Abstract
Kinetochores are large protein assemblies built on chromosomal loci named centromeres. The main functions of kinetochores can be grouped under four modules. The first module, in the inner kinetochore, contributes a sturdy interface with centromeric chromatin. The second module, the outer kinetochore, contributes a microtubule-binding interface. The third module, the spindle assembly checkpoint, is a feedback control mechanism that monitors the state of kinetochore–microtubule attachment to control the progression of the cell cycle. The fourth module discerns correct from improper attachments, preventing the stabilization of the latter and allowing the selective stabilization of the former. In this review, we discuss how the molecular organization of the four modules allows a dynamic integration of kinetochore–microtubule attachment with the prevention of chromosome segregation errors and cell-cycle progression.
Keywords: centromere, chromosome segregation, force generation, kinetochore, spindle assembly checkpoint
An overview of kinetochore function and organization
Conspicuous structures are located at the end and middle of chromosomes, the telomeres and the kinetochores, respectively. Here, we concentrate on the middle structures, the kinetochores. The primary function of kinetochores is to create load-bearing attachments between chromosomes and microtubules in a dividing mother cell. The correct partitioning of sister chromatids to the daughter cells depends on such attachments (Wittmann et al, 2001; Walczak and Heald, 2008). The ability of kinetochores to couple to growing or disassembling microtubules (Rieder and Salmon, 1998) has attracted considerable theoretical interest (e.g. Hill, 1985; Grishchuk et al, 2008a). Low- and high-resolution structural snapshots of several candidate kinetochore–microtubule couplers have revealed a variety of modes of binding and shapes, including ‘rings, bracelets, sleeves and chevrons' and ‘slender fibrils' (Davis and Wordeman, 2007; McIntosh et al, 2008). The relative contribution from these different structures to force generation and chromosome motility is an active area of investigation.
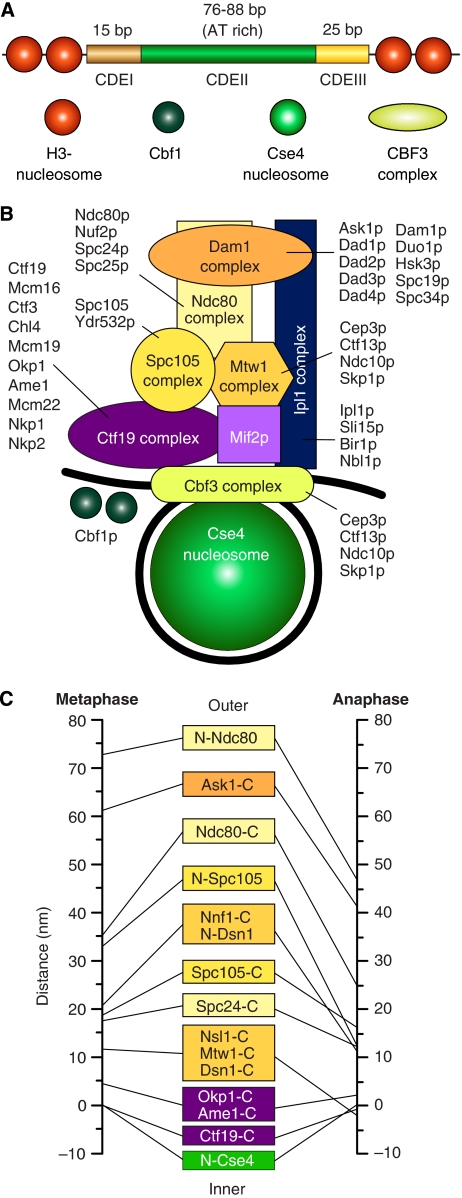
The simplest kinetochores, Saccharomyces cerevisiae's, bind a single microtubule (reviewed in McAinsh et al, 2003; Westermann et al, 2007). They contain approximately 60 proteins, almost 40 of which are clustered in seven different complexes, the CBF3, Ndc80, Mtw1, Spc105, Ctf19, Dam1, and Ipl1 complexes (Figure 1; Supplementary Table I) (McAinsh et al, 2003; Westermann et al, 2007). With few exceptions (most notably the CBF3 and Dam1 complexes), these complexes are conserved from yeast to humans (Figure 2) (Musacchio and Salmon, 2007; Cheeseman and Desai, 2008; Welburn and Cheeseman, 2008).
Figure 1.
The kinetochore of S. cerevisiae. (A) The 125 bp centromere of S. cerevisiae is subdivided in the CDEI, CDEII, and CDEIII regions. The 8 bp CDEI recruits a dimer of Cbf1, a helix-turn-helix protein that runs a parallel life as a transcription factor (Bram and Kornberg, 1987). CDEII, a 76–84 bp AT-rich DNA element, folds around a specialized nuclesome containing Cse4 (Meluh et al, 1998; Keith and Fitzgerald-Hayes, 2000). The four-subunit CBF3 complex is only found in species whose centromeres contain a CDE-III motif (Meraldi et al, 2006). CBF3 binds to the CDE-III motif, an imperfect palyndrome with an approximately 24 bp ‘core' and a less well-conserved CDE-II-distal sequence of 50–60 bp (Lechner and Carbon, 1991). Additionally, at least one CBF3 subunit, Ndc10, is also found in association with CDE-II (Espelin et al, 2003). (B) The Cse4-containing nucleosome wraps around the approximately 125 bp centromeric DNA (black). Mif2p (homologous to CENP-C) is a linker protein creating a connection with the Mtw1, Spc105, and Ndc80 complexes (homologous to Mis12, KNL-1, and Ndc80 complexes of higher eukaryotes). Together with the Dam1 complex, the Ndc80 complex reaches the microtubule-binding region. The Ipl1p complex is equivalent to the chromosome passenger complex (CPC) of higher eukaryotes. The Nbl1p subunit was recently identified as a homologue of the Borealin/DasraB/CSC-1 subunit of higher eukaryotes (Nakajima et al, 2009). It is believed to span from the inner to the outer region of the kinetochore. The kinase activity associated with this complex is directed onto the Ndc80 and Dam1 complexes and regulates the attachment process. Names of constituent subunits are displayed. (C) Average location of kinetochore proteins along the axis of the S. cerevisiae's kinetochore–microtubule attachment in metaphase and late anaphase (Joglekar et al, 2009). N- and C- indicated N- and C-termini.
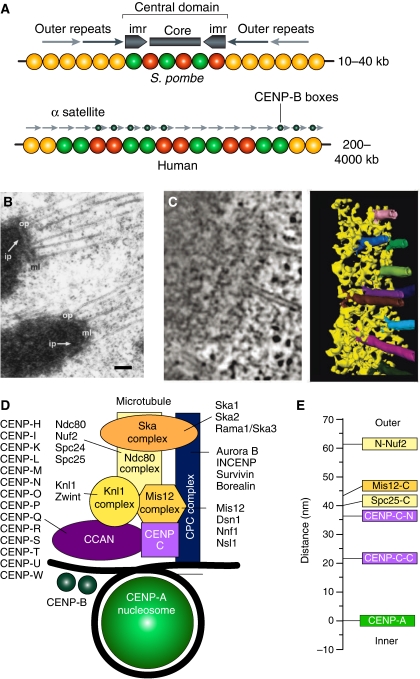
Figure 2.
Organization of regional centromeres and kinetochores. (A) The central domain of the centromere of S. pombe possesses a pair of inverted repeat sequence arrays (marked as imr, for innermost repeat). They flank an unconserved central core sequence. Both CENP-A and H3-containing nucleosomes map to the central domain. The central domain is flanked by the cohesin-rich outer domains, consisting of peri-centromeric heterochromatin. In humans, α-satellite DNA is composed of a core of highly ordered 171 bp repeats termed α-I satellite DNA, which is framed on either side by divergent repetitive sequences and retrotransposons, referred to as α-II satellite DNA. At the outskirts, the centromeric chromatin becomes rich in long interspersed element 1 (LINE-1 elements). On normal human chromosomes, the centromere forms on a small subdomain of the α-I satellite DNA, but there are cases in which the centromere forms on DNA devoid of α-satellite repeats. The α-I satellite DNA contains a sequence known as the CENP-B box, which binds in a sequence-specific manner to the CENP-B protein and facilitates, but is not strictly required for, kinetochore formation. The panel was adapted from Allshire and Karpen (2008) (B) Adjacent kinetochores from a metaphase cell obtained by rapid freezing and freeze substitution (reproduced from ref. McEwen et al, 1998). The prominent outer plate (op) structure stains as heavily as chromatin, and is separated from the underlying inner plate (ip) by a well-defined, translucent, middle layer (ml). Bar represents 200 nm. (C) Electron tomography of the outer plate shows a network of crosslinked fibres, 10 nm in diameter and up to 80–90 nm long, of unknown molecular identity. The long fibres aligned in the plane of the outer plate in the absence of microtubules (not shown), but re-oriented as they bound to the side of microtubules (Dong et al, 2007). (D) A scheme for the outer kinetochore of metazoans analogous to that presented in Figure 1B. (E) Average location of kinetochore proteins along the axis of the kinetochore–microtubule attachment in metaphase in D. melanogaster. N- and C- indicated N- and C-termini.
Conservation of kinetochore constituents suggests that the larger kinetochores of higher eukaryotes, which bind multiple microtubules (kinetochore fibres or K-fibres), are assembled from the repetition of the basic microtubule-binding module of budding yeast (Zinkowski et al, 1991; Blower et al, 2002; Joglekar et al, 2008). This idea is known as the ‘repeat subunit' model. Kinetochores in vertebrates appear as trilaminar plates, with electron dense inner and outer kinetochore plates and an electron lucent middle layer (Figure 2). The inner plate contains kinetochore proteins implicated in the creation of an interface with centromeric chromatin. The outer plate contains kinetochore proteins that interact with the plus ends of microtubules bound ‘end-on'. A fibrous corona, extending outward from the outer plate, is visible in the absence of microtubules and contains microtubule motors, such as CENP-E, and components of the spindle checkpoint, such as the Rod-ZW10-Zwilch (RZZ) complex, both of which only exist in metazoans (reviewed in Cleveland et al, 2003). A recent electron tomography reconstruction of the outer plate revealed a fibrous, flexible network apparently lacking a well-defined organization (Dong et al, 2007) (Figure 2). Although no orderly structure was observed, it is possible that structural work on the microtubule-binding unit will eventually reveal hidden regularities predicted by the ‘repeat subunit' model.
By studying the way in which certain kinetochore proteins influence the recruitment and assembly of other kinetochore proteins, an assembly plan for the inner and outer kinetochore plates has been designed (e.g. Liu et al, 2006; Hori et al, 2008a). In a remarkable recent feat, the position of kinetochore proteins along the inter-kinetochore axis of S. cerevisiae, Drosophila melanogaster's and human kinetochores was mapped with nanometer accuracy (Schittenhelm et al, 2007; Joglekar et al, 2009; Wan et al, 2009). The picture emerging from these analyses is consistent with a model in which kinetochore proteins are piled up according to an inside–out scheme from the centromere towards the microtubule-binding site (Figures 1 and 2). At least two alternative variants of assembly are conceivable, as discussed below.
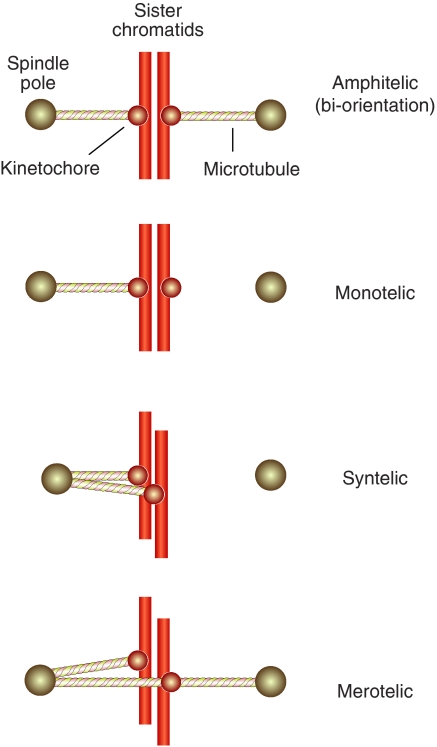
Kinetochores are also involved in at least two fundamental and possibly related feedback mechanisms. The first mechanism allows the discrimination between correct and incorrect kinetochore–microtubule attachments (Pinsky and Biggins, 2005; Kelly and Funabiki, 2009). Correct attachments become stabilized, whereas incorrect attachments are labile and eventually become corrected (Nicklas and Koch, 1969; Li and Nicklas, 1995). The correct configuration of attachment of the sister kinetochores is to opposite spindle poles (bi-orientation or amphitelic orientation). This configuration allows the equational division of sisters to the daughter cells at anaphase (Figure 3). Errors during the phase of attachment, such as syntelic and merotelic attachment (Figure 3), fail to become stabilized and become corrected (e.g. Nicklas and Koch, 1969; Cimini et al, 2003; Lampson et al, 2004).
