Abstract
Blood lipid levels, including low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C) and triglycerides (TG), are highly heritable traits and major risk factors for atherosclerotic cardiovascular disease (CVD). Using individual ancestry estimates at marker locations across the genome, we present a novel quantitative admixture mapping analysis of all three lipid traits in a large sample of African-Americans from the Family Blood Pressure Program. Regression analysis was performed with both total and marker-location-specific European ancestry as explanatory variables, along with demographic covariates. Robust permutation analysis was used to assess statistical significance. Overall European ancestry was significantly correlated with HDL-C (negatively) and TG (positively), but not with LDL-C. We found strong evidence for a novel locus underlying HDL-C on chromosome 8q, which correlated negatively with European ancestry (P = .0014); the same location also showed positive correlation of European ancestry with TG levels. A region on chromosome 14q also showed significant negative correlation between HDL-C levels and European ancestry. On chromosome 15q, a suggestive negative correlation of European ancestry with TG and positive correlation with HDL-C was observed. Results with LDL-C were less significant overall. We also found significant evidence for genome-wide ancestry effects underlying the joint distribution of HDL-C and TG, not fully explained by the locus on chromosome 8. Our results are consistent with a genetic contribution to and may explain the healthier HDL-C and TG profiles found in Blacks versus Whites. The identified regions provide locations for follow-up studies of genetic variants underlying lipid variation in African-Americans and possibly other populations.
INTRODUCTION
Cardiovascular disease (CVD) is a leading cause of morbidity and mortality, making it a major public health concern worldwide (1). The etiology of the disease is multifactorial, with established genetic and environmental risk factors (2–5). Atherosclerosis, which is characterized by lipid accumulation, inflammatory response, cell death and fibrosis in arterial walls, accounts for 75% of all deaths from CVD (6). There is compelling evidence of involvement of blood lipid concentrations in atherosclerosis and CVD (7–9). Blood lipids are complex traits influenced by both genetic and environmental factors (2–5,10). Heritability estimates for lipid concentrations vary [∼40–75% for high-density lipoprotein cholesterol (HDL-C), ∼40–65% for low-density lipoprotein cholesterol (LDL-C) and ∼35–50% for triglycerides (TG)] (11), yet are consistently high for all lipid traits.
The genetic dissection of these complex traits has long been a major challenge, and there have been many studies aimed at finding specific loci for these phenotypes. Various study designs have been employed, ranging from large-scale genome-wide association (12–15) to candidate gene (16–19) and linkage studies (6,20,21). Recently, Willer et al. (15), in a large meta-analysis combining genome-wide scans with ∼9000 individuals of European descent in stage 1 and ∼11 500 individuals of European descent in stage 2, found convincing evidence for 11 loci (genes) previously implicated in lipid metabolism as well as compelling evidence for several newly identified loci (two for HDL-C, three for TG, one for LDL-C and one region encompassing several genes associated with both LDL-C and TG).
At the same time, there are known ethnic trends in lipid levels (22,23). For example, after adjusting for socioeconomic factors, African-Americans generally have healthier lipid levels (lower TG and higher HDL-C) than people of European descent (16,24,25), whereas both have healthier lipid levels than South Asians (26,27). This underscores the importance of undertaking large-scale mapping studies of populations with distinct ethnic backgrounds.
New world admixed populations provide unique opportunities for locating trait loci by genetic admixture mapping (28–33). The African-American population of the United States is typically characterized by admixture of European and African ancestral genomes in different proportions with some spatial variation (34–36). Studies have correlated genome-wide African ancestry in African-Americans with healthier lipid levels (37). In this report, we present results of an admixture mapping analysis examining the correlation of lipid levels (LDL-C, HDL-C and TG) with estimated genome-wide ancestry proportions as well as ancestry estimates at each of 284 marker loci distributed around the genome in 1044 unrelated African-Americans subjects from the Family Blood Pressure Program (FBPP), in a search for locus-specific effects.
RESULTS
Sample demographics for the 1044 individuals are given in Table 1. There were 698 females and 346 males. The subjects from GENOA were on average older (average age 59.6) than the HyperGEN subjects (average age 49.4). Within each network, the average body mass index (BMI), HDL-C and LDL-C were generally higher in females than in males, whereas TG levels showed slightly reversed tendencies. The male and female subjects from GENOA had higher HDL-C and TG levels than their corresponding counterparts from HyperGEN. There was little variation in BMI between networks. Average European ancestry varied modestly between the two networks, as was previously noted (36).
Table 1.
Demographic characteristics of unrelated study participants: Family Blood Pressure Program
| GENOA |
HyperGEN |
|||
|---|---|---|---|---|
| Male | Female | Male | Female | |
| Number of individuals | 102 | 247 | 244 | 451 |
| BMI | 29.25 ± 4.9 | 31.77 ± 6.9 | 29.71 ± 6.3 | 33.79 ± 8.5 |
| Age | 61.83 ± 8.3 | 58.77 ± 9.6 | 49.17 ± 12.3 | 49.53 ± 12.3 |
| HDL-C | 52.96 ± 18.6 | 60.37 ± 18.8 | 50.25 ± 15.3 | 55.98 ± 15.1 |
| LDL-C | 110.06 ± 43.4 | 121.01 ± 38.1 | 119.15 ± 34.4 | 120.97 ± 36.9 |
| TG | 145.12 ± 84.2 | 142.57 ± 67.8 | 116.75 ± 75.7 | 109.24 ± 76.0 |
| European ancestry (%) | 14.2 | 15.57 | 13.35 | 12.16 |
LDL-C levels were modestly correlated with HDL-C and TG levels (correlation coefficient −0.13 with HDL-C and 0.19 with TG). HDL-C and TG, as observed in other studies, showed a stronger inverse correlation (−0.28). The correlation between the traits was virtually unaffected by the transformations. By analysis of variance, all three lipid traits were strongly associated with BMI and age (Table 2). HDL-C was strongly associated with sex. TG and LDL-C differed significantly between networks.
Table 2.
