurrence risk and apparent complex inheritance of most sporadic heterotaxy cases. The E203K and G260R variants appear to represent additional examples of rare functionally deleterious and population-specific genetic alterations in a component of the NODAL signaling pathway. These results are consistent with rare variants in NODAL acting as risk factors or susceptibility alleles for the development of heterotaxy spectrum CVM, and the population specificity is predicted by the rare variant hypothesis due to founder effects resulting from genetic drift (58). Similar results have been found for other genes within the NODAL signal transduction pathway. For example, Bamford et al. (8) previously found the CFC1 R78W mutation in five unrelated African-American heterotaxy cases. This amino acid substitution demonstrated reduced functional activity both in tissue culture and in a zebrafish embryo assay. The allele frequency in an African-American control sample (n = 68) was 0.029 but was not detected in Caucasian controls. Similarly, Selamet-Tierney et al. (10) found missense variants in CFC1 that were not fully penetrant. A recent study showed that variants in FOXH1, CFC1 or TDGF1 are associated with congenital heart disease, including tetralogy of Fallot and laterality spectrum defects and suggested that cumulative impairment of NODAL pathway activity may play a critical role in the development of congenital heart disease (16). Taken together with the current NODAL data, these studies imply that imbalance in dosage-sensitive NODAL signaling is a final common pathway conferring risk for heterotaxy and related CVM.
Studies of Nodal signaling in model organisms support its dosage-sensitive role in the development of LR patterning defects. For example, Smad2/Smad3 carries the Nodal activation signal to the nucleus after heterodimerizing with Smad4. Smad2 null mice die prior to organogenesis (42,59), but chimeric embryos exhibit abnormal turning, ambiguous cardiac looping and anterior midline defects including craniofacial defects and cyclopia (60), consistent with laterality defects. In an elegant study by Lowe et al. (47), conditional hypomorphic Nodal mutants were shown to have a range of phenotypes, depending in part on Nodal dosage. Less severely affected embryos had a high incidence of TGA (100%), reversed heart looping (52%) and right pulmonary isomerism (88%), confirming the involvement of Nodal in LR axis formation and particularly in the looping stage of cardiac development. A variety of complex heterozygotes demonstrate that genetic interaction of mutant or haploinsufficient components of the signaling pathway can cause CVM. For example, Nodal and Smad2 double-heterozygote mice exhibit cardiac and laterality defects such as abnormal heart looping, TGA, right pulmonary isomerism and right-sided stomach (42). ActRIIA and ActRIIB, iv and ActRIIB (43,61) and Nodal and Foxa2 (62) double heterozygotes all provide additional evidence for the dosage sensitivity of Nodal for LR patterning. Alterations in the penetrance of laterality defects in heterozygotes versus double heterozygotes or in hypomorphic versus null mutants strongly imply dose-dependent functions of the Nodal signaling pathway and provide mechanistic insight into the complex affects of rare variant-mediated decreases in NODAL signaling in the development of human CVM. Given the strong evidence that the NODAL pathway plays a role in heterotaxy and CVM, future studies will focus on the gene × gene and gene × environment interactions that impact this developmental signaling system.
MATERIALS AND METHODS
Clinical evaluation
Patients were ascertained by the review of the echocardiogram and catheterization reports of children seen in the Cardiology, Cardiovascular Genetics or Cardiovascular Surgery Clinics at Texas Children's Hospital as well as by direct referral. Inclusion criteria required the presence of CVM typically associated with heterotaxy syndrome or whose suspected mechanism involved abnormal heart tube looping. Individual anatomic descriptions of these CVM were obtained by the review of echocardiography, magnetic resonance imaging and angiography. When available, detailed information about associated congenital anomalies was obtained by the review of the medical records. The complete study data entry form is available upon request. All subjects or their guardians provided written informed consent. The study was approved by the Institutional Review Board at Baylor College of Medicine. Blood samples were obtained after informed consent and utilized for the development of EBV-transformed lymphoblast cell lines. Fibroblasts were available from autopsy specimens in a few cases. Genomic DNA was extracted either directly from whole blood or from cultured cells with Puregene DNA Isolation Kit (Gentra Systems, Plymouth, MN, USA) according to the manufacturer's protocol.