Figure 3.
Bi-orientation, erroneous attachments. A single sister chromosome pair is shown for simplicity. In amphitelic orientation (bi-orientation) each of the two opposing sister kinetochores is bound to microtubules originating from the proximal pole. This is the correct form of attachment. Monotelic attachment is a normal condition during prometaphase before bi-orientation. Premature loss of sister chromatid cohesion at this early stage, for instance as a consequence of a cohesion defect or a mitotic checkpoint defect, can yield aberrant segregation with both sister chromatids distributed to the same daughter cell. Persistent cohesion between chromosomes in anaphase will result in similar errors. In syntelic attachment, both sisters in a pair connect to the same pole. In merotelic attachment, a sister is attached to both poles. This condition occurs quite frequently during mitosis.
The second mechanism works by synchronizing the process of microtubule attachment with the progression of the cell-cycle oscillator. Specifically, loss of cohesion between sister chromatids and mitotic exit through degradation of Cyclin B, two events that are controlled by the cell-cycle machinery, must be coordinated with the completion of kinetochore–microtubule attachment (Peters, 2006). The feedback mechanism responsible for this coordination is named the spindle assembly checkpoint (SAC) (Musacchio and Salmon, 2007).
The foundations: centromeres and associated proteins
As discussed in depth in an accompanying review by Torras-Llort et al (2009), kinetochores are built on chromosomal loci known as centromeres (Cleveland et al, 2003; Vos et al, 2006). Centromeres fall into distinct categories. Point centromeres, which are limited to a subset of fungi, including S. cerevisiae, consist of a defined sequence of approximately 125 base pairs that is sufficient to ‘encode' kinetochore formation (Figure 1A) (McAinsh et al, 2003; Meraldi et al, 2006; Westermann et al, 2007). Invariably, kinetochores built on point centromeres bind a single microtubule. Conversely, regional centromeres extend over much larger DNA regions (Figure 2A) (e.g. 10–40 kb in S. pombe and up to millions of bases in humans) and assemble kinetochores that bind multiple microtubules (Figure 2B and C) (reviewed in Cleveland et al, 2003; Allshire and Karpen, 2008). Holocentric kinetochores created from centromeres that extend all along the chromosome exist in a few organisms, including the nematode Caenorhabditis elegans.
A conserved hallmark of the centromere–kinetochore interface is a specialized nucleosome containing the histone H3 variant CENP-A (also referred to as CenH3, and known as Cse4p in budding yeast) (reviewed in Mellone and Allshire, 2003; Black and Bassett, 2008). There seems to be a single Cse4 nucleosome per chromosome in S. cerevisiae (Figure 1A and B) (Meluh et al, 1998; Furuyama and Biggins, 2007). Recruitment of all inner and outer kinetochore proteins in S. cerevisiae depends on CBF3 and Cse4 (Ortiz et al, 1999; Collins et al, 2005).
Regional centromeres contain multiple CENP-A nucleosomes (Figure 2A) (Blower et al, 2002; Joglekar et al, 2008; Marshall et al, 2008a). Regional centromeres usually consist of long arrays of repetitive DNA sequences (reviewed in Cleveland et al, 2003; Allshire and Karpen, 2008). For instance, human centromeres form on a small subdomain of a highly ordered array containing thousands of copies of a 171-bp repeat sequence known as α-I satellite DNA (Figure 2A). The α-I satellite DNA contains the CENP-B box, a sequence recognized by the CENP-B protein (Earnshaw and Rothfield, 1985; Masumoto et al, 1989). CENP-B is required to establish, but not to maintain, centromeric chromatin, and seems to repress the establishment of ectopic centromeres (Okada et al, 2007).
There is no strict dependency on α-satellite DNA for centromere specification. At times, so-called neo-centromeres form on DNA devoid of α-satellite repeats (reviewed in Marshall et al, 2008b). Thus, although kinetochores are usually assembled on centromeric DNA containing repetitive DNA sequences, they can also form on unrelated, non-repetitive DNA sequences. The main implication is that the specific DNA sequence of centromeres is not strictly required for kinetochore assembly, which in turn hints to the existence of epigenetic mechanisms in the establishment and maintenance of centromere identity (Allshire and Karpen, 2008; Black and Bassett, 2008).
Despite the differences in centromere organization, the composition and overall organization of kinetochores built on point and regional centromeres is similar (Figure 2D and E), as discussed below in more detail.
The epigenetic specification of centromeres
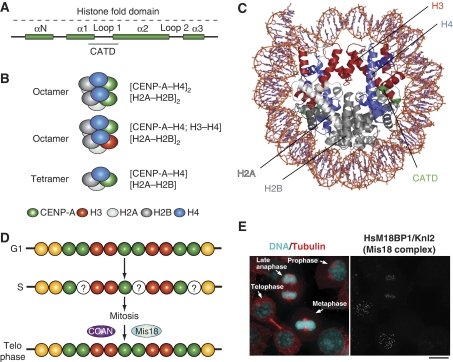
The molecular requirements for epigenetic specification of centromeres are the topic of the review by Torras-Llort et al also contained in this focus review series (Torras-Llort et al, 2009). Besides centromere-specific histone modifications (Sullivan and Karpen, 2004; Carroll and Straight, 2006), CENP-A itself may contribute (reviewed in Carroll and Straight, 2006; Black and Bassett, 2008). A 15-residue sequence of CENP-A, the CENP-A targeting domain (CATD), is key for the propagation of centromere identity through successive cell generations (Figure 4A and B) (Black et al, 2004). When grafted onto H3, the CATD is sufficient to specify centromere localization of the H3CATD chimaera (Black et al, 2004). Furthermore, the H3CATD chimaera performs at least some of the functions normally attributed to CENP-A, such as mediating the recruitment of additional kinetochore and SAC components (Black et al, 2007). Although the exact composition of the CENP-A nucleosome remains controversial (Figure 4B) (Dalal et al, 2007; Allshire and Karpen, 2008), CENP-A forms 2:2 tetramers with histone H4 in vitro (Black et al, 2004) (Figure 4C).
Figure 4.
Epigenetic specification of centromeric chromatin. (A) The histone-fold domain of histone H3 proteins is composed of four α-helical domains (αN and α1–α3). Loop 1 separates α1 and α2. The CENP-A targeting domain (CATD) is sufficient for localization to centromeres when substituted into canonical H3 (the amino acids highlighted in orange are required in Drosophila). The CATD was identified for a 10-fold slowing of hydrogen exchange along the peptide backbone, probably because of increase rigidity of the interface it forms with its histone H4 (Black et al, 2004). (B) In non-centromeric regions, canonical histone H3 assembles into octameric nucleosomes composed of two H2A, H2B, H3, and H4 histone subunits. In centromeric chromatin, CENP-A can assemble into homotypic octamers, in which both H3 subunits are replaced by CENP-A, or into heterotypic octamers, which contain one canonical H3 and one CENP-A subunit. In Drosophila melanogaster, CENP-A has been reported to form half nucleosomes, homotypic tetramers containing one subunit each of H2A, H2B, H4, and CENP-A/CID. (C) Ribbond model of the nucleosome core particle (PDB ID 2CV5). Histone H3 is in red. When grafted onto histone H3, the CATD of CENP-A (green) allows specific and selective incorporation of the H3 chimaera at the centromere. The CENP-A2:H42 tetramers are more compact and rigid than the H32:H42 tetramers (Black et al, 2004). (D) CENP-A is only replenished in telophase. Thus, chromatin entering S phase with a full complement of CENP-A, emerges from DNA replication with half the original levels. The halved levels are retained throughout mitosis. (E) The localization pattern of M18BP1, a subunit of the Mis18 complex. The figure derives from Maddox et al (2007). The dots on the right panel represent centromeres/kinetochores.
Crucial to understanding the mechanism of propagation of centromere identity is the study of CENP-A loading onto chromatin. In metazoans, the levels of CENP-A on the daughter DNA become halved on DNA replication, and are then maintained at the halved levels through mitosis (Figure 4D) (Jansen et al, 2007). CENP-A is incorporated exclusively after exit from mitosis (Jansen et al, 2007; Maddox et al, 2007; Schuh et al, 2007; Hemmerich et al, 2008). A complex containing Mis18, M18BP1/Knl2, and the RbAp46/RbAp48 histone chaperones (related to Mis16 of S. pombe), known as the Mis18 complex, is crucial for chromatin incorporation of CENP-A during mitotic exit (Hayashi et al, 2004; Fujita et al, 2007; Maddox et al, 2007). Consistently with an exclusive role during mitotic exit, the Mis18 complex is recruited to centromeres at anaphase and is then released in G1 (Figure 4E) (Hayashi et al, 2004; Fujita et al, 2007; Maddox et al, 2007). Scm3, a fungal protein with no obvious homologues in higher eukaryotes, has centromere localization dynamics similar to those of Mis16 and Mis18 and acts as a Cnp1/CENP-A binding and loading factor in S. pombe (Pidoux et al, 2009; Williams et al, 2009). Earlier, the Scm3 protein of S. cerevisiae has been proposed to form an unusual hexameric nucleosome with Cse4/CENP-A and H4 (Mizuguchi et al, 2007). The new studies suggest that Scm3 acts as a centromeric receptor for CENP-A incorporation in centromeric nucleosomes (devoid of Scm3).
CENP-A is dispensable for centromeric recruitment of the Mis18 complex or Scm3, whereas Scm3 depends on the Mis18 complex for centromere localization (Fujita et al, 2007; Pidoux et al, 2009; Williams et al, 2009). The localization dependencies depict a hierarchical pathway of recruitment, but the primary chromatin feature recognized by the Mis18 complex is unknown. Its identification is therefore crucial to define the epigenetic marks subtending to centromere propagation. How is CENP-A deposition licensed in a cell-cycle-dependent manner is also uncertain, but three cell-cycle proteins, Cyclin A, RCA1/Emi1, and Cdh1 were recently implicated in CENP-A loading in D. melanogaster (Erhardt et al, 2008).
CENP-A interacts with at least a subset of the subunits of the constitutive centromere-associated network complex (abbreviated as CCAN, and also known as NAC/CAD). The CCAN is probably the most mysterious protein object of the kinetochore. Many of its 14 tentatively assigned subunits (Figure 2D) were identified by proteomics or sequence analysis (Foltz et al, 2006; Izuta et al, 2006; Meraldi et al, 2006; Okada et al, 2006; Hori et al, 2008a). Several CCAN subunits are related to subunits of the Ctf19 (or COMA) and Sim4 complexes of S. cerevisiae and S. pombe, respectively, which can therefore likely be regarded as the homologue of the CCAN (Supplementary Table I) (e.g. Hyland et al, 1999; Ortiz et al, 1999).
Biochemical and functional analyses suggest that rather than forming a single stable complex, CCAN subunits are organized in distinct sub-complexes. A binary CENP-T/W sub-complex contributes to recruiting a CENP-H/I/K sub-complex. The latter, in turn, is required to recruit a third sub-complex containing the CENP-O/P/Q/R/U subunits (Okada et al, 2006; Cheeseman et al, 2008; Hori et al, 2008a, 2008b). Four additional CCAN subunits, CENP-L/M/N/S, also associate to CENP-H/I/K (Okada et al, 2006; Hori et al, 2008a), perhaps as an additional sub-complex.
The structural complexity of the CCAN reflects in complex localization dynamics (Hemmerich et al, 2008). Most CCAN subunits are constitutively present at kinetochores throughout the cell cycle (McClelland et al, 2007; Cheeseman et al, 2008). CENP-N, on the other hand, is abundant at kinetochores in interphase but is largely removed during mitosis (McClelland et al, 2007), not an expected behaviour for a ‘constitutive' kinetochore subunit. Furthermore, though many subunits of the CCAN are probably located within a short distance from CENP-A, CENP-T/W interacts with H3-nucleosomes rather than CENP-A nucleosomes (Hori et al, 2008a).
In S. pombe, certain subunits of the Sim4/CCAN complex, including Mis6/CENP-I, have been implicated in the recruitment of new CENP-A onto chromatin, but not in the maintenance of existing CENP-A (Takahashi et al, 2000; Nishihashi et al, 2002; Pidoux et al, 2003; Okada et al, 2006). The generality of this mechanism, however, is unclear, as the CCAN may not exist in C. elegans and D. melanogaster (A Desai, personal communication). CENP-C, an elongated molecule containing a cupin-like C-terminal domain (Trazzi et al, 2002; Cohen et al, 2008) is deposited at centromeres at the same time as CENP-A, and has also been implicated in CENP-A loading at centromeres (Schuh et al, 2007; Erhardt et al, 2008).