Analysis of variance results on LDL, HDL and TG
| df | F-value | P-value | |
|---|---|---|---|
| LDL-C | |||
| Log (log(BMI)) | 1 | 30.18 | 4.99 × 10−8 |
| Age | 1 | 14.41 | 1.56 × 10−4 |
| Sex | 1 | 0.37 | 0.54 |
| Network | 1 | 6.31 | 0.012 |
| HDL-C | |||
| Log (log(BMI)) | 1 | 39.04 | 6.05 × 10−10 |
| Age | 1 | 31.96 | 2.00 × 10−8 |
| Sex | 1 | 78.39 | <2.2 × 10−16 |
| Network | 1 | 0.20 | 0.66 |
| TG | |||
| Log (log(BMI)) | 1 | 18.03 | 2.37 × 10−05 |
| Age | 1 | 74.66 | <2.2 × 10−16 |
| Sex | 1 | 2.84 | 0.09 |
| Network | 1 | 53.44 | 5.31 × 10−13 |
LDL-C was not found to be correlated with overall European individual ancestry (IA) (P = .80). On the other hand, HDL-C was strongly negatively correlated with European IA (t = −2.76, P=0.0059) and TG was strongly positively correlated with European IA (t = 3.29, P = .0010).
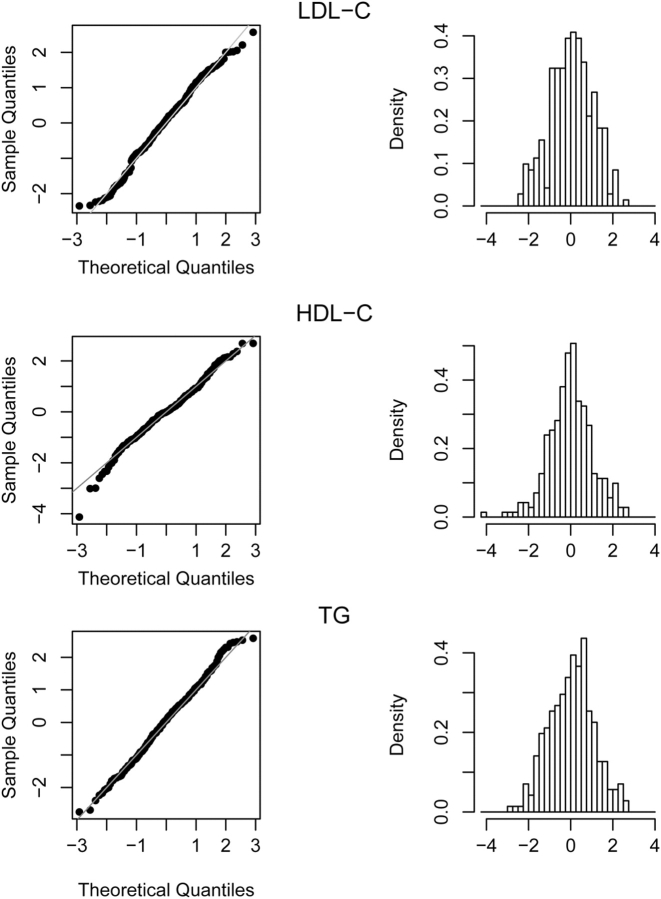
A Q–Q plot of the rl values (standardized regression coefficients) for the 284 loci for LDL-C, TG and HDL-C showed a reasonably good fit to normality (Fig. 1) except for some outlier points, in particular at the low (negative) end of the distribution for HDL-C.
Figure 1.
Q–Q plot comparing rl with a standard normal distribution and the corresponding histograms for the trait LDL-C, HDL-C and TG.
According to normal distribution theory, we expected 14 (∼5% of 284) marker positions to have an absolute standardized regression coefficient |rl| value > 1.96, or seven in each tail. However, we found an excess of points in the tails in each of the three distributions. For LDL-C, there were seven rl values > 1.96 as expected but 10 in the left tail (i.e. values less than −1.96) (Supplementary Material, Table S1). For HDL-C, there were 11 rl values > 1.96 (i.e. in the right tail) and 10 in the left tail (less than −1.96) (Supplementary Material, Table S2). For TG, there were 12 rl values > 1.96 (i.e. in the right tail) and eight in the left tail (Supplementary Material, Table S3).
For LDL-C, the two most promising chromosomal regions were at 3q and 4q, both showing negative correlation with European ancestry. On 3q, five markers covering 64 cM had Z-scores less than −2.0, with a peak of −2.35 at 188.3 cM (marker D3S2427). On chromosome 4q, three markers had Z-scores less than −2.0, with a peak of −2.33 at 78 cM (marker D4S2367).
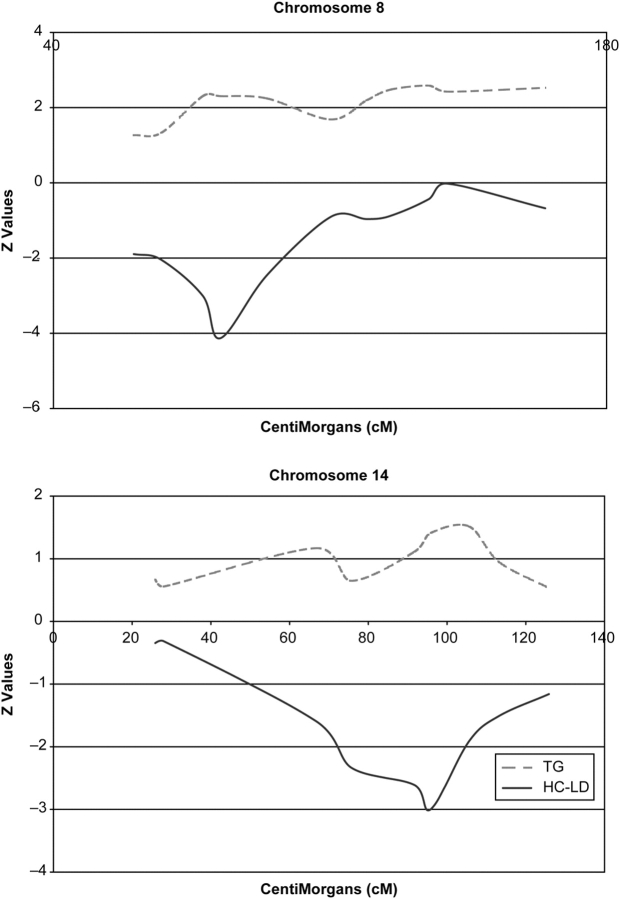
For HDL-C, two significant regions on chromosomes 8q and 14q showed negative correlation with European ancestry, whereas a region on chromosome 9q showed positive correlation with European ancestry. Chromosome 8q had four markers covering 27 cM with Z-scores less than −2.0, with a peak of −4.13 at 82.3 cM (marker D8S1136). On chromosome 14q, three markers had Z-scores less than −2, with a peak of −3.00 at 95.9 cM (marker GATA193A07). On chromosome 9q, there were three markers with Z-scores > 2, with a peak of 2.69 at 110.9 cM.
