Abstract
The conventional DNA polymerase machinery is unable to fully replicate the ends of linear chromosomes. To surmount this problem, nearly all eukaryotes use the telomerase enzyme, a specialized reverse transcriptase that utizes its own RNA template to add short TG-rich repeats to chromosome ends, thus reversing their gradual erosion occurring at each round of replication. This unique, non-DNA templated mode of telomere replication requires a regulatory mechanism to ensure that telomerase acts at telomeres whose TG tracts are too short, but not at those with long tracts, thus maintaining the protective TG repeat ‘cap' at an appropriate average length. The prevailing notion in the field is that telomere length regulation is brought about through a negative feedback mechanism that ‘counts' TG repeat-bound protein complexes to generate a signal that regulates telomerase action. This review summarizes experiments leading up to this model and then focuses on more recent experiments, primarily from yeast, that begin to suggest how this ‘counting' mechanism might work. The emerging picture is that of a complex interplay between the conventional DNA replication machinery, DNA damage response factors, and a specialized set of proteins that help to recruit and regulate the telomerase enzyme.
Keywords: DNA damage response, genome stability, length regulation, telomerase, telomere
Introduction
Long before the basic features of DNA structure and replication were known, the ends of linear eukaryotic chromosomes, called telomeres (literally ‘end parts'), were recognized as possessing special properties (Blackburn, 2006). Pioneering classical genetic studies by Mueller, using the fruit fly Drosophila, and McClintock, who studied maize (Zea mays), showed that native chromosome ends, unlike those arising from breakage at internal chromosome regions, are protected from joining reactions, either with other chromosome ends or with accidental internal breaks. In the absence of this protective function, chromosome ends would be indistinguishable from accidental DNA double-strand breaks (DSBs), with disastrous consequences for chromosome stability. This property of native chromosome ends, often referred to as telomere ‘capping', is still recognized as central to chromosome function, and its molecular basis continues to be the subject of intensive study (see accompanying review; Lydall, 2009).
After the discovery of the double-helical structure of DNA, and the fact that its 5′ to 3′ replication by DNA polymerases requires a short RNA primer for initiation, a second problem posed by chromosome ends became apparent. Because of the absence of an upstream replication fork on the strand constituting the 5′ end of a chromosome, which is generated by lagging-strand synthesis, a single-strand gap will result from the removal of the 5′-most RNA primer that initiated DNA synthesis. This strand-specific loss of information would result in chromosome shortening with each successive cell division, in the absence of some mechanism to restore the lost sequence. Paradoxically, when later discoveries showed that native telomeres, instead of terminating in a blunt end, actually display resection of the 5′ end, and thus a 3′ single-stranded overhang, the problem of sequence loss at telomeres during replication focused instead on the 3′ end, which is produced by leading-strand synthesis (Lingner et al, 1995). The 3′ single-stranded telomere overhang is now recognized as a central feature of the telomere replication mechanism (and also of telomere ‘capping'), as will be discussed below.
Several models proposed in the 1970s suggested that the end-priming conundrum might simply be bypassed by the presence of palindromic hairpin structures at chromosome ends. However, the discovery in the ciliate Tetrahymena that telomeric DNA sequences are actually comprised of simple tandem repeats of a short DNA sequence (Blackburn and Gall, 1978), and the demonstration that terminal hairpins are not sufficient to provide telomere function (Szostak and Blackburn, 1982), indicated that evolution had found a different solution to the ‘end-replication problem'. A landmark study by Szostak and Blackburn (1982) showed that this solution was evolutionarily conserved, by revealing that Tetrahymena telomeric repeat sequences could ‘seed' the formation of functional telomeres in the budding yeast Saccharomyces cerevisiae, thus pointing to a completely novel mechanism for telomere replication and setting the stage for the discovery, a few years later, of the telomerase enzyme (Greider and Blackburn, 1985). Telomerase is a specialized reverse transcriptase that uses a dedicated RNA molecule (Greider and Blackburn, 1987), an integral part of the holoenzyme, as a template for the addition of simple TG-rich repeats to the 3′ ends of chromosomes. The use of the telomerase enzyme to solve the ‘end-replication problem' is remarkably widespread amongst eukaryotes. Oddly enough, though, in Drosophila melanogaster, in which early studies were among the first to reveal the unique properties of telomeres, a retrotransposon-based mechanism is used to replenish the chromosome ends.