Mutation analysis
The probands were screened for mutations in NODAL using PCR-based DHPLC and DNA sequencing. Primers were designed to intron sequences flanking exons of NODAL (Fig. 1 and Supplementary Material, Table 3S). Genomic DNA samples were amplified by PCR using 20 ng genomic DNA in a 30 µl reaction containing 1.5 mm MgCl2, 10 pmol of each primer, 0.5 U platinum-Taq polymerase (Invitrogen, Gaithersburg, MD, USA), and PCR cycling was carried out on a Robo-cycler (Stratagene, La Jolla, CA, USA) using a denaturation cycle at 95°C for 5 min followed by 45 cycles of denaturation at 95°C for 45 s, annealing (Supplementary Material, Table 3S) for 45 s and extension at 72°C for 45 s and a final extension step at 72°C for 5 min. For DHPLC analysis, 50–100 ng of each PCR product was used.
DHPLC and DNA sequence analysis
Heteroduplexes formed by denaturation at 95°C for 5 min, followed by slow cooling to 25°C over a period of 45 min for re-annealing heteroduplexed PCR products, were analyzed by the DHPLC method using the WAVE nucleic acid fragment analysis system (Transgenomic, Omaha, NE, USA) according to the manufacturer's instructions. DNA fragment elution profiles were captured online and visualized using the WAVE MAKER version 4.1 software (Transgenomic). Chromatograms were analyzed and DHPLC peaks were noted. The genomic DNA of the subject were re-amplified, PCR products were purified and sequenced by automated sequencing using a 3100 ABI DNA sequencer (Applied Biosystem) and Big Dye Terminator chemistry (version 3.1), according to the manufacturer's instructions.
Site-directed mutagenesis
A partial IMAGE human NODAL cDNA clone (BC033585.1) in pCMV-SPORT6 vector was purchased from Open Biosystem (Huntsville, AL, USA). The missing 5′ end (approximately 65 bases) was filled by designing long primers to the 5′ end. The primers used for the amplification of the full-length cDNA: forward—5′gccaccatgcacgcccactgcctgcccttccttctgcacgcctggtgggccctactccaggcgggtgctgcgacggtggccactgcgctcctgcgt 3′ and reverse—5′atgtcatcagaggcacccacattcttccac3′.
The PCR product was subcloned into another mammalian cDNA expression vector (pcDNA3.1-V5-6His; Invitrogen, Carlsbad, CA, USA). The anti-V5 epitope tag was used for the detection of the recombinant fusion protein in transfection assays as well as in western blot (data not shown). The identified mutations in the NODAL coding sequence in the gene (E203K, G260R, R275C, V284F, c.700-723delinsTTGACTTCC) were inserted using site-directed mutagenesis (QuickChange®, Stratagene). The mutagenic oligonucleotide primers for both the strands were designed and polyacrylamide gel (PAGE)-purified as suggested by the Kit protocol. The mutagenesis primers used for E203K, G260R, R275C, V284F, c700–723delinsTTGACTTCC are described in Supplementary Material, Table 4S. The mutagenesis protocol was followed per the manufacturer's instruction. PCR was performed with 5–25 ng of wild-type full-length NODAL construct in a 50 µl reaction volume, containing 125 ng of each primer, 200 µm dNTP mix and 2.5 U of Pfu-Ultra DNA polymerase (Stratagene). PCR cycling was carried out on a Robo-cycler (Stratagene) using a denaturation cycle at 95°C for 2 min, followed by 15 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 1 min and extention at 68°C for 12 min. After PCR reaction, the product size was checked on a 0.7% agarose gel and then 10 U of DpnI restriction enzyme was added directly to each amplification reaction and incubated at 37°C for 1 h to digest the parental, non-mutated supercoiled dsDNA. Subsequently, 2 µl of DpnI-treated DNA was used to transform XL1-Blue ultracompetent cells. Mutated clones were confirmed by DNA sequencing using a 3100 ABI DNA sequencer (Applied Biosystems).