Kinetochore–microtubule attachment: an overview
A quarter of a century ago, the ‘search and capture' model laid the foundations for understanding the process of kinetochore–microtubule attachment (Kirschner and Mitchison, 1986). The model incorporated the recently described process of microtubule dynamic instability to propose that mitotic microtubules explore space dynamically and become selectively stabilized once they hit their targets. In mitosis, kinetochores act as targets, and indeed the stabilization of kinetochore-bound microtubules, that is the increase in their half-lives, is a crucial function of kinetochores (Mitchison et al, 1986; Zhai et al, 1995; Rieder and Salmon, 1998).
There is also evidence that kinetochores can nucleate microtubules, or at least, that they can capture and promote the growth of small microtubule stubs generated in their vicinity (Snyder and McIntosh, 1975; Telzer et al, 1975; Witt et al, 1980; Khodjakov et al, 2003; Tulu et al, 2006). The Ran pathway contributes to nucleating microtubules proximally to chromatin and may act as a source of short microtubules for kinetochore capture and elongation (O'Connell and Khodjakov, 2007). Microtubules are polymers of αβ-tubulin dimers. They are polar structures, with plus ends exposing β-tubulin and minus ends exposing α-tubulin. Kinetochore microtubules have their plus ends at the kinetochore (Euteneuer and McIntosh, 1981). The structural polarity of kinetochore-nucleated microtubules has not been formally shown, but it is assumed that these are also oriented with their plus ends at the kinetochore.
A remarkable feature of kinetochores is that they maintain attachment to growing or disassembling microtubules (Mitchison et al, 1986; Mitchison, 1989; Rieder and Salmon, 1998). For instance, kinetochores remain attached during anaphase or during the oscillations about the metaphase plate known as ‘tug-of-war'. Furthermore, kinetochores slide towards the plus end to maintain their position on treadmilling microtubules (also known as microtubule flux), that is microtubules that incorporate tubulin subunits at the plus ends and release them at the minus end without net growth (Mitchison and Salmon, 1992).
How do kinetochores remain coupled to disassembling microtubules? Almost three decades ago it was proposed that kinetochores might maintain attachment to microtubules by encircling the microtubule with a processive sliding collar (Margolis and Wilson, 1981). The protofilaments (PFs) at the plus end of a shrinking microtubule are flared as a result of lattice distortion when the GTP cap is liberated (Mandelkow et al, 1991). The release of mechanical strain from a bending microtubule PF can be harnessed to do mechanical work (Koshland et al, 1988; Grishchuk et al, 2005). A ring could, in principle, be used to propel kinetochores if peeling PFs at a disassembling tip ‘tugged' the side of the ring causing it to slide processively along the microtubule (Grishchuk et al, 2008a) (Figure 5A). The Dam1 complex, discussed in the next paragraph, forms a ring structure around microtubules in vitro.
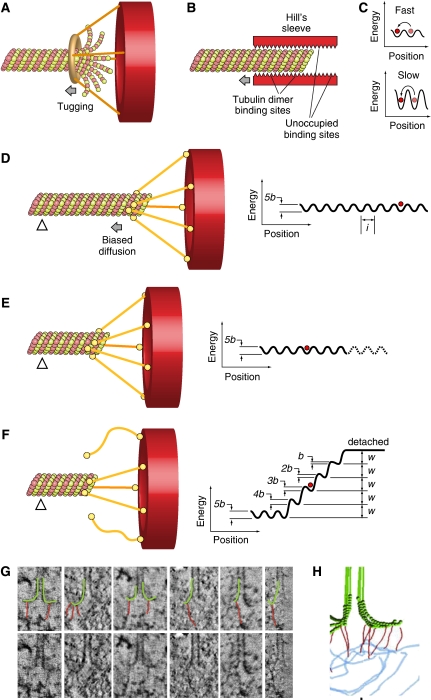
Figure 5.
Biased diffusion. Binding of candidate couplers to microtubule ends can be monitored experimentally by tethering the coupler at the surface of beads, and then monitoring bead motion. Three kinds of tethering to microtubule ends can be distinguished experimentally at this time: (1) Dam1-dependent rings generate high forces. The attached beads do not roll (Grishchuk et al, 2008a). The structure of the Dam1 complex is discussed in Figure 6; (2) Ring-independent Dam1 coupling in which the bead does roll as the MT shortens (Grishchuk et al, 2008b); (3) Motor-dependent tethering in which beads do not roll (Grissom et al, 2009). The mechanism of this coupling is still unknown. Two additional modes of movement have been proposed: (1) biased diffusion, as originally proposed in Hill's model (Hill, 1985) and more recently for the Ndc80 complex (Powers et al, 2009); and (2) power strokes from bending protofilaments acting on non-diffusing, MT-binding fibrils (McIntosh et al, 2008). (A) With a ring coupler encircling a microtubule (inspired by the Dam1 ring, discussed in Figure 6), force may be provided by flared depolymerizing protofilaments, which exercise a pressure against the base of the sleeve. (B) Hill's model depicts the microtubule-binding site of the kinetochore as a ‘sleeve' surrounding the microtubule (Hill, 1985). The microtubule-binding sites are represented by triangles. Maximization of the number of binding sites drives the sliding of the sleeve along the microtubule. The design and theoretical treatment of (B–F) are largely based on earlier work (Joglekar and Hunt, 2002; Powers et al, 2009). (C) The overall activation energy required for sliding along the lattice may cause diffusion to be slow or fast. To be effective, diffusion has to occur with kinetics that must be compatible with the kinetics of microtubule depolymerization. (D) An alternative mechanism for biased diffusion based on the Ndc80 complex was recently proposed (Powers et al, 2009). Kinetochores are shown as red hollow discs. The coupler is an elongated molecule with two globular domains at either end, one for kinetochore binding and one for microtubule binding, and it is inspired by the Ndc80 complex (see Figure 6). Coupling is along the lattice and is mediated by five microtubule-binding elements. The free-energy landscape for this coupler is shown on the right. l denotes spacing of sites. The red circle represents the current position of the coupler on the surface. The energy landscape is corrugated because movement along the filament requires breaking and reforming some bonds (C). b is the activation energy, w is the binding energy. The triangle represents a fiduciary mark along the microtubule. (E) The microtubule has depolymerized and the coupler has diffused on the surface towards the plus end. (F) The release of the coupler (two out of five binding sites have been lost here) implies an increase in free energy because the bond energies, w, must be overcome to move the couple past the filament tip. The heights of the activation energies 5b, 4b,…., b, decrease as the coupler begins to move past the tip. (G) The bottom row shows tomographic slices of kinetochore microtubule ends. The same gallery is also shown in the top row with protofilaments and their associated kinetochore fibrils, indicated by graphic overlays. (H) A tomographic reconstruction of a kinetochore–microtubule interface with associated fibrils. (G, H) are from McIntosh et al (2008).
An alternative to rings, described in the Hill's sleeve model, proposed that the kinetochore may surround the microtubule near the ends, creating a close apposition of the inner surface of a rigid sleeve to the outer surface of the MT, linked by many weak binding sites (Hill, 1985) (Figure 5B). If the translocation from site to site implied relatively small activation energies (i.e. it was fast, Figure 5C) and if the total binding energy was sufficiently large, such a structure may be expected to move by biased diffusion along the microtubule when binding sites are removed from the edge of the binding surface on microtubule disassembly (Figure 5B).
As explained in the next paragraph, our understanding of the structure of the kinetochore–microtubule interface suggests that the kinetochore does not conform to a Hill's sleeve. Recently, microbeads coated with Ndc80 complex, a fibrous component of the KMN network whose function in microtubule binding at the kinetochore is described below, were shown to track the ends of a depolymerizing microtubule (McIntosh et al, 2008; Powers et al, 2009), and were proposed to undergo biased diffusion (Powers et al, 2009) (Figure 5D–F).
A recent EM tomographic reconstruction of kinetochores in PtK1 cells showed the existence of fibrils linking the inner face of flared PFs to the inner plate of the kinetochore (Figure 5G and H) (McIntosh et al, 2008). It was proposed that the fibrils, whose molecular identity is unknown, might restrict the bending of PFs to promote PF stabilization, and could translocate towards the microtubule lattice when coupled to a depolymerizing microtubule. Thus, slender fibrils might provide a synthesis between Hill's thermal ratchet model and the harnessing of force by microtubule depolymerization.
The molecular machinery of kinetochore–microtubule attachment
Several microtubule-associated proteins (MAPs), including EB1, CLASP, Ch-TOG/XMAP215, APC (adenomatous polyposis coli), Clip170, Nde1/Ndel1, and Lis1 and the kinesin-13 kinesins Kif2a and MCAK, which are devoid of microtubule motor activity but rather act as microtubule de-stabilizers, have been implicated in the control of kinetochore microtubule dynamics (reviewed in Maiato et al, 2004). On the other hand, none of the MAPs identified at mitotic kinetochores seems to be essential for forming load-bearing kinetochore–microtubule attachments (Cheeseman and Desai, 2008).
Although ATP-powered molecular motors could, in principle, couple kinetochores to disassembly microtubule tips (Lombillo et al, 1995; Grissom et al, 2009), most if not all chromosome movement after metaphase alignment, and in particular poleward movement at anaphase, is due to the ability of kinetochores to remain attached to assembling or disassembling microtubules (Koshland et al, 1988; Coue et al, 1991). Consistently, minus end directed motors are dispensable for poleward chromosome translocation in yeast (Grishchuk and McIntosh, 2006; Tanaka et al, 2007).
The dispensability of MAPs and motors for generating load-bearing attachment indicates that kinetochores contain specialized machinery to deal with microtubule binding (Maiato et al, 2004; Davis and Wordeman, 2007). The KMN network complex (an acronym for Knl-1, Mis12, Ndc80) has emerged as a crucial components of such machinery (reviewed in Cheeseman and Desai, 2008). The KMN is a conserved 10-subunit assembly gathering three distinct sub-complexes, known as Knl1, Mis12, and Ndc80 (Figures 1B and 2D; Supplementary Table I) (De Wulf et al, 2003; Desai et al, 2003; Nekrasov et al, 2003; Pinsky et al, 2003; Westermann et al, 2003; Cheeseman et al, 2004; Obuse et al, 2004; Liu et al, 2005; Przewloka et al, 2007). Preventing kinetochore recruitment of the microtubule-binding component of the KMN network by RNAi or other methods results in a kinetochore-null phenotype, that is load-bearing kinetochore–microtubule attachments cannot be formed and kinetochores exhibit only residual, motor-driven motility (Wigge and Kilmartin, 2001; Desai et al, 2003; McCleland et al, 2003; Cheeseman et al, 2004, 2006; Kerres et al, 2004; DeLuca et al, 2005, 2006; Emanuele et al, 2005; Kline, 2006; Vorozhko et al, 2008).
The approximately 170 kDa Ndc80 complex contains four subunits: Ndc80 (also known as Hec1), Nuf2, Spc24, and Spc25 (Figure 6A). It is a stable sub-complex of the KMN network (Ciferri et al, 2005; Wei et al, 2005) and it adopts an approximately 60 nm dumbbell shape that crosses the kinetochore vertically from the inner to the outer plate (Ciferri et al, 2005, 2008; Wei et al, 2005; DeLuca et al, 2006; Schittenhelm et al, 2007; Joglekar et al, 2009; Wan et al, 2009). Two sub-complexes, containing the Spc24:Spc25 and Nuf2:Ndc80 subunits, respectively, occupy opposite ends of the dumbbell (Ciferri et al, 2005; Wei et al, 2005). Globular domains in each of these sub-complexes flank extended coiled-coil regions that meet in a tetramerization domain within the central shaft (Figure 6A and B).
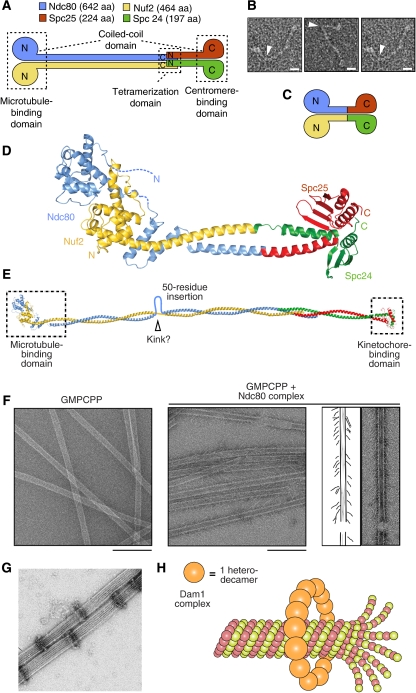
Figure 6.