The identification of mutants defective in telomerase components in yeast (Lundblad and Szostak, 1989; Singer and Gottschling, 1994; Lendvay et al, 1996), or knockout of the telomerase RNA gene in mouse (Blasco et al, 1997), proved that telomerase is indeed required to prevent the slow erosion of chromosome ends, and thus for cell and, ultimately, whole organism viability. Nevertheless, a small fraction of telomerase-minus yeast cells escape senescence and survive through the activation of recombination-based telomere maintenance mechanisms. A similar telomerase-independent survival mechanism, referred to as the ALT (alternative lengthening of telomeres) pathway, is also observed in a small percentage of human tumours (reviewed in Cesare and Reddel, 2008). For further discussion of recombination-based mechanisms for regulation of telomere stability and telomere repeat tract length, the reader is referred to recent reviews (Lustig, 2003; Bhattacharyya and Lustig, 2006).
Recognizing the telomere length regulation problem: a case of localized feedback?
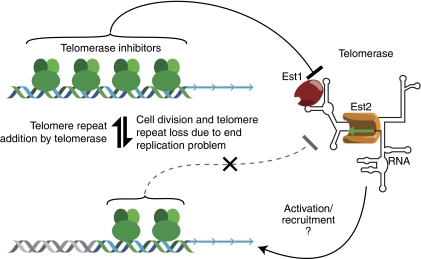
The short repeated sequences found at telomeres (irregular TG1−3 repeats in yeast, regular T2AG3 ones in vertebrates; generally referred to hereafter as TG repeats) vary considerably in length between organisms (∼300 bp in yeast, 5–10 kb in humans) but are centred about a fixed average length characteristic of a given species. This implies that the telomerase enzyme is instructed in some way as to when it is appropriate to add TG repeats to a given chromosome end. Formally, one could imagine an alternative hypothesis in which exonucleolytic processing of telomeres would be activated in proportion to TG tract length. However, subsequent experiments (Marcand et al, 1999) would clearly show that the telomerase pathway, and not the end attrition that occurs in its absence, is regulated according to TG tract length.
Murray et al (1988) recognized this problem early on, and, based on their studies of de novo telomere formation in yeast, they proposed that ‘(t)he constant average length of yeast telomeres implies a feedback mechanism that senses the length of telomeric DNA and reduces the extent of non-template-directed DNA synthesis when the telomeric DNA exceeds a certain length.' This proposal of a feedback mechanism regulating telomerase that can sense the length of telomeric DNA eventually gained experimental support. In budding yeast, the duplex portion of the irregular TG1−3 repeats at telomeres is bound directly by a tandem Myb domain containing protein called Rap1 (Shore, 1994). In the course of studying the effect of Rap1 on telomeric gene silencing, Marcand et al (1997) noticed that tethering of hybrid proteins containing the Rap1 C-terminus adjacent to a single-telomeric TG tract would reduce the length of that telomere, in a manner roughly proportional to the number of targeted Rap1 C-termini, without affecting other telomeres in the same cell. They interpreted this result in terms of a negative feedback mechanism for telomerase regulation involving the Rap1 C-terminus (see Figure 1). At the same time, studies from the de Lange laboratory led to a similar proposal, namely that in human cells the telomere repeat binding protein TRF1 controls telomere length in cis by inhibiting telomerase action at individual telomeres (van Steensel and de Lange, 1997). Thus, overexpression of TRF1, which might be expected to increase its binding at telomeres, leads to telomere shortening, whereas expression of a dominant-negative allele that interferes with DNA binding leads to telomere elongation.
Figure 1.
The ‘protein-counting' model for telomerase regulation (see text for details). Telomeric repeat DNA is indicated by the blue (TG-rich 3′ end strand) and green (CA-rich 5′ end strand) ribbons, terminating in a TG-rich 3′ overhang whose length varies according to organism and cell-cycle position. Proteins bound directly and/or indirectly to the duplex TG repeat tract generate an inhibitory signal that blocks telomerase action. (Telomerase in budding yeast consists of the catalytic subunit, Est2, the template RNA, TLC1, and essential regulatory subunits Est1, which interacts with TLC1, and Est3 (not indicated). In the absence of telomerase-based telomere extension, incomplete conventional replication leads to repeat loss and a gradual reduction in telomerase inhibition. Subsequent telomerase action returns the system to a more inhibitory state. At equilibrium in S. cerevisiae, the duplex TG repeat tract is ∼250–350 bp in length and is thought to bind directly ∼15–20 Rap1 molecules.