Nodal signaling assays
Undifferentiated P19 (mouse embryonic carcinoma) cells which are known to be responsive to Nodal signaling (63) were used. These cells were cultured in α-minimal essential medium containing 7.5% calf serum and 2.5% fetal bovine serum. The cells were seeded overnight at 25–35% confluence in six-well plates and then co-transfected with NODAL expression constructs (wild type, E203K, G260R, R275C, V284F, c.700–723del and c.700–723delinsTTGACTTCC) and luciferase expression reporters using FuGENE 6 transfection reagent (Roche, Indianapolis, IN, USA). The luciferase reporter plasmid pAR3-lux (responsive to TGF-β, Activin and Nodal signaling) was obtained as a gift from Dr Jeffery Wrana (Mount Sinai Hospital, Toronto, Canada). Two other luciferase reporter plasmids, p(SBE)4, responsive to general TGF-β signaling, and p(CAGA)12, specifically responsive to Smad3, were obtained as gifts from Dr Michael R. Kuehn (NCI, MD, USA). As a control, for cell number and transfection efficiency, 100–150 ng of pRL-TK (TK promoter and Renilla luciferase, Promega, Madison, WI, USA) was also included in the transfection mix (one-tenth of total DNA used per transfection). All transfections were done in triplicate with a total amount of 1–1.5 µg DNA per well. Forty-eight hours after transfection, luciferase activity was analyzed using the Dual-Luciferase Reporter Assay System (Promega) in a Monolight-3010 Luminometer (Pharmingen, San Jose, CA, USA).
Western blot
P19 cells were plated in 10 cm plates (106 cells) in 10 ml media. Cells were serum-starved for 12 h prior to transfection. The following day, the cells were transfected with either wild-type NODAL or variant constructs. At 16–18 h post-transfection, the cells were washed in cold 1× PBS containing protease inhibitors cocktail (Complete, Mini; EDTA-free, Roche) plus phosphatase inhibitors [1 mm sodium orthovanadate (Na3VO4) and 1 mm sodium fluoride] and then lysed in RIPA buffer containing protease and phosphatase inhibitors. The protein concentration was determined using BCA Protein Assay Kit (Pierce). In Laemmli sample buffer, 30–50 µg of sample was diluted (Bio-Rad, Hercules, CA, USA), boiled for 5 min and then fractionated by 4–12% gradient NuPAGE® Novex SDS–PAGE (Invitrogen, Carlsbad, CA, USA). After transferring to nitrocellulose membranes, the membranes were briefly washed in TBST, incubated in 5% w/v non-fat dried milk in TBST for 1 h at room temperature, and washed three times with TBST. Western blot analysis was performed using the phospho-specific Smad2 (Ser465/467) antibodies (Cell Signaling Technology Inc., Danvers, MA, USA) by incubating overnight at 4°C (1:1000 dilution) followed by staining (1 h at room temperature) with a horseradish peroxidase anti-rabbit secondary antibody (1:3000 dilution, Vectastain ABC Mouse IgG Kit: Novocastra, Newcastle upon Tyne, UK) and chemiluminescent detection using an ECL kit (Amersham-Pharmacia Biotech, Inc., Piscataway, NJ, USA).