The two most promising locations for TG included one extended region positively correlated with European ancestry on chromosome 8q and another negatively correlated with European ancestry on chromosome 15q. On chromosome 15q, the peak Z-score of −2.75 occurred at 90.0 cM (marker D15S652), whereas two neighboring markers also had Z-scores less than −2. On chromosome 8q, an extended region of 87 cM stretching from 77.9 to 164.5 cM contained seven markers with Z-scores > 2. Two separate peaks occurred in this interval, one at 135.1 cM with a Z-score of 2.58 (marker D8S1179) and the other at 77.9–82.3 cM (markers D8S1113 and D8S1136) with a Z-score of 2.31. We note that this is precisely the same location harboring the most significant result for HDL-C (marker D8S1136).
To determine the statistical significance of our findings, we employed a permutation test (see Materials and Methods). For LDL-C and TG, none of the markers was significant at P < 0.05. For HDL-C, however, in 5000 permutations, the minimum of 284 rl scores crossed −4.13 only seven times, −3.01 seventy-three times and −2.99 ninety times. Hence, the result associated with the marker D8S1136 is highly significant on a genome-wide level (P = 0.0014), whereas that for D8S1113 and GATA193A07 are also formally significant (P < 0.02).
We also looked at possible significant interactions of BMI, age, sex, network, usage of lipid-lowering medication, and hypertension status with local ancestry at the significant genomic marker locations for all three lipid traits and did not find any. The Z-scores for HDL-C and TG across chromosomes 8 and 14 are plotted in Figure 2, which also gives a visual impression of the inverse relationship between the two traits. We also examined the possibility of outlier points driving the Z-scores for the three significant loci (Supplementary Material, Fig. S1) but found no evidence of outliers.
Figure 2.
Overview of the Z-scores and its inverse relationship for HDL-C and TG across chromosomes 8 and 14.
Furthermore, as another check on the robustness of our conclusions, we examined results of multiple additional analyses based on different randomly selected sets of unrelated individuals from the African-American families in our study. The results for these subsets were essentially identical to those we obtained in the original analysis, indicating that our results were not dependent on the original selection of unrelated individuals from these families.
Our results also indicated an overall genome-wide ancestry deviation in opposite directions for HDL-C and TG. To determine whether multiple loci are influential in the joint distribution of these lipid traits, we looked at the correlation between the regression coefficients of the 284 markers for the two lipids. If a common set of genes is responsible, for example, for both high HDL-C and low TG values, then we would observe a large negative correlation between the ancestry regression coefficients.
For the 284 markers, the correlation between the regression coefficients for HDL-C and TG was −0.55. To determine statistical significance of this correlation, we performed 5000 permutations (as described in Materials and Methods), and for each permutation calculated the pairwise correlation between the ancestry regression coefficients for the two lipid traits. Out of 5000 permutations, the correlation exceeded −0.55 only 70 times, which corresponds to a P-value of .014. After removing the two chromosomes eight markers with highly negative rl scores for HDL-C and highly positive rl scores for TG from the analysis, the correlation declined only to −0.53, which was still statistically significant (P = 0.028). After removal of the markers on chromosomes 8q and 15q, the correlation still trended in the same direction but was no longer significant (P = .063). To assess whether the regressions of HDL-C and TG on total European IA could be attributed solely to ancestry at the peak location on chromosome 8q, we performed a regression analysis of HDL-C (and TG) including both 8q locus-specific European ancestry and total European ancestry in the model. For both lipids, total European IA was no longer significant (and regression coefficients were close to 0), once the locus-specific European ancestry was included in the model. Hence, the overall ancestry effect we observed for both lipids could be explained simply by a locus on chromosome 8q, although the situation may be more complex, with other loci contributing.
DISCUSSION
Numerous genes have been directly implicated in the variation of lipid traits such as apolipoproteins (APOA1, APOA2, APOA4, APOE), ATP-binding cassette sub-family A member 1 (ABCA1), lecithin cholesterol acyltransferase (LCAT) and lipoprotein lipase (LPL), among others (16,38–43). All these genes have been extensively studied and well-characterized biochemically. With the exception of APOE, variants in the above-mentioned genes and a few others have been found mostly in Mendelian disorders involving lipid metabolism. Identifying more common and/or less penetrant variants affecting lipid levels has been a more challenging endeavor.
Among the three lipid traits we studied, LDL-C had the least significant results. Although no points for LDL-C were formally significant, we did note a modest increase in markers correlated with excess African ancestry on chromosomes 3q and 4q. The region on chromosome 3q was previously identified in a linkage study with 470 subjects in 10 pedigrees with a maximum LOD score of 4.1 (44), but not in other linkage and association studies. On chromosome 4q, the marker D4S2367, which had the second lowest rl value, is close to the SNP rs10518072. This SNP was associated with LDL-C levels in the Framingham Heart genome-wide association study (P = 1.4 × 10−4) (13) but was not reported by Willer et al. (15) study.
In contrast, the results for HDL-C were more significant. Most of the markers in the extreme left tail (negative correlation with European ancestry) are on chromosomes 8q and 14q. The peak region on chromosome 8q (at 82 cM), our most significant finding overall, also showed evidence of positive correlation of European ancestry with TG. Deviations in the other tail of the Z-score distribution were more subtle and less significant. Three consecutive markers on chromosome 9q showed up in this tail, with a peak between 104 and 111 cM.
Of interest, the region on chromosomes 9q22–q32 that we identified harbors the ABC1 (ATP-binding cassette) gene at 9q31. ABC1 has long been associated with studies involving HDL-C and is regarded as a candidate gene for HDL-C deficiency and other related phenotypes (45–48). Mutations in ABC1 can cause Tangier disease, a genetic disorder of cholesterol characterized by very low levels of HDL-C (46,47,49–52). Hence, ABC1 would be a candidate for an HDL-C quantitative trait locus (QTL) in our analysis as well.