The molecular machinery of kinetochore–microtubule attachment. (A) Topology of the Ndc80 complex. Ndc80 and Nuf2 engage in a dimer. They contain N-terminal CH domains followed by a coiled-coil region that mediates inter-subunit interactions. Spc24 and Spc25 have N-terminal coiled-coils that mediate inter-subunit interactions, followed by globular domains that are responsible for binding to the Mis12 complex. Tetramerization engages the C-terminal region of the Ndc80:Nuf2 dimer and the N-terminal region of the Spc24:Spc25 dimer. aa, amino acids. N and C indicate the N- and C-termini, respectively. (A, D) were reproduced from Ciferri et al (2008). (B) Gallery of three individual Ndc80 complexes. Arrowheads mark a prominent kink along the shaft. The scale bar corresponds to 10 nm. The images are reproduced from Wang et al (2008). (C) By fusing the C-termini of the Ndc80 and Nuf2 subunits to the N-termini of the Spc25 and Spc24 subunits, respectively, a ‘bonsai' version of the Ndc80 complex was created. Most of the coiled-coil in the central shaft was deleted. The resulting complex retains the ability to bind microtubules in vitro and to localize to kinetochores when injected into living cells (Ciferri et al, 2008). (D) Overall view of the 2.9 Å crystal structure of the bonsai-Ndc80 complex (PDB ID 2VE7). The two CH domains pack in a tight dimeric assembly. An 80-residue N-terminal disordered segment in the Ndc80 subunit escaped structure determination (dashed line). Together with the globular region of Ndc80:Nuf2, this segment contributes to microtubule binding. (E) A model of the full length Ndc80 complex. The model is based on earlier electron microscopy work on the Ndc80 complex (Wei et al, 2005; Wang et al, 2008) and on a crosslinking and mass spectrometry analysis that identified the register of coiled-coil interaction within the central shaft (Maiolica et al, 2007). The regions contained in the crystal structure of bonsai-Ndc80 are boxed. The coiled-coil is interrupted by a 50-residue insertion in the Ndc80 sequence that increases the overall flexibility of the Ndc80 rod. (F) Left: negatively stained control microtubules stabilized with GMPCPP, a non-hydrolysable GTP analogue that stabilizes the microtubule lattice. Middle: negatively stained GMPCPP microtubules in the presence of 5 μM Ndc80 complex (C. elegans). The Ndc80 complex forms angled rod-like projections on the microtubule lattice. Right: traces of the EM images depicting the angled rod-like complexes bound to the lattice. Scale bars represent 200 nm. The panel was reproduced from Cheeseman et al (2006). (G) Negative stain electron microscopy of Dam1 rings assembled around microtubules in vitro. Bar=50 nm. The panel reproduced from Westermann et al (2005). (H) The Dam1 complexes are heterodecamers. They contain one copy each of 10 essential budding yeast proteins. Dam1 rings form by oligomerization of individual complexes around microtubules.
The Spc24:Spc25 dimer binds to the Mis12 and Knl1 complexes near the inner plate (Kiyomitsu et al, 2007; Schittenhelm et al, 2007; Joglekar et al, 2009; Wan et al, 2009). The Nuf2:Ndc80 dimer, on the other hand, points outward and binds microtubules directly (Cheeseman et al, 2006; Wei et al, 2007; Ciferri et al, 2008). Structural work, including a structure of a ‘bonsai' Ndc80 complex (Figure 6C and D) showed that the microtubule-binding domain of Ndc80:Nuf2 combines an 80-residue unstructured basic region of Ndc80 (pI approximately 10.8) and two tightly packed calponin-homology (CH) domains, one in each chain (Figures 6C and D) (Wei et al, 2007; Ciferri et al, 2008). Lysine residues in the two CH domains contribute to high-affinity microtubule binding (Cheeseman et al, 2006; Wei et al, 2007; Ciferri et al, 2008). On microtubules, the acidic C-terminal tails of tubulin subunits (so called E-hooks) are important for high-affinity binding to the Ndc80 complex (Wei et al, 2007; Ciferri et al, 2008; Powers et al, 2009).
Despite these advances, the exact mode of binding of microtubules by the KMN network remains unclear. A comparison of the crystal structure of the Ndc80:Nuf2 globular regions and three-dimensional EM maps obtained by helical reconstruction of Ndc80:Nuf2 bound to microtubules contended that a binding mechanism involving both the CH domains of Ndc80 and Nuf2 is unlikely (Wilson-Kubalek et al, 2008). Other studies indicated that the basic N-terminal tail of Ndc80 might be sufficient for high-affinity microtubule binding, even in the absence of CH domains (Guimaraes et al, 2008; Miller et al, 2008). Finally, Knl1 may also contain a microtubule-binding region, but the boundaries of the region responsible are unknown (Cheeseman et al, 2006).
At high concentrations, the Ndc80 complex binds along the microtubule lattice of microtubules stabilized with taxol or non-hydrolysable GTP analogues, adopting a 20–60° angle relative to the microtubule long axis (Figure 6F) (Wei et al, 2007; Cheeseman and Desai, 2008; Ciferri et al, 2008; Wilson-Kubalek et al, 2008). At low concentration, the Ndc80 complex shows a modest preference for depolymerizing plus ends of dynamic microtubules, a preference that is greatly enhanced when the Ndc80 complexes are crosslinked with antibodies (Powers et al, 2009). Beads coated with the Ndc80 complex undergo biased diffusion towards the minus end of a depolymerizing microtubule and can resist 0.5–2.5 pN of tensile force (McIntosh et al, 2008; Powers et al, 2009). As explained above, these observations suggest that Ndc80 acts as a Hill's coupler. By quantitative fluorescence microscopy of GFP-tagged kinetochore proteins in S. cerevisiae and S. pombe, it was found that there are 6–8 copies of the KMN network per microtubule-attachment site, whereas approximately 30 KMN complexes per microtubule-attachment site are found at kinetochores in Xenopus laevis extracts (Emanuele et al, 2005; Joglekar et al, 2006, 2008).
Besides the KMN network, other kinetochore-bound complexes have attracted considerable attention as microtubule-coupling devices at the kinetochore. Most notably, the Dam1 complex, an essential hetero-decameric complex of S. cerevisiae, has been extensively studied for its ability to form rings around microtubules (Figure 6G and H) (Miranda et al, 2005; Westermann et al, 2005) and more generally for its support to the process of chromosome segregation (e.g. Cheeseman et al, 2001; Asbury et al, 2006; Westermann et al, 2006; Franck et al, 2007; Tanaka et al, 2007; Grishchuk et al, 2008a). Approximately 16 hetero-decameric complexes have been predicted to account for a full ring around the microtubule, and this is also approximately the number of Dam1 complexes present at one microtubule-binding site in this organism (Joglekar et al, 2006; Westermann et al, 2006). Rings, however, have not been observed in electron tomograms of the S. cerevisiae's kinetochore–microtubule interface and are not required for processive attachment of the Dam complex to microtubules (O'Toole et al, 1999; McIntosh, 2005; Gestaut et al, 2008). A bead coated with the Dam1 complex undergoes assembly- and disassembly-driven motility and remains coupled to a disassembling microtubule against a force of 0.5–3 pN (Asbury et al, 2006). Furthermore, high tension applied to the Dam1 complex stabilizes the microtubule plus end, an essential function of kinetochores as explained above (Franck et al, 2007). However, the generality of these findings is questioned by the observation that the Dam1 complex is conserved but is not essential in fission yeast (Sanchez-Perez et al, 2005; Gachet et al, 2008), and that homologues of the Dam1 complex have not been identified in higher eukaryotes.
The 3-subunit Ska complex (Figure 2D) was recently identified as a new microtubule-binding activity at metazoan kinetochores (Hanisch et al, 2006; Gaitanos et al, 2009; Raaijmakers et al, 2009; Theis et al, 2009; Welburn et al, 2009). Ablation of the Ska complex by RNAi leads to a very severe attachment phenotype that is reminiscent of the kinetochore-null phenotype observed with Ndc80 complex depletions (Gaitanos et al, 2009; Raaijmakers et al, 2009; Theis et al, 2009; Welburn et al, 2009). As the Ska complex is recruited to kinetochores through the Ndc80 complex, the effects from inhibiting Ndc80 by RNA interference may represent the convolution of two phenotypes caused by loss of the Ndc80 complex as well as of the Ska complex. The Ska complex does not associate tightly with the Ndc80 complex and its association with kinetochores might be stabilized by microtubules (Hanisch et al, 2006; Gaitanos et al, 2009; Raaijmakers et al, 2009; Theis et al, 2009). It has been proposed that the Ska complex is a functional homologue of the Dam1 that can form rings around microtubules (Welburn et al, 2009), but this contention may require further evaluation.
Sli15p and Bir1p of S. cerevisiae, respectively, homologous to INCENP and Survivin in higher eukaryotes, are part of a complex that is commonly referred to as the chromosome passenger complex (CPC), and that also includes the Ipl1/Aurora B kinase and Nbl1p/Borealin/DasraB/CSC-1 (Vader et al, 2006; Ruchaud et al, 2007). Indeed, the components of this complex are not mere passengers riding chromosomes to perform their essential functions at anaphase, as originally proposed (Earnshaw and Bernat, 1991). Rather, they perform essential functions on chromosomes all along mitosis (Ruchaud et al, 2007). Thus, the term ‘chromosomal passenger' is a misnomer, but its use has become so customary in the literature that we refrain from proposing an alternative here. Sli15p and Bir1p possibly provide for an additional kinetochore–microtubule coupling mechanism (Sandall et al, 2006). Budding yeast centromeric (CEN) DNA binds to microtubules in a CBF3-dependent manner after incubation in a cell extract (Kingsbury and Koshland, 1991; Hyman et al, 1992; Sorger et al, 1994; Severin et al, 1997). However, CBF3 is not sufficient, indicating that other factors are necessary to link CBF3–CEN DNA to microtubules (Sorger et al, 1994). The Bir1p:Sli15p complex was identified as a potential additional factor in linking the CBF3–CEN DNA complex to microtubules in vitro (Sandall et al, 2006). Indeed, Sli15p/INCENP contains a microtubule-binding site in its C-terminal region (Sandall et al, 2006).
Although molecular motors are dispensable for anaphase chromosome movement, they have an important auxiliary function in the initial side-on capture of microtubules and in the congression of chromosomes to the metaphase plate. These functions require cytoplasmic Dynein, a minus end directed motor, and CENP-E, a plus end directed motor, respectively, (e.g. see Alexander and Rieder, 1991; Kapoor et al, 2006). The RZZ complex interacts with the KMN network to recruit Spindly, Dynein, and the SAC proteins Mad1 and Mad2 to kinetochores (Civril and Musacchio, 2008). The coiled-coil protein, Spindly, is important for the coordination of the conversion of side-on to end-on attachments, but the molecular details of this process are still unknown (Griffis et al, 2007; Civril and Musacchio, 2008; Gassmann et al, 2008; Yamamoto et al, 2008).
Vertical and horizontal kinetochores
The architecture of the kinetochore, and most notably the relationship between the inner and outer plates, remains elusive. Our understanding of kinetochore assembly derives from proteomic analyses describing the composition of the more tightly interacting complexes and sub-complexes (see above). Furthermore, the effects from depleting certain kinetochore proteins on the (mis)localization of other kinetochore proteins have been extensively studied (e.g. Liu et al, 2006; McClelland et al, 2007; Cheeseman et al, 2008; Hori et al, 2008a). Although the results cannot always be univocally interpreted, they support a map of ‘epistatic' relationships in which the inner kinetochore components are indeed required for the localization of the outer kinetochore components (Figure 7A). For instance, CENP-A, CENP-T/W, CENP-C, and the CCAN CENP-H/I/K proteins all contribute, to different extents, to the recruitment of the KMN network and associated proteins (e.g. Liu et al, 2003; Hayashi et al, 2004; Mikami et al, 2005; Saitoh et al, 2005; Liu et al, 2006; Okada et al, 2006; McClelland et al, 2007; Cheeseman et al, 2008; Hori et al, 2008a). In the absence of accurate physical maps of kinetochores, it is unknown whether the relationships described in Figure 7A correspond to actual physical contacts between complexes. Alternatively, the inner kinetochore proteins may contribute to an organization of the centromere–kinetochore interface that promotes the recruitment of the outer kinetochore proteins, for instance by mechanisms based on post-translational modification.