Immunocytochemistry
Cultured P19 cells were plated at 1 × 104 cells per well in a two-well chambered slides (Nalge-Nunc International, Naperville, IL, USA) in 1 ml media. Cells were serum-starved 8–12 h before transfection. Cells were transfected with 0.5 μg of plasmid DNA containing either wild-type NODAL or the constructs carrying NODAL variants using FuGENE 6 (Roche). At 18 h post-transfection, the cells were washed in cold 1× PBS, fixed with 4% paraformaldehyde in PBS, permeabilized with 0.5% Triton X-100 for 5 min, washed and then blocked with 3% BSA plus 10% normal horse/goat serum for 1 h. Cells were incubated with a 1: 200 dilution of rabbit antiphospho-Smad2 antibody (Cell Signaling Technology Inc.) in 1% BSA in TBS overnight at 4°C. This was followed by incubation with Alexa Fluor 488 goat anti-rabbit IgG, diluted 1:500 (Molecular Probes, Eugene, OR, USA) for 1 h at room temperature. After extensive washing, nuclei were stained with 4′,6′-diamidino-2-phenylindole, dihydrochloride (DAPI) (Molecular Probes) diluted 1:1000 in PBS for 5 min at room temperature. The immunoreactivity was visualized by fluorescence microscopy using a Zeiss Axioplan 2 epifluorescence microscope equipped with a CoolSNAP HQ CCD camera.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by NIH HL088639 (S.M.W.) and NIH HD39056 (J.W.B.).
ACKNOWLEDGEMENTS
We would like to thank all the families and patients for their participation as well as referring genetic counselors and physicians.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Ai D., Liu W., Ma L., Dong F., Lu M.F., Wang D., Verzi M.P., Cai C., Gage P.J., Evans S., et al. Pitx2 regulates cardiac left–right asymmetry by patterning second cardiac lineage-derived myocardium. Dev. Biol. 2006;296:437–449. doi: 10.1016/j.ydbio.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chhin B., Hatayama M., Bozon D., Ogawa M., Schon P., Tohmonda T., Sassolas F., Aruga J., Valard A.G., Chen S.C., et al. Elucidation of penetrance variability of a ZIC3 mutation in a family with complex heart defects and functional analysis of ZIC3 mutations in the first zinc finger domain. Hum. Mutat. 2007;28:563–570. doi: 10.1002/humu.20480. [DOI] [PubMed] [Google Scholar]
- 3.Fritz B., Kunz J., Knudsen G.P., Louwen F., Kennerknecht I., Eiben B., Orstavik K.H., Friedrich U., Rehder H. Situs ambiguus in a female fetus with balanced (X;21) translocation—evidence for functional nullisomy of the ZIC3 gene? Eur. J. Hum. Genet. 2005;13:34–40. doi: 10.1038/sj.ejhg.5201213. [DOI] [PubMed] [Google Scholar]
- 4.Gebbia M., Ferrero G.B., Pilia G., Bassi M.T., Aylsworth A., Penman-Splitt M., Bird L.M., Bamforth J.S., Burn J., Schlessinger D., et al. X-linked situs abnormalities result from mutations in ZIC3. Nat. Genet. 1997;17:305–308. doi: 10.1038/ng1197-305. [DOI] [PubMed] [Google Scholar]
- 5.Tzschach A., Hoeltzenbein M., Hoffmann K., Menzel C., Beyer A., Ocker V., Wurster G., Raynaud M., Ropers H.H., Kalscheuer V., et al. Heterotaxy and cardiac defect in a girl with chromosome translocation t(X;1)(q26;p13.1) and involvement of ZIC3. Eur. J. Hum. Genet. 2006;14:1317–1320. doi: 10.1038/sj.ejhg.5201707. [DOI] [PubMed] [Google Scholar]
- 6.Ware S.M., Peng J., Zhu L., Fernbach S., Colicos S., Casey B., Towbin J., Belmont J.W. Identification and functional analysis of ZIC3 mutations in heterotaxy and related congenital heart defects. Am. J. Hum. Genet. 2004;74:93–105. doi: 10.1086/380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Megarbane A., Salem N., Stephan E., Ashoush R., Lenoir D., Delague V., Kassab R., Loiselet J., Bouvagnet P. X-linked transposition of the great arteries and incomplete penetrance among males with a nonsense mutation in ZIC3. Eur. J. Hum. Genet. 2000;8:704–708. doi: 10.1038/sj.ejhg.5200526. [DOI] [PubMed] [Google Scholar]