The three markers in our study, which were clearly significant after permutation testing, are D8S1113 and D8S1136 from the region 8q11.23–q21.11 and GATA193A07 from the region 14q24.1–q31. The region 8q11.23–q21.11 is the location of the CYP7A1 gene at 8q12.1. This gene encodes a member of the cytochrome P450 superfamily of enzymes which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. This region also harbors the candidate gene CYP7B1 (at 8q12.3; 65.6 Mb from pter) between the markers D8S1113 and D8S1136. Little other evidence in the literature implicates this region of 8q in HDL-C or TG levels, suggesting our results may be indicating a novel locus. In a study of lipid levels among 113 Hispanic families, the GATA193A07 marker on chromosome 14 showed modest evidence of linkage for TG (two point LOD score 2.12) and for the ratio of TG to HDL-C (two point LOD score 2.46) (52). Weak evidence of linkage to the locus was also obtained in a genome scan of QTLs influencing HDL-C levels (19,53).
Analysis of TG did not reveal any formally significant loci, although there was an overall excess of observations in the right tail of the Z-score distribution. This excess was due to a preponderance of loci on chromosome 8. Three of these loci overlapped with our most significant region for HDL-C at 8q11–q21, whereas four other markers mapped to a non-overlapping, more distal location between 8q23 and q24. We do note, however, that rs7007075, the SNP most strongly associated (P < 7.7 × 10−6) with TG levels in the Framingham Heart genome-wide association study, lies in this region. The SNP rs17321515 (8q24.13) at 3′ downstream of the gene TRIB1 has been one of the most significant findings (P=4 × 10−17) of genome-wide association studies (12). This study also reported five independently associated SNPs in this region, all of which lie within the two consecutive markers D8S592 at 8q24.11 and D8S1179 at 8q24.13, included in our study. The rl value corresponding to the marker D8S1179 (2.59) and D8S592 (2.48) for TG are the highest and the third highest that we found in our study (Supplementary Material, Table S3).
As noted by us and others, the various lipid fractions are modestly correlated within individuals. The largest correlation is the negative association between HDL-C and TG. Prior studies have suggested this correlation is, in part, genetically determined (44,54–62). Thus, it is appropriate in any genomic analysis to search for genes underlying the joint distribution of these traits.
In our admixture mapping analysis, we therefore searched for locations potentially underlying multiple traits. Examination of the joint distribution of regression coefficients for HDL-C and TG did indeed demonstrate a correlation (−0.55) that was statistically significant based on a permutation analysis. The excess correlation over expected was due partially but not exclusively to markers on chromosomes 8q and 15q, suggesting a possible multi-genic contribution to this correlation.
Admixture mapping analysis is complementary to other gene mapping approaches such as QTL linkage analysis and genome-wide association studies. The power of the method depends both on a measurable effect influencing the trait of interest and on a sizeable difference in allele frequency between ancestral populations, in this case Europeans and Africans. Some of our more significant findings overlapped with prior linkage and genome-wide association studies, and these indicate chromosome regions for more detailed study in follow-up. However, the degree of overlap among these approaches depends on a number of factors, including the underlying allele frequencies and degree of linkage disequilibrium between trait-associated alleles and marker SNPs.
The power of admixture mapping is also dependent upon the accuracy of the estimates of locus-specific ancestry. Ancestry informativeness of the marker set that we have used is on an average 50% and never falls < 40% (33).
Of note, the direction of our significant findings for HDL-C and TG and lack of significant findings for LDL-C are consistent with a genetic contribution to the known ethnic difference for HDL-C and TG. Specifically, the regression coefficient we obtained for European IA on (untransformed) HDL-C (−8.18) can fully explain the observed Black–White differences in HDL-C. If we assume that African-Americans have 15% European ancestry, on average (36), then the expected difference between Whites and African-Americans would be 85% of −8.18 or −6.95. In our sample, the standard deviation of HDL-C was 16.80, so the projected difference of −6.95 units corresponds ∼0.4 standard deviations, a figure comparable to the ethnic difference observed in two large studies (24,25). Similarly, the regression coefficient of European IA on (untransformed) TG levels was 64.47. The standard deviation of TG in our sample was 76.36. Therefore, the predicted ethnic difference from our analysis would be 85% of 64.47 = 54.80, which corresponds ∼0.7 standard deviations, again comparable to the differences observed for US Blacks and Whites (24,25).
To our knowledge, this is the largest quantitative admixture mapping effort of blood lipid levels in terms of sample size and marker locus involvement. Overall, our findings are encouraging and provide regions for follow-up analyses of genes influencing HDL-C and TG, and possibly LDL-C in these and other African-American individuals.
MATERIALS AND METHODS
Subjects
The FBPP is a large multicenter genetic study of high blood pressure and related traits in multiple racial/ethnic groups, including European-Americans, African-Americans, Mexican Americans and Asians and Asian Americans (63). It includes four component networks: GenNet, GENOA, HyperGEN and SAPPHIRE. GenNet, GENOA and HyperGEN who independently collected samples from European-American and African-American families.
All the individuals we included in this study were unrelated self-identified African-Americans from the field centers of GENOA and HyperGEN for whom fasting plasma lipid concentrations were available. To maximize the number of unrelated individuals in our sample, whenever possible we selected unrelated founder individuals; otherwise we chose at random one individual per family. Our final sample of 1044 individuals consisted of 349 individuals sampled by the GENOA network and 695 individuals sampled by the HyperGEN network. Out of these 1044 individuals, 32 did not have their LDL-C on record, so all our findings for LDL-C are based on 1012 individuals, whereas those for HDL-C and TG are based on 1044 individuals. There were 57 individuals (52 for LDL-C) who were noted to be taking lipid-lowering medication. The two networks included in this study measured fasting plasma lipids using standard enzymatic methods in laboratories participating in the CDC standardization program. All individuals who participated in the FBPP gave informed consent; the Institutional Review Board at each clinic site approved all protocols, and a Certificate of Confidentiality was obtained from the Federal Government for this study.
Genotyping
DNA was extracted from whole blood by standard methods by each of the four FBPP networks and was sent to the US National Heart, Lung and Blood Institute’s Mammalian genotyping service in Marshfield, Wisconsin, for genotyping. Screening set 8 (372 highly polymorphic microsattelite markers with an average inter-marker map distance of 10 cM) was used.