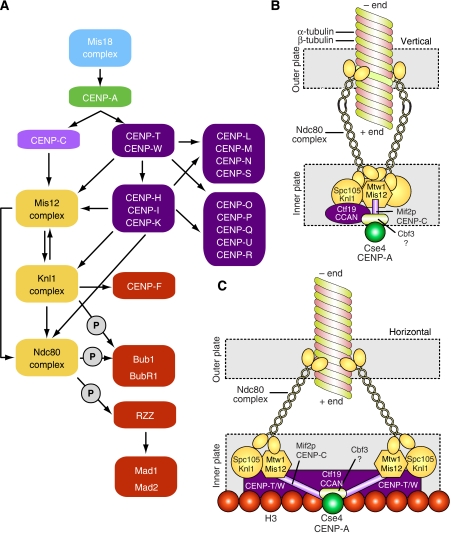
Figure 7.
Models of kinetochore assembly. (A) ‘Epistatic' relationships between kinetochore proteins. Arrows indicate a dependency for localization, where the pointed end indicates a protein(s) that requires proteins at the barbed end for kinetochore localization. The list of proteins shown here is not comprehensive. The circles enclosing a ‘P' indicate post-translational modifications. (B) The vertical layout. Kinetochore proteins ultimately converge on a single Cse4p/CENP-A nucleosome (e.g. Joglekar et al, 2009). Given that there are 6–8 KMN network complexes per Cse4/CENP-A nucleosome, it is sensible to assume that this special nucleosome is placed directly below the microtubule, approximately on the same axis, with the different KMN network surrounding the microtubule roughly equidistantly (only two KMN complexes are shown here). (C) The horizontal model. Rather than being placed along an idealized vertical line from the inner to the outer kinetochore, the kinetochore components are distributed horizontally. Specifically, the KMN network components are linked to the kinetochore core by Mif2p/CENP-C, but are also establishing specific contacts with H3 nucleosomes through CENP-T/W.
We hypothesize two alternative designs for kinetochores, both of which are compatible with the super-resolution microscopic analyses described in Figures 1C and 2E (Schittenhelm et al, 2007; Joglekar et al, 2009; Wan et al, 2009). In discussing these kinetochore designs, which we name ‘vertical' and ‘horizontal', we refer to an archetypical single microtubule-binding unit. The kinetochore of S. cerevisiae provides a useful framework for such a unit, but we implicitly adopt the idea that kinetochores-binding multiple microtubules are at least in part modular and that they contain an array of equivalent units (see below).
In the ‘vertical' kinetochore (Figure 7B), the components of the inner and outer kinetochore are recruited sequentially onto the CENP-A platform along a vertical plan of assembly. In this model, CENP-A provides the physical basis for the recruitment of all additional kinetochore proteins, starting from the inner kinetochore (CCAN and CENP-C) and continuing with the KMN network. In this model, strong physical contacts between the inner and outer kinetochore layers are probably necessary, because the forces exercised by bound microtubules converge directly, through the outer kinetochore, on the single specialized CENP-A nucleosome and associated CENP-C and CCAN. Indeed, CENP-C (Mif2p in S. cerevisiae) has been identified as a low-abundance component of KMN precipitates, as well as a binding partner of Cse4/CENP-A (Ando et al, 2002; Westermann et al, 2003; Cheeseman et al, 2004), and may therefore act as a linker between inner and outer kinetochores. A puzzling aspect is that with only 1–2 molecules per Cse4 nucleosome, CENP-C is significantly sub-stoichiometric with respect to KMN network complexes (Joglekar et al, 2006).
Another possible linkage between the inner and outer kinetochore engages Nuf2 and CENP-H (Mikami et al, 2005). However, linkages involving CCAN subunits are unlikely to be essential for outer kinetochore assembly, because the ablation of the CCAN subunits partially affects but never abolishes the recruitment of outer kinetochore components, including KMN network subunits, and the resulting phenotypes are clearly distinct (Liu et al, 2003, 2006; Hayashi et al, 2004; Mikami et al, 2005; Saitoh et al, 2005; Okada et al, 2006; McClelland et al, 2007; Cheeseman et al, 2008). For instance, though Ndc80-depleted cells are unable to form a metaphase plate, cells depleted of CCAN subunits have milder chromosome congression and segregation phenotypes and can form stable attachments (Fukagawa et al, 2001; Nishihashi et al, 2002; Liu et al, 2003; Minoshima et al, 2005; Foltz et al, 2006; Okada et al, 2006; McClelland et al, 2007).
An objection to the vertical model is that force exercised by a bound microtubule through the KMN network components converges onto a single Cse4p/CENP-A nucleosome, rather than being distributed over a larger attachment site. A related prediction is that the microtubule (25 nm diameter) connects to the Cse4p/CENP-A nucleosome, a much smaller structure (10 nm or less) (Bloom et al, 2006). If the single Cse4p nucleosome broadly lies along the microtubule's long axis, the KMN complexes would have to radiate from this central point outward to be able to bind to the external wall of the microtubule (Figure 7B). It is difficult to reconcile this geometry with that observed on reconstitution of the interaction of recombinant Ndc80 complexes with microtubules in vitro (Figure 6F) (Cheeseman et al, 2006; Wilson-Kubalek et al, 2008). If the binding mode observed in the in vitro studies existed in cells, the Spc24:Spc25 globular regions would project onto the kinetochore at a distance of 20–40 nm from the microtubule axis (and thus from the Cse4p/CENP-A nucleosome, if its position coincided with the microtubule axis).
In the ‘horizontal' model, this geometric limitation is resolved by placing the KMN complexes away from the ‘central' CENP-A nucleosome, anchoring them to H3 nucleosomes surrounding the CENP-A nucleosome (Figure 7C). A desirable feature of this design is that microtubule-generated pulling forces are distributed over several distinct contact points rather than on a single point as in the vertical model.
The CENP-T:CENP-W dimer has been recently shown to contribute to the stability of the outer plate, as observed earlier for the Ndc80 complex (DeLuca et al, 2005; Hori et al, 2008a). CENP-T and CENP-W are homologous proteins showing sequence similarity to the Negative Cofactor 2 (NC2) complex, which contains a histone-fold domain. Budding yeast homologues of these proteins have not been observed. CENP-T was originally purified using CENP-A as bait (Foltz et al, 2006), but it has been suggested that co-purification with CENP-A was due to partial micrococcal nuclease cleavage. On more stringent analyses, the CENP-T:CENP-W dimer revealed an association with H3 nucleosomes (Hori et al, 2008a). CENP-C was also found in contact with H3 nucleosomes, in agreement with its role in recruiting the KMN network. Direct association between CENP-C and the CENP-T:CENP-W complex, however, has not been identified (Hori et al, 2008a). In summary, the CENP-T:CENP-W complex may contribute to creating a binding site for KMN network and associated proteins on H3 nucleosomes surrounding CENP-A. An indirect confirmation of this model derives from the observation that the KMN network interacts with HP1 (heterochromatin protein 1), a protein that binds to the methylated form of Lys9 of histone H3 (Obuse et al, 2004; Przewloka et al, 2007).
The organization of kinetochores that bind multiple microtubules
The question whether regional centromere/kinetochores containing multiple microtubule-binding sites (Figure 8A and B) are built from the repetition of a simpler functional unit remains open. The existence of a regularly repeated microtubule-binding unit has not emerged from tomographic reconstructions of the outer plate, which instead depicted the microtubule-binding interface of the outer kinetochore as a disorganized ‘velcro' or ‘spider's web' for microtubule attachment (Figure 2C) (Dong et al, 2007).
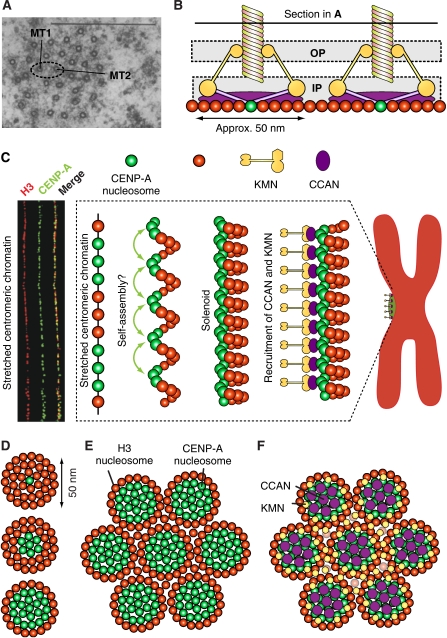
Figure 8.
‘Repeat subunit' models. (A) A transverse section through the K-fibre of a metaphase PtK1 cell, showing multiple microtubules. Bar=0.5 μM, magnification × 60000. Source of figure is from Rieder (1981). (B) Horizontal clustering of modules (only two are shown) may explain the distribution of microtubules in the K-fibre shown in (A). (C) The solenoid model. Left: centromere stretching experiment indicating that the array of CENP-A nucleosomes, coalesced in three-dimensional space, are not contiguous along the DNA but are interrupted by spacers containing blocks of H3-containing nucleosomes. The image was reproduced from Blower et al (2002). Right: CENP-A nucleosome coalescence could be entirely self-directed, or alternatively, it might necessitate the action of bridging factors—perhaps components of the CCAN—to organize into the array that forms the foundation of the mitotic kinetochore. The panel is an adaptation from Black and Bassett (2008). (D) Three distinct hypothetical patterns of CENP-A and H3 nucleosomes with different ratios of H3 to CENP-A. CENP-A is always shown at the centre, and is surrounded by H3. (E) The pattern at the bottom of (D) is now shown to ‘coalesce' in a larger assembly. (F) Speculative pattern of deposition of CCAN and KMN modules on the pattern shown in (E). CCAN is on CENP-A nucleosomes, whereas KMN goes to H3 nucleosomes.
On the other hand, similarity in composition, abundance ratios, and epistatic relationships of kinetochore complexes with relatively minor differences from yeast to humans, suggests that at least the hierarchical relationship between different kinetochore layers is maintained from point to regional centromere/kinetochores. For instance, the ratio between KMN components and the core subunits of the kinetochore (e.g. CENP-A) is conserved in yeasts with point and regional centromeres, and has lead to suggest that the kinetochore of S. pombe contains 3–5 units modelled on the single microtubule-binding site of S. cerevisiae (Joglekar et al, 2008). Furthermore, kinetochore proteins occupy relative analogous positions within the kinetochore layouts of the point centromere/kinetochore of S. cerevisiae and the regional centromere/kinetochore of D. melanogaster (Figures 1C and 2E) (Schittenhelm et al, 2007; Joglekar et al, 2009; Wan et al, 2009).
Regional centromere/kinetochores can disassemble into smaller ‘units' if their connection with centromeric chromatin is artificially loosened (Zinkowski et al, 1991; O'Connell et al, 2008). The actual structural organization of the ‘units' has not been elucidated. However, chromatin fibre analyses of centromeric chromatin in humans and flies suggest that CENP-A comes in discrete blocks alternating with H3-containing blocks (Figure 8C) (Blower et al, 2002). It has been proposed that CENP-A and H3 might be sorted on different faces of an ‘amphipathic' super-helical arrangement of centromeric chromatin, a solenoid in which the CENP-A-containing face will be facing outward towards the kinetochore, and the H3-containing face will be embedded in the centromere (Figure 8C) (Zinkowski et al, 1991; Blower et al, 2002; Marshall et al, 2008a).
The solenoid model neglects the emerging role of H3 in the assembly of the outer kinetochore (see above). An alternative speculative model is that CENP-A nucleosomes are surrounded by H3 nucleosomes to create the centromeric inner kinetochore moiety of a microtubule-binding unit. Three possible examples of this organization, with progressively larger numbers of CENP-A nucleosomes, are illustrated in Figure 8D. The functional units, in turn, might coalesce into a larger array (Figure 8E). If the KMN network is recruited to H3 nucleosomes, this type of construction in the inner kinetochore might be directing the KMN network complexes to the edges of each microtubule-binding unit (Figure 8F). As there are 6–8 KMN complexes per microtubule-binding site (Joglekar et al, 2006, 2008), the speculative configuration of the centromere/inner kinetochore in Figure 8E would position the KMN complexes at the appropriate distance from the microtubule-binding site. The latter would be identified as a ‘hole' in the distribution of the KMN network complexes in correspondence of the CENP-A/CCAN complexes in the underlying chromatin (Figure 8F). The ‘holes' would allow microtubules to penetrate deeply within the outer kinetochore surface, allowing the KMN complexes to surround the microtubule to stabilize the end-on configuration. As the KMN network complexes are elongated, flexible fibrous structures, it may be difficult to visualize the ‘holes' in tomographic reconstructions of the outer plate in the absence of microtubules (Dong et al, 2007).