- 8.Bamford R.N., Roessler E., Burdine R.D., Saplakoglu U., dela Cruz J., Splitt M., Goodship J.A., Towbin J., Bowers P., Ferrero G.B., et al. Loss-of-function mutations in the EGF-CFC gene CFC1 are associated with human left–right laterality defects. Nat. Genet. 2000;26:365–369. doi: 10.1038/81695. [DOI] [PubMed] [Google Scholar]
- 9.Goldmuntz E., Bamford R., Karkera J.D., dela Cruz J., Roessler E., Muenke M. CFC1 mutations in patients with transposition of the great arteries and double-outlet right ventricle. Am. J. Hum. Genet. 2002;70:776–780. doi: 10.1086/339079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selamet Tierney E.S., Marans Z., Rutkin M.B., Chung W.K. Variants of the CFC1 gene in patients with laterality defects associated with congenital cardiac disease. Cardiol Young. 2007;17:268–274. doi: 10.1017/S1047951107000455. [DOI] [PubMed] [Google Scholar]
- 11.Kosaki R., Gebbia M., Kosaki K., Lewin M., Bowers P., Towbin J.A., Casey B. Left–right axis malformations associated with mutations in ACVR2B, the gene for human activin receptor type IIB. Am. J. Med. Genet. 1999;82:70–76. doi: 10.1002/(sici)1096-8628(19990101)82:1<70::aid-ajmg14>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 12.Kosaki K., Bassi M.T., Kosaki R., Lewin M., Belmont J., Schauer G., Casey B. Characterization and mutation analysis of human LEFTY A and LEFTY B, homologues of murine genes implicated in left–right axis development. Am. J. Hum. Genet. 1999;64:712–721. doi: 10.1086/302289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirayama-Yamada K., Kamisago M., Akimoto K., Aotsuka H., Nakamura Y., Tomita H., Furutani M., Imamura S., Takao A., Nakazawa M., et al. Phenotypes with GATA4 or NKX2.5 mutations in familial atrial septal defect. Am. J. Med. Genet. A. 2005;135:47–52. doi: 10.1002/ajmg.a.30684. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe Y., Benson D.W., Yano S., Akagi T., Yoshino M., Murray J.C. Two novel frameshift mutations in NKX2.5 result in novel features including visceral inversus and sinus venosus type ASD. J. Med. Genet. 2002;39:807–811. doi: 10.1136/jmg.39.11.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson S.W., Morris C.D., Goldmuntz E., Reller M.D., Jones M.A., Steiner R.D., Maslen C.L. Missense mutations in CRELD1 are associated with cardiac atrioventricular septal defects. Am. J. Hum. Genet. 2003;72:1047–1052. doi: 10.1086/374319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roessler E., Ouspenskaia M.V., Karkera J.D., Velez J.I., Kantipong A., Lacbawan F., Bowers P., Belmont J.W., Towbin J.A., Goldmuntz E., et al. Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am. J. Hum. Genet. 2008;83:18–29. doi: 10.1016/j.ajhg.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karkera J.D., Lee J.S., Roessler E., Banerjee-Basu S., Ouspenskaia M.V., Mez J., Goldmuntz E., Bowers P., Towbin J., Belmont J.W., et al. Loss-of-function mutations in growth differentiation factor-1 (GDF1) are associated with congenital heart defects in humans. Am. J. Hum. Genet. 2007;81:987–994. doi: 10.1086/522890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peeters H., Debeer P., Bairoch A., Wilquet V., Huysmans C., Parthoens E., Fryns J.P., Gewillig M., Nakamura Y., Niikawa N., et al. PA26 is a candidate gene for heterotaxia in humans: identification of a novel PA26-related gene family in human and mouse. Hum. Genet. 2003;112:573–580. doi: 10.1007/s00439-003-0917-5. [DOI] [PubMed] [Google Scholar]