Statistical analysis
We used the computer program Structure (64,65) to estimate genome-wide, as well as site-specific IAs in all African-American participants. The linkage model was used, with genetic distance between markers specified according to the Marshfield map. In each analysis, the Markov Chain Monte Carlo algorithm was run for 100 000 steps of burn-in followed by another 100 000 steps. We assumed a model with two ancestral populations. We included 1378 unrelated non-Hispanic White participants from the FBPP to represent the European ancestral population as well as 127 unrelated sub-Saharan African individuals from the Human Genome Diversity Project to represent the African ancestral population (66). The latter set of individuals had been genotyped at more than 300 short tandem repeat at the time of our analysis, and we included genotypes at 284 markers which were overlapping with the FBPP genotypes.
The distributions of all three lipid traits for the 1044 (1012 for LDL-C) unrelated individuals were non-normal due to positive skewness. The best normalization of the LDL-C data was obtained using the Box–Cox power transformation (67) with parameter (λ) 0.69, whereas the value of λ was −0.33 and −0.14, respectively, for HDL-C and TG.
We first calculated regression coefficients for each of the transformed lipid traits on overall European IA, allowing for covariates. Then, for each individual at each locus, a European ancestry deviation xl was defined as the estimated ancestry at locus l minus the background ancestry estimated from the genome-wide markers for that individual, as previously described (68). The variable xl was then used as the primary independent variable in a linear regression model with the quantitative trait value of LDL-C or HDL-C or TG as the dependent variable. Log–log transformed BMI, age, sex and network were included as covariates in this analysis, if significant. The use of lipid-lowering medication was not found to be significant for any of the traits, possibly due to the small number of individuals (n = 57) on medication. Hypertension status was also included as a covariate, but was not significant for any of the traits. We also checked for possible two factor interactions and none was significant. All non-significant covariates and interactions were dropped from the model in subsequent analyses. The standardized regression coefficient of rl, defined as, was assumed to be distributed as asymptotically normal and was used to assess statistical significance. To account for multiple testing (284 markers), we performed a permutation analysis in which we randomly reassigned the vector of genetic ancestry estimates for the 284 marker locations to individuals whose LDL-C, HDL-C, TG and covariate data remained intact. This procedure preserved the correlation structure of the markers and the correlation structure of the traits and covariates, but dissociated the relationship between the markers and phenotypes. For each permuted dataset, we performed the same regression analysis of the traits on excess ancestry at each marker location, as was done for the original data, and obtained the most extreme values (positive and negative) of rl (the Z-score statistics). Five thousand permutations were performed. To derive P-values adjusted for multiple testing, we determined the percentage of times out of 5000 permutations that an observed value of rl was exceeded in the permuted data analysis.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by grants awarded to the Family Blood Pressure Program, which is supported by a series of cooperative agreements from the National Heart, Lung and Blood Institute to GenNet, HyperGEN, GENOA and SAPPHIRe.
ACKNOWLEDGEMENTS
We thank Mark Kvale for his assistance with analysis. The following investigators are associated with the Family Blood Pressure Program: GenNet Network: Alan B. Weder (Network Director), Lillian Gleiberman (Network Coordinator), Anne E. Kwitek, Aravinda Chakravarti, Richard S. Cooper, Carolina Delgado, Howard J. Jacob and Nicholas J. Schork. GENOA Network: Eric Boerwinkle (Network Director), Tom Mosley, Alanna Morrison, Kathy Klos, Craig Hanis, Sharon Kardia and Stephen Turner. HyperGEN Network: Steven C. Hunt (Network Director), Janet Hood, Donna Arnett, John H. Eckfeldt, R. Curtis Ellison, Chi Gu, Gerardo Heiss, Paul Hopkins, Aldi T. Kraja, Jean-Marc Lalouel, Mark Leppert, Albert Oberman, Michael A. Province, D.C. Rao, Treva Rice and Robert Weiss. SAPPHIRe Network: David Curb (Network Director), David Cox, Timothy Donlon, Victor Dzau, John Grove, Kamal Masaki, Richard Myers, Richard Olshen, Richard Pratt, Tom Quertermous, Neil Risch and Beatriz Rodriguez. National Heart, Lung and Blood Institute: Dina Paltoo and Cashell E. Jaquish. Web Site: http://www.biostat.wustl.edu/fbpp/FBPP.shtml
Conflict of Interest statement. None declared.
REFERENCES
- 1.Mackay J., Mensah G.A. The Atlas of Heart Disease and Stroke. World Health Organization, 1211 Geneva 27, Switzerland; 2004. [Google Scholar]
- 2.Ordovas J.M. Genetic interactions with diet influence the risk of cardiovascular disease. Am. J. Clin. Nutr. 2006;83:443S–446S. doi: 10.1093/ajcn/83.2.443S. [DOI] [PubMed] [Google Scholar]
- 3.Freeman D.J., Griffin B.A., Holmes A.P., Lindsay G.M., Gaffney D., Packard C.J., Shepherd J. Regulation of plasma HDL cholesterol and subfraction distribution by genetic and environmental factors. associations between the TaqI B RFLP in the CETP gene and smoking and obesity. Arterioscler. Thromb. 1994;14:336–344. doi: 10.1161/01.atv.14.3.336. [DOI] [PubMed] [Google Scholar]