The molecular bases of feedback control of kinetochores: error correction
The ability to discriminate between correct and incorrect microtubule attachments, selectively stabilizing the former and preventing the stabilization of the latter, is crucial for chromosome stability during cell division (Nicklas and Koch, 1969; Li and Nicklas, 1995). Attachment errors, such as syntelic and merotelic attachments, can be artificially stabilized in high numbers if the activity of the Aurora B kinase is inhibited with a small molecule inhibitor (e.g. Ditchfield et al, 2003; Hauf et al, 2003; Lampson et al, 2004; Cimini et al, 2006). In a revealing assay, re-activation of Aurora B results in the correction of improper attachments after inhibitor washout (Lampson et al, 2004). A similar accumulation of attachment errors is generated when temperature-sensitive mutants of Ipl1, the only Aurora kinase of S. cerevisiae, are exposed to the non-permissive temperature (Tanaka et al, 2002). These studies implicate Ipl1/Aurora B as an essential component of the error correction mechanism required to prevent the stabilization of improper attachments.
The exact molecular details of the correction mechanism are elusive, but the regulation of microtubule-binding factors at the kinetochore is probably crucial (Kelly and Funabiki, 2009). For instance, Aurora B phosphorylates the basic N-terminal tail of Ndc80, neutralizing the positive charge and lowering the affinity of Ndc80 for microtubules (Cheeseman et al, 2006; DeLuca et al, 2006; Ciferri et al, 2008). Aurora B also controls the activity of MCAK and Kif2a, two kinesin-13 family members that are implicated in the regulation of the stability of kinetochore microtubule (Ohi et al, 2003; Andrews et al, 2004; Lan et al, 2004; Knowlton et al, 2006, 2009; Huang et al, 2007; Zhang et al, 2007; Bakhoum et al, 2009). Overall, these interactions may modulate the binding affinity of kinetochores for microtubules, as well as the dynamics of the microtubule plus end.
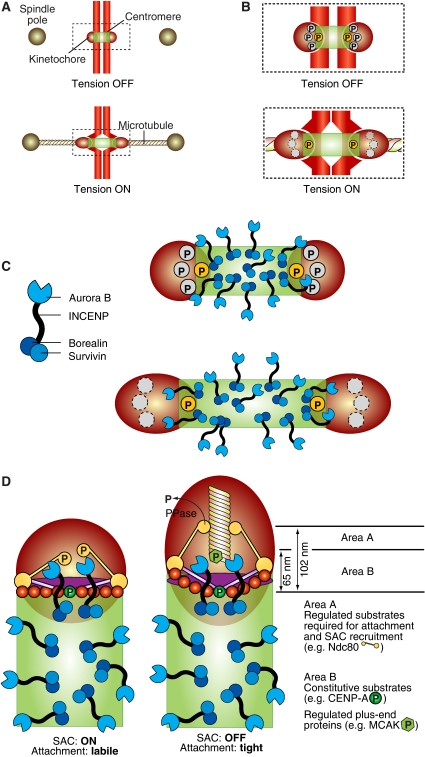
How does Aurora B distinguish correct from incorrect attachments? How is its activity differentially regulated at correct and incorrect attachments? Bi-oriented sister chromatids are under tension, that is they experience a force that tends to part the sisters, stretching centromeric chromatin as well as the kinetochore (Skibbens et al, 1993; Waters et al, 1996; Maresca and Salmon, 2009; Uchida et al, 2009). Incompletely (monotelic or even unbound) or incorrectly (syntelic) attached sisters, on the other hand, are not under tension (e.g. Ditchfield et al, 2003; Liu et al, 2009). As (1) the distance between centromeres and kinetochores increases when the sisters are under tension, and (2) the CPC is located at the centromere, it was proposed that the ability of Ipl1/Aurora B to reach its substrates in the kinetochore may be reduced or eliminated when tension builds up (Figure 9A and B) (Tanaka et al, 2002). Recently, this hypothesis was corroborated by elegant experiments in which an Aurora B substrate docked within the kinetochore at a sufficiently large distance from the centromere became dephosphorylated as microtubule attachment ensued (Liu et al, 2009). Substrates closer to the centromere, on the other hand, were constitutively phosphorylated with or without microtubule attachment. Overall, these results suggest that Aurora B delivers constitutive levels of phosphorylation during the attachment phase, and that the regulation of attachment depends on the accessibility of the substrates (Figure 9B) (Liu et al, 2009).
Figure 9.
Error correction and the spindle checkpoint. (A) Schematic description of the geometry of the centromere–kinetochore interface in the absence and presence of tension. (B) The boxed area in (A) enlarged. Phosphorylation of certain substrates at the centromere–kinetochore interface is constitutive (the yellow circle marked by ‘P'), that is the substrate is phosphorylated with or without tension. Other substrates are only phosphorylated in the absence of tension, because their separation from the centromere exceeds a threshold value when tension is present. (C) Left: schematic description of the CPC complex. Right: the CPC occupies the centromere, and only a subset of complexes is located near the centromere–kinetochore interface. (D) A comprehensive model of checkpoint control and error correction. In the absence of tension, either substrate like Ndc80 become phosphorylated by Aurora B or by other kinases whose activation requires Aurora B. This creates a condition for SAC activation through the recruitment of SAC proteins (Ditchfield et al, 2003; Hauf et al, 2003). On the other hand, the phosphorylation of Ndc80 decreases the binding affinity for microtubules (Cheeseman et al, 2006; DeLuca et al, 2006; Ciferri et al, 2008). This creates a state of labile attachment that will become corrected unless a force is applied. The removal of Ndc80 and possibly other substrates from the reach of Aurora B stabilizes the attachment through the action of a phosphatase.
Aurora B is tethered, through the INCENP linker, to a Borealin:Survivin complex embedded in the centromere (Figure 9C) (Vader et al, 2006; Ruchaud et al, 2007). As the inter-kinetochore centromeric region extends for 1 μM or more in vertebrates, most CPC complexes tethered within this domain are expected to be unable to reach substrates in the kinetochore, and that only a subset of Aurora B molecules located near the centromere–kinetochore interface, can target the kinetochore. If this subset was tethered and was only able to reach as far as a certain distance from the point of tethering, kinetochore stretching on microtubule attachment might indeed result in the separation of Aurora B from its substrates (Figure 9D). In agreement with this model, two recent papers showed that kinetochores become stretched during kinetochore–microtubule attachment. For instance, the distance between the C-terminus of Ndc80 and CENP-A is approximately 102 or 65 nm when chromosomes are or are not under tension, respectively (Maresca and Salmon, 2009; Uchida et al, 2009).
The fact that Aurora B is active in the presence of unattached kinetochores poses a conceptual difficulty. It suggests that a model in which Aurora B activity is required to destabilize tensionless kinetochore–microtubule attachments is probably simplistic. As unattached kinetochores are also tensionless, the destabilization model predicts that they would be targeted by Aurora B and would be permanently prevented from attaching. Rather, Aurora B may function by preventing premature stabilization of the attachments, that is by creating an initial condition of labile attachment that will be corrected unless microtubules pulled in the right direction and enforced tension, subtracting kinetochore substrates from the Aurora B kinase and making them become stabilized (Figure 9D). The correction mechanism remains obscure. The intrinsic instability of microtubules might be sufficient to release improperly attached microtubules whose attachment remained labile. On the other hand, the model in Figure 9D might have interesting implications for the regulation of microtubule plus end dynamics by centromere-associated proteins.
The molecular bases of feedback control of kinetochores: the spindle checkpoint
We will not dwell on the molecular mechanism of the SAC, which has been recently reviewed (Musacchio and Salmon, 2007) and that constitutes the topic of a review by Ciliberto and Shah in this issue of the EMBO journal. We will rather discuss the relationship between the microtubule-binding machinery, the error correction mechanism, and the SAC. Indeed, the challenge of studies on feedback control at kinetochores is to explain its dynamic relationship with the molecular machinery controlling microtubule attachment, a task now made easier by the identification of the likely key players of kinetochore–microtubule attachment (reviewed in Cheeseman and Desai, 2008; Tanaka and Desai, 2008). Emphasizing the tight relationship between feedback control mechanisms and microtubule attachment, the majority of SAC proteins are recruited to the Knl1/Mis12/Ndc80 (KMN) complex (as discussed in Burke and Stukenberg, 2008).
Since the early days, the relationship of the error correction machinery with the SAC has proved a great intellectual challenge and a topic of speculation (McIntosh, 1991; Rieder and Palazzo, 1992). It is widely believed that Aurora B has an indirect role in SAC control. Specifically, Aurora B may elicit SAC signalling when, by destabilizing improper tensionless kinetochore–microtubule attachments, it creates unattached kinetochores that in turn recruit bona fide checkpoint proteins such as the products of the MAD and BUB genes, which then combine to halt cell-cycle progression (Pinsky and Biggins, 2005; Pinsky et al, 2006). As observed above, however, unattached kinetochores are also tensionless, and Aurora B is active at kinetochores of nocodazole-treated cells, which lack any attachment (Liu et al, 2009).
This raises the question whether Aurora B activity is directly implicated in SAC control. In agreement with this hypothesis, Aurora B is required for kinetochore recruitment of SAC proteins in the presence of microtubule-depolymerizing drugs (Ditchfield et al, 2003; Hauf et al, 2003), which in turn is an absolute requirement for SAC activation (e.g. Meraldi et al, 2004). Overall, these observations suggest a direct involvement of Aurora B in SAC control, reinforcing the link between error correction and SAC control. Evidence that Aurora B is required to maintain the SAC from unattached kinetochores is available in fission yeast and Xenopus (Kallio et al, 2002; Petersen and Hagan, 2003). In other organisms, it has been difficult to show an SAC override when inhibiting Aurora B (as discussed in Pinsky and Biggins, 2005; Kelly and Funabiki, 2009). However, this may be a consequence of residual kinase activity on incomplete depletion or inactivation of Aurora B.
As the ability of Aurora B to correct improper attachments may rely on increased distance from its kinetochore substrates, it is logical to ask whether its function in the SAC is regulated in the same manner. In agreement with this idea, it was shown recently that intra-kinetochore stretching is crucial for determining the state of checkpoint signalling as well as the state of kinetochore phosphorylation (Figure 9D) (Maresca and Salmon, 2009; Uchida et al, 2009). In addition, in the case of the SAC, the regulatory mechanism is consistent with a model in which Aurora B substrates are progressively separated from the kinase as attachment ensues (Liu et al, 2009).
The discovery that intra-kinetochore stretching controls the SAC promises to change the way we think about the interaction of the checkpoint components with kinetochores. Changes in the separation of kinetochore proteins of 35–40 nm are sufficient to control the state of checkpoint activation (Maresca and Salmon, 2009). The relative displacements of kinetochore proteins on the establishment of tension have been measured, providing a clear physical correlate to this model (Maresca and Salmon, 2009; Wan et al, 2009).
In summary, SAC activation and error correction could respond to the same molecular logic. When Aurora B is able to phosphorylate its outer kinetochore substrates, the SAC is on and attachment is labile. When the substrates are removed from the kinase, attachment becomes stabilized and the SAC is concomitantly turned off (Figure 9D). This model of attachment and SAC control predicts the existence of a crucial phosphatase activity to revert the initial state of kinetochore phosphorylation when the distance of kinetochore substrates from Aurora B increases.
A look into the future
Although it is early to formulate a general theory of kinetochore function and regulation, our understanding of kinetochore's processes has been propelled forward by tremendous recent progress. In this review, we have discussed possible formulations of the static organization of kinetochores, as well as those aspects of dynamic regulation that may subtend to the stabilization of kinetochore–microtubule attachment and to SAC control.
Unveiling the static organization of kinetochores will eventually require high-resolution structural investigations on progressively more complex portions of the kinetochore. Recombinant reconstitution is expected to provide crucial support to structural analysis, and will also allow characterizing the physical interactions between kinetochore modules. The combination of improved sample preservation approaches and advancements in the field of electron tomography and high-resolution fluorescence microscopy are expected to enlighten kinetochore organization at increasing resolution. Being able to distinguish between the ‘vertical' and ‘horizontal' models of the kinetochore, discussed above, as well as to investigate the possible existence of a repeating kinetochore module are crucial goals for future research. Gaining a better understanding of the organization of centromeric chromatin and of its interactions and modifications may significantly contribute to this goal.