- 19.Peeters H., Voz M.L., Verschueren K., De Cat B., Pendeville H., Thienpont B., Schellens A., Belmont J.W., David G., Van De Ven W.J., et al. Sesn1 is a novel gene for left–right asymmetry and mediating nodal signaling. Hum. Mol. Genet. 2006;15:3369–3377. doi: 10.1093/hmg/ddl413. [DOI] [PubMed] [Google Scholar]
- 20.Peeters H., Devriendt K. Human laterality disorders. Eur. J. Med. Genet. 2006;49:349–362. doi: 10.1016/j.ejmg.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Conlon F.L., Barth K.S., Robertson E.J. A novel retrovirally induced embryonic lethal mutation in the mouse: assessment of the developmental fate of embryonic stem cells homozygous for the 413.d proviral integration. Development. 1991;111:969–981. doi: 10.1242/dev.111.4.969. [DOI] [PubMed] [Google Scholar]
- 22.Conlon F.L., Lyons K.M., Takaesu N., Barth K.S., Kispert A., Herrmann B., Robertson E.J. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development. 1994;120:1919–1928. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- 23.Lowe L.A., Supp D.M., Sampath K., Yokoyama T., Wright C.V., Potter S.S., Overbeek P., Kuehn M.R. Conserved left–right asymmetry of nodal expression and alterations in murine situs inversus. Nature. 1996;381:158–161. doi: 10.1038/381158a0. [DOI] [PubMed] [Google Scholar]
- 24.Zhou X., Sasaki H., Lowe L., Hogan B.L., Kuehn M.R. Nodal is a novel TGF-beta-like gene expressed in the mouse node during gastrulation. Nature. 1993;361:543–547. doi: 10.1038/361543a0. [DOI] [PubMed] [Google Scholar]
- 25.Feldman B., Gates M.A., Egan E.S., Dougan S.T., Rennebeck G., Sirotkin H.I., Schier A.F., Talbot W.S. Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature. 1998;395:181–185. doi: 10.1038/26013. [DOI] [PubMed] [Google Scholar]
- 26.Jones C.M., Kuehn M.R., Hogan B.L., Smith J.C., Wright C.V. Nodal-related signals induce axial mesoderm and dorsalize mesoderm during gastrulation. Development. 1995;121:3651–3662. doi: 10.1242/dev.121.11.3651. [DOI] [PubMed] [Google Scholar]
- 27.Sampath K., Cheng A.M., Frisch A., Wright C.V. Functional differences among Xenopus nodal-related genes in left–right axis determination. Development. 1997;124:3293–3302. doi: 10.1242/dev.124.17.3293. [DOI] [PubMed] [Google Scholar]
- 28.Hamada H., Meno C., Watanabe D., Saijoh Y. Establishment of vertebrate left–right asymmetry. Nat. Rev. Genet. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- 29.Levin M., Pagan S., Roberts D.J., Cooke J., Kuehn M.R., Tabin C.J. Left/right patterning signals and the independent regulation of different aspects of situs in the chick embryo. Dev. Biol. 1997;189:57–67. doi: 10.1006/dbio.1997.8662. [DOI] [PubMed] [Google Scholar]
- 30.Schier A.F. Nodal signaling in vertebrate development. Annu. Rev. Cell Dev. Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- 31.Schier A.F., Shen M.M. Nodal signalling in vertebrate development. Nature. 2000;403:385–389. doi: 10.1038/35000126. [DOI] [PubMed] [Google Scholar]
- 32.Whitman M. Nodal signaling in early vertebrate embryos: themes and variations. Dev. Cell. 2001;1:605–617. doi: 10.1016/s1534-5807(01)00076-4. [DOI] [PubMed] [Google Scholar]
- 33.Mercola M., Levin M. Left–right asymmetry determination in vertebrates. Annu. Rev. Cell Dev. Biol. 2001;17:779–805. doi: 10.1146/annurev.cellbio.17.1.779. [DOI] [PubMed] [Google Scholar]
- 34.Harvey R.P. Links in the left/right axial pathway. Cell. 1998;94:273–276. doi: 10.1016/s0092-8674(00)81468-3. [DOI] [PubMed] [Google Scholar]