Even before the emergence of this feedback (‘counting') model for telomere length regulation, the ability to carry out sophisticated genetic analysis in yeast had already provided a clear foundation for the model as well as some hints regarding underlying mechanisms. Indeed, genetic experiments had already suggested that Rap1 might be a negative regulator of telomere elongation: point mutations in the Rap1 C-terminus had been shown to lead to telomere elongation and to a new equilibrium set point (Sussel and Shore, 1991), and complete deletion of a C-terminal domain of the protein was found to cause severe telomere elongation and an absence of any apparent regulation (Kyrion et al, 1992). These studies had thus already shown that Rap1 might have a key function in the proposed counting mechanism leading to feedback regulation of telomerase action. However, other work suggested that Rap1 has a more complex function in telomere regulation. Rap1 is encoded by an essential gene in yeast, probably due to its involvement in gene activation in other contexts (Shore, 1994), and the first temperature-sensitive (ts) lethal mutations in RAP1 displayed a telomere shortening phenotype at semi-permissive temperatures (Lustig et al, 1990).
Further study of the Rap1 C-terminus revealed two additional proteins with direct roles in telomere length regulation. The first hint that the Rap1 C-terminus might interact with other proteins to negatively regulate telomerase action came from experiments showing that overexpression of this domain, in the absence of the centrally located DNA-binding domain, causes telomere elongation (Conrad et al, 1990). These experiments were interpreted to mean that Rap1 interacts with one (or more) negative regulators of telomere elongation, present in limiting amounts in the cell, which can be titrated away from telomeres by overexpression of the Rap1 C-terminal domain. The identity of two such negative regulators, Rif1 (Rap1-interacting factor 1) (Hardy et al, 1992) and Rif2 (Wotton and Shore, 1997), soon came to light through two-hybrid screens carried out with the Rap1 C-terminus as bait. Deletion of either RIF1 or RIF2 causes telomere elongation, ∼600 bp in the case of rif1-Δ and less (∼150 bp) for rif2-Δ. Thus, both single mutants would seem to retain some form of feedback regulation on telomerase action, albeit with an alteration in the TG tract length set point at equilibrium. Significantly, though, cells in which both RIF1 and RIF2 are deleted display extremely elongated telomeres (Wotton and Shore, 1997), with no apparent equilibrium length, similar to that observed in RAP1 C-terminus deletion mutants. These findings suggested that Rif1 and Rif2 might operate through different mechanisms, which together are essential for feedback regulation on telomerase. Indeed, tethering experiments carried out with Rif1 and Rif2, similar to those described above for the Rap1 C-terminus, have shown that Rap1 counting is in fact Rif1 and Rif2 counting (Levy and Blackburn, 2004).
Roles for DNA replication and end processing
At about the same time that the evidence for a Rap1-based feedback mechanism of telomere length regulation was accumulating, other studies in yeast were adding a somewhat bewildering list of additional proteins to the picture. The first mutants to be identified with altered telomere length regulation would turn out to highlight the important role of both conventional DNA replication and the DNA damage checkpoint in this process. Lustig and Petes, who used Southern blotting to screen for mutations with altered telomere length, identified the aptly named TEL1 gene (Lustig and Petes, 1986). It took several years to clone the gene (which is toxic in Escherichia coli) and discover it to be the yeast orthologue of mammalian ATM (ataxia telangesia mutated), a PI3-kinase-like protein with a key role in DNA damage repair (Greenwell et al, 1995). This turned out to be just the first of a sudden flurry of reports connecting DNA repair and DNA damage checkpoint proteins to telomere length regulation, as it soon became clear that mutations in genes encoding the yeast Ku heterodimer (a DNA end-binding protein involved in non-homologous end joining, NHEJ), as well as the three components of the MRX (Mre11, Rad50, Xrs2) complex, also required for NHEJ, all display extremely short telomeres, similar to those of tel1 mutants (Boulton and Jackson, 1996; Gravel et al, 1998; Laroche et al, 1998; Nugent et al, 1998; Polotnianka et al, 1998; Mishra and Shore, 1999). These studies, in addition to indicating the complex nature of telomere length regulation, also raised a striking paradox, namely the direct involvement of DNA repair proteins at telomeres, sites in which DNA end joining must be strictly repressed to avoid generation of di-centric chromosomes. The molecular solution to this puzzle is still the subject of active investigation (see accompanying review; Lydall, 2009).