Dynamic kinetochore regulation reflects the interaction of the kinetochore's different modules. The realization of the importance of the KMN network, as the regulatory hub for kinetochore–microtubule stabilization as well as SAC control is a crucial recent advancement. Future studies will have to address the structural bases through which Aurora B acts as a ‘nanoruler' within the kinetochore to regulate kinetochore–microtubule attachment stability as well as the spindle checkpoint cascade. Identifying the phosphorylation sites on the KMN network responsible for the coordination of these processes is a key goal for future studies. We expect that the manipulation of these sites will be crucial for validation of different models of dynamic kinetochore control.
Supplementary Material
Supplementary Table I
Acknowledgments
We apologize to all those authors whose work could not be cited because of space restrains. We thank the members of the Musacchio laboratory for many helpful discussions, and J Richard McIntosh for critical reading of the manuscript. Work in the Musacchio laboratory is generously funded by the Association for International Cancer Resarch (AICR), the Telethon Foundation, the European Commission's FP6 program contracts 3D-Repertoire and Mitocheck and the FP7 European Research Council grant KINCON, the Italian Association for Cancer Research (AIRC), the Fondo di Investimento per la Ricerca di Base (FIRB), the Italian Ministry of Health, and the Cariplo Foundations. SS is a graduate student of the European School of Molecular Medicine.
References
- Alexander SP, Rieder CL (1991) Chromosome motion during attachment to the vertebrate spindle: initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J Cell Biol 113: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Karpen GH (2008) Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 9: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando S, Yang H, Nozaki N, Okazaki T, Yoda K (2002) CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol 22: 2229–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR (2004) Aurora B regulates MCAK at the mitotic centromere. Dev Cell 6: 253–268 [DOI] [PubMed] [Google Scholar]
- Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN (2006) The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci USA 103: 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Thompson SL, Manning AL, Compton DA (2009) Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol 11: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Bassett EA (2008) The histone variant CENP-A and centromere specification. Curr Opin Cell Biol 20: 91–100 [DOI] [PubMed] [Google Scholar]
- Black BE, Brock MA, Bédard S, Woods VL, Cleveland DW (2007) An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci USA 104: 5008–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Cleveland DW (2004) Structural determinants for generating centromeric chromatin. Nature 430: 578–582 [DOI] [PubMed] [Google Scholar]
- Bloom K, Sharma S, Dokholyan NV (2006) The path of DNA in the kinetochore. Curr Biol 16: R276–R278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2: 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram RJ, Kornberg RD (1987) Isolation of a Saccharomyces cerevisiae centromere DNA-binding protein, its human homolog, and its possible role as a transcription factor. Mol Cell Biol 7: 403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke DJ, Stukenberg PT (2008) Linking kinetochore-microtubule binding to the spindle checkpoint. Dev Cell 14: 474–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Straight AF (2006) Centromere formation: from epigenetics to self-assembly. Trends Cell Biol 16: 70–78 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates JR, Huffaker TC, Drubin DG, Barnes G (2001) Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol 155: 1137–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127: 983–997 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A (2008) Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 9: 33–46 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Hori T, Fukagawa T, Desai A (2008) KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol Biol Cell 19: 587–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, Oegema K, Desai A (2004) A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev 18: 2255–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, De Luca J, Monzani S, Ferrari KJ, Ristic D, Wyman C, Stark H, Kilmartin J, Salmon ED, Musacchio A (2005) Architecture of the human Ndc80-Hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem 280: 29088–29095 [DOI] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dosreis G, Maiolica A, Polka J, Deluca J, Dewulf P (2008) Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Moree B, Canman JC, Salmon ED (2003) Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci 116: 4213–4225 [DOI] [PubMed] [Google Scholar]
- Cimini D, Wan X, Hirel CB, Salmon ED (2006) Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol 16: 1711–1718 [DOI] [PubMed] [Google Scholar]
- Civril F, Musacchio A (2008) Spindly attachments. Genes Dev 22: 2302–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- Cohen RL, Espelin CW, De Wulf P, Sorger PK, Harrison SC, Simons KT (2008) Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol Biol Cell 19: 4480–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins KA, Castillo AR, Tatsutani SY, Biggins S (2005) De novo kinetochore assembly requires the centromeric histone H3 variant. Mol Biol Cell 16: 5649–5660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coue M, Lombillo VA, McIntosh JR (1991) Microtubule depolymerization promotes particle and chromosome movement in vitro. J Cell Biol 112: 1165–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y, Furuyama T, Vermaak D, Henikoff S (2007) Structure, dynamics, and evolution of centromeric nucleosomes. Proc Natl Acad Sci USA 104: 15974–15981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TN, Wordeman L (2007) Rings, bracelets, sleeves, and chevrons: new structures of kinetochore proteins. Trends Cell Biol 17: 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P, McAinsh AD, Sorger PK (2003) Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev 17: 2902–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF (2005) Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell 16: 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED (2006) Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127: 969–982 [DOI] [PubMed] [Google Scholar]
- Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Shevchenko A, Hyman A, Oegema K (2003) KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev 17: 2421–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS (2003) Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol 161: 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF (2007) The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol 9: 516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Bernat RL (1991) Chromosomal passengers: toward an integrated view of mitosis. Chromosoma 100: 139–146 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91: 313–321 [DOI] [PubMed] [Google Scholar]
- Emanuele MJ, McCleland ML, Satinover DL, Stukenberg PT (2005) Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol Biol Cell 16: 4882–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF (2008) Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol 183: 805–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin CW, Simons KT, Harrison SC, Sorger PK (2003) Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10p to CDEII. Mol Biol Cell 14: 4557–4568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer U, McIntosh JR (1981) Structural polarity of kinetochore microtubules in PtK1 cells. J Cell Biol 89: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, Cleveland DW (2006) The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 8: 458–469 [DOI] [PubMed] [Google Scholar]
- Franck AD, Powers AF, Gestaut DR, Gonen T, Davis TN, Asbury CL (2007) Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol 9: 832–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M (2007) Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell 12: 17–30 [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Mikami Y, Nishihashi A, Regnier V, Haraguchi T, Hiraoka Y, Sugata N, Todokoro K, Brown W, Ikemura T (2001) CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J 20: 4603–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S, Biggins S (2007) Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci USA 104: 14706–14711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet Y, Reyes C, Courtheoux T, Goldstone S, Gay G, Serrurier C, Tournier S (2008) Sister kinetochore recapture in fission yeast occurs by two distinct mechanisms, both requiring Dam1 and Klp2. Mol Biol Cell 19: 1646–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA (2009) Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J 28: 1442–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R, Essex A, Hu J, Maddox P, Motegi F, Sugimoto A, O'rourke S, Bowerman B, Mcleod I, Yates JR III, Oegema K, Cheeseman IM, Desai A (2008) A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev 22: 2385–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut DR, Graczyk B, Cooper J, Widlund PO, Zelter A, Wordeman L, Asbury CL, Davis TN (2008) Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol 10: 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Stuurman N, Vale RD (2007) Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol 177: 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, Efremov AK, Volkov VA, Spiridonov IS, Gudimchuk N, Westermann S, Drubin D, Barnes G, McIntosh JR, Ataullakhanov FI (2008a) The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc Natl Acad Sci USA 105: 15423–15428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, McIntosh JR (2006) Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J 25: 4888–4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR (2005) Force production by disassembling microtubules. Nature 438: 384–388 [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, Spiridonov IS, Volkov VA, Efremov A, Westermann S, Drubin D, Barnes G, Ataullakhanov FI, McIntosh JR (2008b) Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc Natl Acad Sci USA 105: 6918–6923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissom PM, Fiedler T, Grishchuk EL, Nicastro D, West RR, McIntosh JR (2009) Kinesin-8 from fission yeast: a heterodimeric, plus-end-directed motor that can couple microtubule depolymerization to cargo movement. Mol Biol Cell 20: 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes G, Dong Y, Mcewen B, Deluca J (2008) Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol 18: 1778–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A, Sillje HH, Nigg EA (2006) Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J 25: 5504–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM (2003) The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol 161: 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M (2004) Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729 [DOI] [PubMed] [Google Scholar]
- Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S (2008) Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol 180: 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TL (1985) Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci USA 82: 4404–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, Cheeseman IM, Fukagawa T (2008a) CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135: 1039–1052 [DOI] [PubMed] [Google Scholar]
- Hori T, Okada M, Maenaka K, Fukagawa T (2008b) CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol Biol Cell 19: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Feng J, Famulski J, Rattner JB, Liu ST, Kao GD, Muschel R, Chan GK, Yen TJ (2007) Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J Cell Biol 177: 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland KM, Kingsbury J, Koshland D, Hieter P (1999) Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J Cell Biol 145: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Middleton K, Centola M, Mitchison TJ, Carbon J (1992) Microtubule-motor activity of a yeast centromere-binding protein complex. Nature 359: 533–536 [DOI] [PubMed] [Google Scholar]
- Izuta H, Ikeno M, Suzuki N, Tomonaga T, Nozaki N, Obuse C, Kisu Y, Goshima N, Nomura F, Nomura N, Yoda K (2006) Comprehensive analysis of the ICEN (interphase centromere complex) components enriched in the CENP-A chromatin of human cells. Genes Cells 11: 673–684 [DOI] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW (2007) Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon E, Bloom K (2008) Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol 181: 587–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bloom K, Salmon ED (2009) In Vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol 19: 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED (2006) Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol 8: 581–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Hunt AJ (2002) A simple, mechanistic model for directional instability during mitotic chromosome movements. Biophys J 83: 42–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio MJ, McCleland ML, Stukenberg PT, Gorbsky GJ (2002) Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr Biol 12: 900–905 [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A (2006) Chromosomes can congress to the metaphase plate before biorientation. Science 311: 388–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith KC, Fitzgerald-Hayes M (2000) CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere DNA around a cse4p variant nucleosome. Genetics 156: 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A, Funabiki H (2009) Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr Opin Cell Biol 21: 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerres A, Vietmeier-Decker C, Ortiz J, Karig I, Beuter C, Hegemann J, Lechner J, Fleig U (2004) The fission yeast kinetochore component Spc7 associates with the EB1 family member Mal3 and is required for kinetochore-spindle association. Mol Biol Cell 15: 5255–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Copenagle L, Gordon MB, Compton DA, Kapoor TM (2003) Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J Cell Biol 160: 671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury J, Koshland D (1991) Centromere-dependent binding of yeast minichromosomes to microtubules in vitro. Cell 66: 483–495 [DOI] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T (1986) Beyond self-assembly: from microtubules to morphogenesis. Cell 45: 329–342 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T, Obuse C, Yanagida M (2007) Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell 13: 663–676 [DOI] [PubMed] [Google Scholar]
- Kline S (2006) The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol 173: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton AL, Lan W, Stukenberg PT (2006) Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol 16: 1705–1710 [DOI] [PubMed] [Google Scholar]
- Knowlton AL, Vorozhko VV, Lan W, Gorbsky GJ, Stukenberg PT (2009) ICIS and Aurora B Coregulate the microtubule depolymerase Kif2a. Curr Biol 19: 758–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland DE, Mitchison TJ, Kirschner MW (1988) Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature 331: 499–504 [DOI] [PubMed] [Google Scholar]
- Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM (2004) Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol 6: 232–237 [DOI] [PubMed] [Google Scholar]
- Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, Hunt DF, Walczak CE, Stukenberg PT (2004) Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol 14: 273–286 [DOI] [PubMed] [Google Scholar]
- Lechner J, Carbon J (1991) A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64: 717–725 [DOI] [PubMed] [Google Scholar]
- Li X, Nicklas RB (1995) Mitotic forces control a cell-cycle checkpoint. Nature 373: 630–632 [DOI] [PubMed] [Google Scholar]
- Liu D, Vader G, Vromans M, Lampson M, Lens S (2009) Sensing chromosome bi-orientation by spatial separation of Aurora B kinase from kinetochore substrates. Science 323: 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ST, Hittle JC, Jablonski SA, Campbell MS, Yoda K, Yen TJ (2003) Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat Cell Biol 5: 341–345 [DOI] [PubMed] [Google Scholar]