- 35.Massague J. TGF-beta signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 36.Attisano L., Wrana J.L. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 37.Saijoh Y., Adachi H., Sakuma R., Yeo C.Y., Yashiro K., Watanabe M., Hashiguchi H., Mochida K., Ohishi S., Kawabata M., et al. Left–right asymmetric expression of lefty2 and nodal is induced by a signaling pathway that includes the transcription factor FAST2. Mol. Cell. 2000;5:35–47. doi: 10.1016/s1097-2765(00)80401-3. [DOI] [PubMed] [Google Scholar]
- 38.Bianco C., Adkins H.B., Wechselberger C., Seno M., Normanno N., De Luca A., Sun Y., Khan N., Kenney N., Ebert A., et al. Cripto-1 activates nodal- and ALK4-dependent and -independent signaling pathways in mammary epithelial cells. Mol. Cell. Biol. 2002;22:2586–2597. doi: 10.1128/MCB.22.8.2586-2597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan Y.T., Liu J.J., Luo Y., Chaosu E., Haltiwanger R.S., Abate-Shen C., Shen M.M. Dual roles of Cripto as a ligand and coreceptor in the nodal signaling pathway. Mol. Cell. Biol. 2002;22:4439–4449. doi: 10.1128/MCB.22.13.4439-4449.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding J., Yang L., Yan Y.T., Chen A., Desai N., Wynshaw-Boris A., Shen M.M. Cripto is required for correct orientation of the anterior–posterior axis in the mouse embryo. Nature. 1998;395:702–707. doi: 10.1038/27215. [DOI] [PubMed] [Google Scholar]
- 41.Gritsman K., Zhang J., Cheng S., Heckscher E., Talbot W.S., Schier A.F. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell. 1999;97:121–132. doi: 10.1016/s0092-8674(00)80720-5. [DOI] [PubMed] [Google Scholar]
- 42.Nomura M., Li E. Smad2 role in mesoderm formation, left–right patterning and craniofacial development. Nature. 1998;393:786–790. doi: 10.1038/31693. [DOI] [PubMed] [Google Scholar]
- 43.Oh S.P., Li E. The signaling pathway mediated by the type IIB activin receptor controls axial patterning and lateral asymmetry in the mouse. Genes Dev. 1997;11:1812–1826. doi: 10.1101/gad.11.14.1812. [DOI] [PubMed] [Google Scholar]
- 44.Shen M.M., Wang H., Leder P. A differential display strategy identifies Cryptic, a novel EGF-related gene expressed in the axial and lateral mesoderm during mouse gastrulation. Development. 1997;124:429–442. doi: 10.1242/dev.124.2.429. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto M., Meno C., Sakai Y., Shiratori H., Mochida K., Ikawa Y., Saijoh Y., Hamada H. The transcription factor FoxH1 (FAST) mediates Nodal signaling during anterior–posterior patterning and node formation in the mouse. Genes Dev. 2001;15:1242–1256. doi: 10.1101/gad.883901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pogoda H.M., Solnica-Krezel L., Driever W., Meyer D. The zebrafish forkhead transcription factor FoxH1/Fast1 is a modulator of nodal signaling required for organizer formation. Curr. Biol. 2000;10:1041–1049. doi: 10.1016/s0960-9822(00)00669-2. [DOI] [PubMed] [Google Scholar]
- 47.Lowe L.A., Yamada S., Kuehn M.R. Genetic dissection of nodal function in patterning the mouse embryo. Development. 2001;128:1831–1843. doi: 10.1242/dev.128.10.1831. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen K.B., Sorensen S., Cartegni L., Corydon T.J., Doktor T.K., Schroeder L.D., Reinert L.S., Elpeleg O., Krainer A.R., Gregersen N., et al. Seemingly neutral polymorphic variants may confer immunity to splicing-inactivating mutations: a synonymous SNP in exon 5 of MCAD protects from deleterious mutations in a flanking exonic splicing enhancer. Am. J. Hum. Genet. 2007;80:416–432. doi: 10.1086/511992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parmley J.L., Chamary J.V., Hurst L.D. Evidence for purifying selection against synonymous mutations in mammalian exonic splicing enhancers. Mol. Biol. Evol. 2006;23:301–309. doi: 10.1093/molbev/msj035. [DOI] [PubMed] [Google Scholar]