- 4.Sigurdsson G., Jr, Gudnason V., Sigurdsson G., Humphries S.E. Interaction between a polymorphism of the apo A-I promoter region and smoking determines plasma levels of HDL and apo A-I. Arterioscler. Thromb. 1992;12:1017–1022. doi: 10.1161/01.atv.12.9.1017. [DOI] [PubMed] [Google Scholar]
- 5.Kauma H., Savolainen M.J., Heikkila R., Rantala A.O., Lilja M., Reunanen A., Kesaniemi Y.A. Sex difference in the regulation of plasma high density lipoprotein cholesterol by genetic and environmental factors. Hum. Genet. 1996;97:156–162. doi: 10.1007/BF02265258. [DOI] [PubMed] [Google Scholar]
- 6.Bielinski S.J., Tang W., Pankow J.S., Miller M.B., Mosley T.H., Boerwinkle E., Olshen R.A., Curb J.D., Jaquish C.E., Rao D.C., et al. Genome-wide linkage scans for loci affecting total cholesterol, HDL-C, and triglycerides: the family blood pressure program. Hum. Genet. 2006;120:371–380. doi: 10.1007/s00439-006-0223-0. [DOI] [PubMed] [Google Scholar]
- 7.Law M.R., Wald N.J., Rudnicka A.R. Quantifying effect of statins on low density lipoprotein cholesterol, ischaemic heart disease, and stroke: systematic review and meta-analysis. Br. Med. J. 2003;326:1423. doi: 10.1136/bmj.326.7404.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuulasmaa K., Tunstall-Pedoe H., Dobson A., Fortmann S., Sans S., Tolonen H., Evans A., Ferrario M., Tuomilehto J. Estimation of contribution of changes in classic risk factors to trends in coronary-event rates across the WHO MONICA project populations. Lancet. 2000;355:675–687. doi: 10.1016/s0140-6736(99)11180-2. [DOI] [PubMed] [Google Scholar]
- 9.Clarke R., Emberson J.R., Parish S., Palmer A., Shipley M., Linksted P., Sherliker P., Clark S., Armitage J., Fletcher A., et al. Cholesterol fractions and apolipoproteins as risk factors for heart disease mortality in older men. Arch. Intern. Med. 2007;167:1373–1378. doi: 10.1001/archinte.167.13.1373. [DOI] [PubMed] [Google Scholar]
- 10.Breslow J.L. Genetic markers for coronary heart disease. Clin. Cardiol. 2001;24(Suppl. II):14–17. doi: 10.1002/clc.4960240706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss L.A., Pan L., Abney M., Ober C. The sex-specific genetic architecture of quantitative traits in humans. Nat. Genet. 2006;38:218–222. doi: 10.1038/ng1726. [DOI] [PubMed] [Google Scholar]
- 12.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N.P., Rieder M.J., Cooper G.M., Roos C., Voight B.F., Havulinna A.S., et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kathiresan S., Manning A.K., Demissie S., D’Agostino R.B., Surti A., Guiducci C., Gianniny L., Burtt N.P., Melander O., Orho-Melander M., et al. A genome-wide association study for blood lipid phenotypes in the Framingham heart study. BMC Med. Genet. 2007;8(Suppl. 1):S17. doi: 10.1186/1471-2350-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., et al. and Diabetes Genetics Initiative of Broad Institute of Harvard MIT, Lund University, Novartis Institutes of BioMedical Research. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 15.Willer C.J., Sanna S., Jackson A.U., Scuteri A., Bonnycastle L.L., Clarke R., Heath S.C., Timpson N.J., Najjar S.S., Stringham H.M., et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen J., Pertsemlidis A., Kotowski I.K., Graham R., Garcia C.K., Hobbs H.H. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 17.Bosse Y., Feitosa M.F., Despres J.P., Lamarche B., Rice T., Rao D.C., Bouchard C., Perusse L., Vohl M.C. Detection of a major gene effect for LDL peak particle diameter and association with apolipoprotein H gene haplotype. Atherosclerosis. 2005;182:231–239. doi: 10.1016/j.atherosclerosis.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Bosse Y., Despres J.P., Chagnon Y.C., Rice T., Rao D.C., Bouchard C., Perusse L., Vohl M.C. Quantitative trait locus on 15q for a metabolic syndrome variable derived from factor analysis. Obesity (Silver Spring) 2007;15:544–550. doi: 10.1038/oby.2007.577. [DOI] [PubMed] [Google Scholar]
- 19.Gagnon F., Jarvik G.P., Badzioch M.D., Motulsky A.G., Brunzell J.D., Wijsman E.M. Genome scan for quantitative trait loci influencing HDL levels: evidence for multilocus inheritance in familial combined hyperlipidemia. Hum. Genet. 2005;117:494–505. doi: 10.1007/s00439-005-1338-4. [DOI] [PubMed] [Google Scholar]
- 20.Bosse Y., Chagnon Y.C., Despres J.P., Rice T., Rao D.C., Bouchard C., Perusse L., Vohl M.C. Genome-wide linkage scan reveals multiple susceptibility loci influencing lipid and lipoprotein levels in the quebec family study. J. Lipid Res. 2004;45:419–426. doi: 10.1194/jlr.M300401-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Bosse Y., Chagnon Y.C., Despres J.P., Rice T., Rao D.C., Bouchard C., Perusse L., Vohl M.C. Compendium of genome-wide scans of lipid-related phenotypes: adding a new genome-wide search of apolipoprotein levels. J. Lipid Res. 2004;45:2174–2184. doi: 10.1194/jlr.R400008-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.France M.W., Kwok S., McElduff P., Seneviratne C.J. Ethnic trends in lipid tests in general practice. QJM. 2003;96:919–923. doi: 10.1093/qjmed/hcg154. [DOI] [PubMed] [Google Scholar]
- 23.Balarajan R. Ethnic differences in mortality from ischaemic heart disease and cerebrovascular disease in England and Wales. Br. Med,, J. 1991;302:560–564. doi: 10.1136/bmj.302.6776.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haffner S.M., D’Agostino R., Jr, Goff D., Howard B., Festa A., Saad M.F., Mykkanen L. LDL size in African Americans, Hispanics, and non-Hispanic Whites: the insulin resistance atherosclerosis study. Arterioscler. Thromb. Vasc. Biol. 1999;19:2234–2240. doi: 10.1161/01.atv.19.9.2234. [DOI] [PubMed] [Google Scholar]