- Liu ST, Rattner JB, Jablonski SA, Yen TJ (2006) Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol 175: 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, McLeod I, Anderson S, Yates JR III, He X (2005) Molecular analysis of kinetochore architecture in fission yeast. EMBO J 24: 2919–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo VA, Stewart RJ, McIntosh JR (1995) Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature 373: 161–164 [DOI] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A (2007) Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol 176: 757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, DeLuca J, Salmon ED, Earnshaw WC (2004) The dynamic kinetochore-microtubule interface. J Cell Sci 117: 5461–5477 [DOI] [PubMed] [Google Scholar]
- Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J (2007) Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol Cell Proteomics 6: 2200–2211 [DOI] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E, Milligan RA (1991) Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J Cell Biol 114: 977–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ, Salmon ED (2009) Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol 184: 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis RL, Wilson L (1981) Microtubule treadmills—possible molecular machinery. Nature 293: 705–711 [DOI] [PubMed] [Google Scholar]
- Marshall O, Marshall A, Choo K (2008a) Three-dimensional localization of CENP-A suggests a complex higher order structure of centromeric chromatin. J Cell Biol 183: 1193–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KH (2008b) Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 82: 261–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T (1989) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 109: 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh AD, Tytell JD, Sorger PK (2003) Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol 19: 519–539 [DOI] [PubMed] [Google Scholar]
- McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT (2003) The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev 17: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland SE, Borusu S, Amaro AC, Winter JR, Belwal M, McAinsh AD, Meraldi P (2007) The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. EMBO J 26: 5033–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Hsieh CE, Mattheyses AL, Rieder CL (1998) A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma 107: 366–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR (1991) Structural and mechanical control of mitotic progression. Cold Spring Harb Symp Quant Biol 56: 613–619 [DOI] [PubMed] [Google Scholar]
- McIntosh JR (2005) Rings around kinetochore microtubules in yeast. Nat Struct Mol Biol 12: 210–212 [DOI] [PubMed] [Google Scholar]
- McIntosh JR, Grishchuk EL, Morphew M, Efremov AK, Zhudenkov K, Volkov VA, Cheeseman I, Desai A, Mastronarde D, Ataullakhanov FI (2008) Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell 135: 322–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone BG, Allshire RC (2003) Stretching it: putting the CEN(P-A) in centromere. Curr Opin Genet Dev 13: 191–198 [DOI] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM (1998) Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94: 607–613 [DOI] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, Sorger PK (2004) Timing and checkpoints in the regulation of mitotic progression. Dev Cell 7: 45–60 [DOI] [PubMed] [Google Scholar]
- Meraldi P, McAinsh AD, Rheinbay E, Sorger PK (2006) Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol 7: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami Y, Hori T, Kimura H, Fukagawa T (2005) The functional region of CENP-H interacts with the Nuf2 complex that localizes to centromere during mitosis. Mol Cell Biol 25: 1958–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Johnson M, Stukenberg P (2008) Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80Hec1. Curr Biol 18: 1785–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoshima Y, Hori T, Okada M, Kimura H, Haraguchi T, Hiraoka Y, Bao YC, Kawashima T, Kitamura T, Fukagawa T (2005) The constitutive centromere component CENP-50 is required for recovery from spindle damage. Mol Cell Biol 25: 10315–10328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda JJ, De Wulf P, Sorger PK, Harrison SC (2005) The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol 12: 138–143 [DOI] [PubMed] [Google Scholar]
- Mitchison T, Evans L, Schulze E, Kirschner M (1986) Sites of microtubule assembly and disassembly in the mitotic spindle. Cell 45: 515–527 [DOI] [PubMed] [Google Scholar]
- Mitchison TJ (1989) Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J Cell Biol 109: 637–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Salmon ED (1992) Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J Cell Biol 119: 569–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C (2007) Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 129: 1153–1164 [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Tyers RG, Wong CC, Yates JR III, Drubin DG, Barnes G (2009) Nbl1p: a Borealin/Dasra/CSC-1-like protein essential for Aurora/Ipl1 complex function and integrity in Saccharomyces cerevisiae. Mol Biol Cell 20: 1772–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov VS, Smith MA, Peak-Chew S, Kilmartin JV (2003) Interactions between centromere complexes in Saccharomyces cerevisiae. Mol Biol Cell 14: 4931–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB, Koch CA (1969) Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J Cell Biol 43: 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihashi A, Haraguchi T, Hiraoka Y, Ikemura T, Regnier V, Dodson H, Earnshaw WC, Fukagawa T (2002) CENP-I is essential for centromere function in vertebrate cells. Dev Cell 2: 463–476 [DOI] [PubMed] [Google Scholar]
- O'Connell CB, Khodjakov AL (2007) Cooperative mechanisms of mitotic spindle formation. J Cell Sci 120: 1717–1722 [DOI] [PubMed] [Google Scholar]
- O'Connell CB, Loncarek J, Hergert P, Kourtidis A, Conklin DS, Khodjakov A (2008) The spindle assembly checkpoint is satisfied in the absence of interkinetochore tension during mitosis with unreplicated genomes. J Cell Biol 183: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole ET, Winey M, McIntosh JR (1999) High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell 10: 2017–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C, Iwasaki O, Kiyomitsu T, Goshima G, Toyoda Y, Yanagida M (2004) A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat Cell Biol 6: 1135–1141 [DOI] [PubMed] [Google Scholar]
- Ohi R, Coughlin ML, Lane WS, Mitchison TJ (2003) An inner centromere protein that stimulates the microtubule depolymerizing activity of a KinI kinesin. Dev Cell 5: 309–321 [DOI] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, Desai A, Fukagawa T (2006) The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol 8: 446–457 [DOI] [PubMed] [Google Scholar]
- Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H (2007) CENP-B controls centromere formation depending on the chromatin context. Cell 131: 1287–1300 [DOI] [PubMed] [Google Scholar]
- Ortiz J, Stemmann O, Rank S, Lechner J (1999) A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev 13: 1140–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM (2006) The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol 7: 644–656 [DOI] [PubMed] [Google Scholar]
- Petersen J, Hagan IM (2003) S. pombe aurora kinase/survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr Biol 13: 590–597 [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, Allshire RC (2009) Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell 33: 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Richardson W, Allshire RC (2003) Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J Cell Biol 161: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Biggins S (2005) The spindle checkpoint: tension versus attachment. Trends Cell Biol 15: 486–493 [DOI] [PubMed] [Google Scholar]
- Pinsky BA, Kung C, Shokat KM, Biggins S (2006) The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol 8: 78–83 [DOI] [PubMed] [Google Scholar]
- Pinsky BA, Tatsutani SY, Collins KA, Biggins S (2003) An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev Cell 5: 735–745 [DOI] [PubMed] [Google Scholar]
- Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL (2009) The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell 136: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka MR, Zhang W, Costa P, Archambault V, D'Avino PP, Lilley KS, Laue ED, McAinsh AD, Glover DM (2007) Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS ONE 2: e478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, Tanenbaum ME, Maia AF, Medema RH (2009) RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J Cell Sci (in press) [DOI] [PubMed] [Google Scholar]
- Rieder CL (1981) The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma 84: 145–158 [DOI] [PubMed] [Google Scholar]
- Rieder CL, Palazzo RE (1992) Colcemid and the mitotic cycle. J Cell Sci 102(Part 3): 387–392 [DOI] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED (1998) The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol 8: 310–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC (2007) Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol 8: 798–812 [DOI] [PubMed] [Google Scholar]
- Saitoh S, Ishii K, Kobayashi Y, Takahashi K (2005) Spindle checkpoint signaling requires the mis6 kinetochore subcomplex, which interacts with mad2 and mitotic spindles. Mol Biol Cell 16: 3666–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, Franco-Sanchez A, Fleig U, Toda T, Millar JB (2005) The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J 24: 2931–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandall S, Severin F, McLeod IX, Yates JR III, Oegema K, Hyman A, Desai A (2006) A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell 127: 1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm RB, Heeger S, Althoff F, Walter A, Heidmann S, Mechtler K, Lehner CF (2007) Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma 116: 385–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S (2007) Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol 17: 237–243 [DOI] [PubMed] [Google Scholar]
- Severin FF, Sorger PK, Hyman AA (1997) Kinetochores distinguish GTP from GDP forms of the microtubule lattice. Nature 388: 888–891 [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Skeen VP, Salmon ED (1993) Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol 122: 859–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JA, McIntosh JR (1975) Initiation and growth of microtubules from mitotic centers in lysed mammalian cells. J Cell Biol 67: 744–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Severin FF, Hyman AA (1994) Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J Cell Biol 127: 995–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M (2000) Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288: 2215–2219 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kitamura E, Kitamura Y, Tanaka TU (2007) Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol 178: 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Desai A (2008) Kinetochore-microtubule interactions: the means to the end. Curr Opin Cell Biol 20: 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Gálová M, Schiebel E, Stark MJ, Nasmyth K (2002) Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108: 317–329 [DOI] [PubMed] [Google Scholar]
- Telzer BR, Moses MJ, Rosenbaum JL (1975) Assembly of microtubules onto kinetochores of isolated mitotic chromosomes of HeLa cells. Proc Natl Acad Sci USA 72: 4023–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis M, Slabicki M, Junqueira M, Paszkowski-Rogacz M, Sontheimer J, Kittler R, Heninger AK, Glatter T, Kruusmaa K, Poser I, Hyman AA, Pisabarro MT, Gstaiger M, Aebersold R, Shevchenko A, Buchholz F (2009) Comparative profiling identifies C13orf3 as a component of the Ska complex required for mammalian cell division. EMBO J 28: 1453–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torras-Llort M, Moreno-Moreno O, Azorín F (2009) Focus on the center: the role of chromatin on the regulation of centromere identity and function. EMBO J, (e-pub ahead of print 23 July 2009; doi:10.1038/emboj.2009.174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trazzi S, Bernardoni R, Diolaiti D, Politi V, Earnshaw WC, Perini G, Della Valle G (2002) In vivo functional dissection of human inner kinetochore protein CENP-C. J Struct Biol 140: 39–48 [DOI] [PubMed] [Google Scholar]
- Tulu US, Fagerstrom C, Ferenz NP, Wadsworth P (2006) Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr Biol 16: 536–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida KS, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T (2009) Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol 184: 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G, Medema RH, Lens SM (2006) The chromosomal passenger complex: guiding Aurora-B through mitosis. J Cell Biol 173: 833–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorozhko VV, Emanuele MJ, Kallio MJ, Stukenberg PT, Gorbsky GJ (2008) Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin. Chromosoma 117: 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos L, Famulski JK, Chan GK (2006) How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly. Biochem Cell Biol 84: 619–639 [DOI] [PubMed] [Google Scholar]
- Walczak CE, Heald R (2008) Mechanisms of mitotic spindle assembly and function. Int Rev Cytol 265: 111–158 [DOI] [PubMed] [Google Scholar]
- Wan X, O'Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, Stukenberg PT, Desai A, Salmon ED (2009) Protein architecture of the human kinetochore microtubule attachment site. Cell 137: 672–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E (2008) Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol 383: 894–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Skibbens RV, Salmon ED (1996) Oscillating mitotic newt lung cell kinetochores are, on average, under tension and rarely push. J Cell Sci 109 (Part 12): 2823–2831 [DOI] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC (2007) The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol 14: 54–59 [DOI] [PubMed] [Google Scholar]
- Wei RR, Sorger PK, Harrison SC (2005) Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA 102: 5363–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J, Cheeseman I (2008) Toward a molecular structure of the eukaryotic kinetochore. Dev Cell 15: 645–655 [DOI] [PubMed] [Google Scholar]
- Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR III, Cheeseman IM (2009) The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell 16: 374–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G (2005) Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol Cell 17: 277–290 [DOI] [PubMed] [Google Scholar]
- Westermann S, Cheeseman IM, Anderson S, Yates JR, Drubin DG, Barnes G (2003) Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol 163: 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Drubin DG, Barnes G (2007) Structures and functions of yeast kinetochore complexes. Annu Rev Biochem 76: 563–591 [DOI] [PubMed] [Google Scholar]
- Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G (2006) The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature 440: 565–569 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kilmartin JV (2001) The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol 152: 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P (2009) Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell 33: 287–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA (2008) Orientation and structure of the Ndc80 complex on the microtubule lattice. J Cell Biol 182: 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt PL, Ris H, Borisy GG (1980) Origin of kinetochore microtubules in Chinese hamster ovary cells. Chromosoma 81: 483–505 [DOI] [PubMed] [Google Scholar]
- Wittmann T, Hyman A, Desai A (2001) The spindle: a dynamic assembly of microtubules and motors. Nat Cell Biol 3: E28–E34 [DOI] [PubMed] [Google Scholar]
- Yamamoto TG, Watanabe S, Essex A, Kitagawa R (2008) SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. J Cell Biol 183: 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Borisy GG (1995) Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol 131: 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lan W, Ems-McClung SC, Stukenberg PT, Walczak CE (2007) Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol Biol Cell 18: 3264–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski RP, Meyne J, Brinkley BR (1991) The centromere-kinetochore complex: a repeat subunit model. J Cell Biol 113: 1091–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table I