- 50.Bassi M.T., Kosaki K., Belmont J., Casey B. NODAL, LEFTY, and HNF3b changes associated with complex heart defects and other features of left–right axis malformations (abstract no. 8) Am. J. Hum. Genet. 1997;61(suppl.):A4. [Google Scholar]
- 51.Faure S., Lee M.A., Keller T., ten Dijke P., Whitman M. Endogenous patterns of TGFbeta superfamily signaling during early Xenopus development. Development. 2000;127:2917–2931. doi: 10.1242/dev.127.13.2917. [DOI] [PubMed] [Google Scholar]
- 52.Cui X.M., Chai Y., Chen J., Yamamoto T., Ito Y., Bringas P., Shuler C.F. TGF-beta3-dependent SMAD2 phosphorylation and inhibition of MEE proliferation during palatal fusion. Dev. Dyn. 2003;227:387–394. doi: 10.1002/dvdy.10326. [DOI] [PubMed] [Google Scholar]
- 53.Cui X.M., Shiomi N., Chen J., Saito T., Yamamoto T., Ito Y., Bringas P., Chai Y., Shuler C.F. Overexpression of Smad2 in Tgf-beta3-null mutant mice rescues cleft palate. Dev. Biol. 2005;278:193–202. doi: 10.1016/j.ydbio.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 54.Beck S., Le Good J.A., Guzman M., Ben Haim N., Roy K., Beermann F., Constam D.B. Extraembryonic proteases regulate Nodal signalling during gastrulation. Nat. Cell Biol. 2002;4:981–985. doi: 10.1038/ncb890. [DOI] [PubMed] [Google Scholar]
- 55.Cui Y., Hackenmiller R., Berg L., Jean F., Nakayama T., Thomas G., Christian J.L. The activity and signaling range of mature BMP-4 is regulated by sequential cleavage at two sites within the prodomain of the precursor. Genes Dev. 2001;15:2797–2802. doi: 10.1101/gad.940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Good J.A., Joubin K., Giraldez A.J., Ben-Haim N., Beck S., Chen Y., Schier A.F., Constam D.B. Nodal stability determines signaling range. Curr. Biol. 2005;15:31–36. doi: 10.1016/j.cub.2004.12.062. [DOI] [PubMed] [Google Scholar]
- 57.Thomsen G.H., Melton D.A. Processed Vg1 protein is an axial mesoderm inducer in Xenopus. Cell. 1993;74:433–441. doi: 10.1016/0092-8674(93)80045-g. [DOI] [PubMed] [Google Scholar]
- 58.Bodmer W., Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weinstein M., Yang X., Li C., Xu X., Gotay J., Deng C.X. Failure of egg cylinder elongation and mesoderm induction in mouse embryos lacking the tumor suppressor smad2. Proc. Natl Acad. Sci. USA. 1998;95:9378–9383. doi: 10.1073/pnas.95.16.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heyer J., Escalante-Alcalde D., Lia M., Boettinger E., Edelmann W., Stewart C.L., Kucherlapati R. Postgastrulation Smad2-deficient embryos show defects in embryo turning and anterior morphogenesis. Proc. Natl Acad. Sci. USA. 1999;96:12595–12600. doi: 10.1073/pnas.96.22.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oh S.P., Yeo C.Y., Lee Y., Schrewe H., Whitman M., Li E. Activin type IIA and IIB receptors mediate Gdf11 signaling in axial vertebral patterning. Genes Dev. 2002;16:2749–2754. doi: 10.1101/gad.1021802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Collignon J., Varlet I., Robertson E.J. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 1996;381:155–158. doi: 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- 63.Kumar A., Novoselov V., Celeste A.J., Wolfman N.M., ten Dijke P., Kuehn M.R. Nodal signaling uses activin and transforming growth factor-beta receptor-regulated Smads. J. Biol. Chem. 2001;276:656–661. doi: 10.1074/jbc.M004649200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.