- 25.Sorlie P.D., Sharrett A.R., Patsch W., Schreiner P.J., Davis C.E., Heiss G., Hutchinson R. The relationship between lipids/lipoproteins and atherosclerosis in African Americans and Whites: the atherosclerosis risk in communities study. Ann. Epidemiol. 1999;9:149–158. doi: 10.1016/s1047-2797(98)00063-5. [DOI] [PubMed] [Google Scholar]
- 26.Enas E.A., Yusuf S., Mehta J.L. Prevalence of coronary artery disease in Asian Indians. Am. J. Cardiol. 1992;70:945–949. doi: 10.1016/0002-9149(92)90744-j. [DOI] [PubMed] [Google Scholar]
- 27.Pais P., Pogue J., Gerstein H., Zachariah E., Savitha D., Jayprakash S., Nayak P.R., Yusuf S. Risk factors for acute myocardial infarction in Indians: a case–control study. Lancet. 1996;348:358–363. doi: 10.1016/s0140-6736(96)02507-x. [DOI] [PubMed] [Google Scholar]
- 28.Chakraborty R., Weiss K.M. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc. Natl Acad. Sci. USA. 1988;85:9119–9123. doi: 10.1073/pnas.85.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Risch N. Mapping genes for complex diseases using association studies in admixed populations. Am. J. Hum. Genet. 1992;51:13. [Google Scholar]
- 30.Patterson N., Hattangadi N., Lane B., Lohmueller K.E., Hafler D.A., Oksenberg J.R., Hauser S.L., Smith M.W., O’Brien S.J., Altshuler D., et al. Methods for high-density admixture mapping of disease genes. Am. J. Hum. Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith M.W., Patterson N., Lautenberger J.A., Truelove A.L., McDonald G.J., Waliszewska A., Kessing B.D., Malasky M.J., Scafe C., Le E., et al. A high-density admixture map for disease gene discovery in African Americans. Am. J. Hum. Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shriver M.D., Parra E.J., Dios S., Bonilla C., Norton H., Jovel C., Pfaff C., Jones C., Massac A., Cameron N., et al. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum. Genet. 2003;112:387–399. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- 33.Zhu X., Luke A., Cooper R.S., Quertermous T., Hanis C., Mosley T., Gu C.C., Tang H., Rao D.C., Risch N., et al. Admixture mapping for hypertension loci with genome-scan markers. Nat. Genet. 2005;37:177–181. doi: 10.1038/ng1510. [DOI] [PubMed] [Google Scholar]
- 34.Parra E.J., Marcini A., Akey J., Martinson J., Batzer M.A., Cooper R., Forrester T., Allison D.B., Deka R., Ferrell R.E., et al. Estimating African American admixture proportions by use of population-specific alleles. Am. J. Hum. Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parra E.J., Kittles R.A., Argyropoulos G., Pfaff C.L., Hiester K., Bonilla C., Sylvester N., Parrish-Gause D., Garvey W.T., Jin L., et al. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am. J. Phys. Anthropol. 2001;114:18–29. doi: 10.1002/1096-8644(200101)114:1<18::AID-AJPA1002>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 36.Tang H., Jorgenson E., Gadde M., Kardia S.L., Rao D.C., Zhu X., Schork N.J., Hanis C.L., Risch N. Racial admixture and its impact on BMI and blood pressure in African and Mexican Americans. Hum. Genet. 2006;119:624–633. doi: 10.1007/s00439-006-0175-4. [DOI] [PubMed] [Google Scholar]
- 37.Reiner A.P., Carlson C.S., Ziv E., Iribarren C., Jaquish C.E., Nickerson D.A. Genetic ancestry, population sub-structure, and cardiovascular disease-related traits among African-American participants in the CARDIA study. Hum. Genet. 2007;121:565–575. doi: 10.1007/s00439-007-0350-2. [DOI] [PubMed] [Google Scholar]
- 38.Breslow J.L. Genetics of lipoprotein abnormalities associated with coronary artery disease susceptibility. Annu. Rev. Genet. 2000;34:233–254. doi: 10.1146/annurev.genet.34.1.233. [DOI] [PubMed] [Google Scholar]
- 39.Ordovas J.M., Litwack-Klein L., Wilson P.W., Schaefer M.M., Schaefer E.J. Apolipoprotein E isoform phenotyping methodology and population frequency with identification of apoE1 and apoE5 isoforms. J. Lipid Res. 1987;28:371–380. [PubMed] [Google Scholar]
- 40.Sing C.F., Davignon J. Role of the apolipoprotein E polymorphism in determining normal plasma lipid and lipoprotein variation. Am. J. Hum. Genet. 1985;37:268–285. [PMC free article] [PubMed] [Google Scholar]
- 41.Guerra R., Wang J., Grundy S.M., Cohen J.C. A hepatic lipase (LIPC) allele associated with high plasma concentrations of high density lipoprotein cholesterol. Proc. Natl Acad. Sci. USA. 1997;94:4532–4537. doi: 10.1073/pnas.94.9.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wittrup H.H., Tybjaerg-Hansen A., Nordestgaard B.G. Lipoprotein lipase mutations, plasma lipids and lipoproteins, and risk of ischemic heart disease. A meta-analysis. Circulation. 1999;99:2901–2907. doi: 10.1161/01.cir.99.22.2901. [DOI] [PubMed] [Google Scholar]
- 43.Lai C.Q., Demissie S., Cupples L.A., Zhu Y., Adiconis X., Parnell L.D., Corella D., Ordovas J.M. Influence of the APOA5 locus on plasma triglyceride, lipoprotein subclasses, and CVD risk in the Framingham heart study. J. Lipid Res. 2004;45:2096–2105. doi: 10.1194/jlr.M400192-JLR200. [DOI] [PubMed] [Google Scholar]
- 44.Rainwater D.L., Almasy L., Blangero J., Cole S.A., VandeBerg J.L., MacCluer J.W., Hixson J.E. A genome search identifies major quantitative trait loci on human chromosomes 3 and 4 that influence cholesterol concentrations in small LDL particles. Arterioscler. Thromb. Vasc. Biol. 1999;19:777–783. doi: 10.1161/01.atv.19.3.777. [DOI] [PubMed] [Google Scholar]
- 45.Brunham L.R., Singaraja R.R., Hayden M.R. Variations on a gene: rare and common variants in ABCA1 and their impact on HDL cholesterol levels and atherosclerosis. Annu. Rev. Nutr. 2006;26:105–129. doi: 10.1146/annurev.nutr.26.061505.111214. [DOI] [PubMed] [Google Scholar]
- 46.Oram J.F., Lawn R.M. ABCA1. The gatekeeper for eliminating excess tissue cholesterol. J. Lipid Res. 2001;42:1173–1179. [PubMed] [Google Scholar]
- 47.Oram J.F., Vaughan A.M. ABCA1-mediated transport of cellular cholesterol and phospholipids to HDL apolipoproteins. Curr. Opin. Lipidol. 2000;11:253–260. doi: 10.1097/00041433-200006000-00005. [DOI] [PubMed] [Google Scholar]
- 48.Shioji K., Nishioka J., Naraba H., Kokubo Y., Mannami T., Inamoto N., Kamide K., Takiuchi S., Yoshii M., Miwa Y., et al. A promoter variant of the ATP-binding cassette transporter A1 gene alters the HDL cholesterol level in the general Japanese population. J. Hum. Genet. 2004;49:141–147. doi: 10.1007/s10038-004-0124-9. [DOI] [PubMed] [Google Scholar]
- 49.Oram J.F. Tangier disease and ABCA1. Biochim. Biophys. Acta. 2000;1529:321–330. doi: 10.1016/s1388-1981(00)00157-8. [DOI] [PubMed] [Google Scholar]
- 50.Kolovou G.D., Mikhailidis D.P., Anagnostopoulou K.K., Daskalopoulou S.S., Cokkinos D.V. Tangier disease four decades of research: a reflection of the importance of HDL. Curr. Med. Chem. 2006;13:771–782. doi: 10.2174/092986706776055580. [DOI] [PubMed] [Google Scholar]
- 51.Soro-Paavonen A., Naukkarinen J., Lee-Rueckert M., Watanabe H., Rantala E., Soderlund S., Hiukka A., Kovanen P.T., Jauhiainen M., Peltonen L., et al. Common ABCA1 variants, HDL levels, and cellular cholesterol efflux in subjects with familial low HDL. J. Lipid Res. 2007;48:1409–1416. doi: 10.1194/jlr.P600012-JLR200. [DOI] [PubMed] [Google Scholar]
- 52.Malhotra A., Wolford J.K. and American Diabetes Association GENNID Study Group. Analysis of quantitative lipid traits in the genetics of NIDDM (GENNID) study. Diabetes. 2005;54:3007–3014. doi: 10.2337/diabetes.54.10.3007. [DOI] [PubMed] [Google Scholar]
- 53.Tang W., Miller M.B., Rich S.S., North K.E., Pankow J.S., Borecki I.B., Myers R.H., Hopkins P.N., Leppert M., Arnett D.K., et al. Linkage analysis of a composite factor for the multiple metabolic syndrome: the national heart, lung, and blood institute family heart study. Diabetes. 2003;52:2840–2847. doi: 10.2337/diabetes.52.11.2840. [DOI] [PubMed] [Google Scholar]
- 54.Nettleton J.A., Steffen L.M., Ballantyne C.M., Boerwinkle E., Folsom A.R. Associations between HDL-cholesterol and polymorphisms in hepatic lipase and lipoprotein lipase genes are modified by dietary fat intake in African American and White adults. Atherosclerosis. 2007;194:e131–e140. doi: 10.1016/j.atherosclerosis.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brisson D., St-Pierre J., Santure M., Hudson T.J., Despres J.P., Vohl M.C., Gaudet D. Genetic epistasis in the VLDL catabolic pathway is associated with deleterious variations on triglyceridemia in obese subjects. Int. J. Obes. (Lond) 2007;31:1325–1333. doi: 10.1038/sj.ijo.0803586. [DOI] [PubMed] [Google Scholar]
- 56.Knoblauch H., Muller-Myhsok B., Busjahn A., Ben Avi L., Bahring S., Baron H., Heath S.C., Uhlmann R., Faulhaber H.D., Shpitzen S., et al. A cholesterol-lowering gene maps to chromosome 13q. Am. J. Hum. Genet. 2000;66:157–166. doi: 10.1086/302704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soro A., Pajukanta P., Lilja H.E., Ylitalo K., Hiekkalinna T., Perola M., Cantor R.M., Viikari J.S., Taskinen M.R., Peltonen L. Genome scans provide evidence for low-HDL-C loci on chromosomes 8q23, 16q24.1–24.2, and 20q13.11 in finnish families. Am. J. Hum. Genet. 2002;70:1333–1340. doi: 10.1086/339988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deeb S.S., Zambon A., Carr M.C., Ayyobi A.F., Brunzell J.D. Hepatic lipase and dyslipidemia: interactions among genetic variants, obesity, gender, and diet. J. Lipid Res. 2003;44:1279–1286. doi: 10.1194/jlr.R200017-JLR200. [DOI] [PubMed] [Google Scholar]
- 59.Silverman D.I., Ginsburg G.S., Pasternak R.C. High-density lipoprotein subfractions. Am. J. Med. 1993;94:636–645. doi: 10.1016/0002-9343(93)90217-d. [DOI] [PubMed] [Google Scholar]
- 60.Kwiterovich P.O., Jr The metabolic pathways of high-density lipoprotein, low-density lipoprotein, and triglycerides: a current review. Am. J. Cardiol. 2000;86:5L–10L. doi: 10.1016/s0002-9149(00)01461-2. [DOI] [PubMed] [Google Scholar]
- 61.Manninen V., Tenkanen L., Koskinen P., Huttunen J.K., Manttari M., Heinonen O.P., Frick M.H. Joint effects of serum triglyceride and LDL cholesterol and HDL cholesterol concentrations on coronary heart disease risk in the Helsinki heart study. Implications for treatment. Circulation. 1992;85:37–45. doi: 10.1161/01.cir.85.1.37. [DOI] [PubMed] [Google Scholar]
- 62.Brinton E.A., Eisenberg S., Breslow J.L. Human HDL cholesterol levels are determined by apoA-I fractional catabolic rate, which correlates inversely with estimates of HDL particle size. Effects of gender, hepatic and lipoprotein lipases, triglyceride and insulin levels, and body fat distribution. Arterioscler. Thromb. 1994;14:707–720. doi: 10.1161/01.atv.14.5.707. [DOI] [PubMed] [Google Scholar]
- 63.FBPP Investigators. Multi-center genetic study of hypertension: the family blood pressure program (FBPP) Hypertension. 2002;39:3–9. doi: 10.1161/hy1201.100415. [DOI] [PubMed] [Google Scholar]
- 64.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Falush D., Stephens M., Pritchard J.K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosenberg N.A., Pritchard J.K., Weber J.L., Cann H.M., Kidd K.K., Zhivotovsky L.A., Feldman M.W. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 67.Box G.E.P., Cox D.R. An analysis of transformations. Journal of Royal Statistical Society, Series B. 1964;26:211–246. [Google Scholar]
- 68.Basu A., Tang H., Arnett D., Gu C., Mosley T., Kardia S., Luke A., Tayo B., Cooper R., Zhu X., et al. 2009 doi: 10.1038/oby.2009.24. Admixture Mapping of Quantitative Trait Loci for BMI in African Americans. Evidence for Loci on Chromosomes 3q, 5f, and 15q. Obesity, doi: 10.1038/oby.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.