Even before the identification of TEL1, Hartwell's group had shown that ts mutations of CDC17, later found to encode DNA polymerase α (DNA Pol1), cause severe telomere elongation at semi-permissive temperatures (Carson and Hartwell, 1985). The DNA polymerase α/primase complex is responsible for synthesizing the RNA primer required for initiation of DNA synthesis, and this fact suggests that some aspect of lagging-strand synthesis at telomeres might have an important function in telomerase regulation. Subsequent studies showed that (ts) mutations in most other DNA replication-related genes do not cause telomere length changes, with the exception of CDC44 (RFC1), which encodes the large subunit of replication factor C, involved in the transfer of lagging-strand synthesis from DNA polymerase α/primase to DNA polymerase δ (Adams and Holm, 1996). The specificity of the DNA polymerase α/primase complex to telomere length regulation was more recently underscored by the identification of alleles of POL12, which encodes the essential B subunit of this complex, that also cause telomere elongation (Grossi et al, 2004).
Telomere replication in the context of the cell cycle
The significance of the largely genetic observations described above came into focus following a series of key findings that clarified some of the molecular events involved in telomere replication. Chief among these was the discovery by Wellinger, Zakian, and co-workers that TG-rich single-stranded DNA at telomeres increases in length as cells pass through S phase, possibly due to exonucleolytic resection of the telomeric 5′, CA-rich strand (Wellinger et al, 1993, 1996). The fact that single-stranded DNA is required for telomerase activity in vitro immediately suggested that this hypothesized processing reaction might be directly responsible for telomerase regulation. As will be discussed below, however, telomeric G-rich single-stranded DNA is also a binding site for the Cdc13–Stn1–Ten1 (CST) protein complex, which itself has an essential function in telomerase action. One attractive way to unify these two sets of observations is a model in which resection of telomere ends, carried out or regulated by Tel1 and the MRX complex, generates a binding site for the CST complex, which in turn recruits and regulates telomerase at chromosome ends (see below).
The results of Wellinger and co-workers established TG single-stranded overhang generation as a new step in telomere replication and begged the question of its relationship to conventional replication of chromosome ends. This issue was addressed directly by an elegant experiment in which a unique DNA replication origin was removed from a linear plasmid in yeast. Under these conditions, tail formation did not occur, suggesting that end processing is dependent on, and normally follows replication of telomeres by the conventional DNA polymerase machinery (Dionne and Wellinger, 1998). Similar experiments, using a site-specific recombination approach to remove (or not) a unique replication origin plus the internal portion of a telomeric TG tract (thus shortening the TG tract), showed that telomerase action, which occurs preferentially at a shortened TG tract (see below), is restricted to S phase and stimulated by DNA replication (Marcand et al, 2000). Consistent with this latter finding, Diede and Gottschling (1999) showed that resection and efficient generation of a telomere at an induced DSB flanked by a ‘seed' of TG repeat sequence requires both DNA polymerase α and DNA polymerase δ, thus tightly linking telomerase elongation of the TG-rich strand with replication of the CA-rich (lagging) strand. More recently, resection and elongation in de novo telomere formation has been shown to require cyclin-dependent kinase (CDK) activity (Frank et al, 2006), paralleling observations made at non-telomeric DSBs (Ira et al, 2004; Aylon and Kupiec, 2005). Taken together, these experiments support a scenario in which conventional DNA replication precedes and is required for an end-processing reaction that generates a TG-rich single-stranded overhang structure at telomeres. This processing reaction, mechanistically related to that which occurs at accidental DSBs as a prelude to homologous recombination, instead prepares the telomeric DNA substrate for subsequent telomerase action, which is itself strictly limited to the period in S phase after replication fork arrival (Raghuraman et al, 2001). Finally, the action of telomerase may be tightly coupled to ‘fill-in' synthesis of the CA-rich strand to complete a telomerase-based cycle of end elongation.
It is worth pointing out here that lagging-strand synthesis at the telomere will inevitably lead to a 3′ single-stranded overhang, due to RNA primer removal, whose length will presumably depend also on the precise initiation point of telomere-terminal lagging-strand synthesis. As such, it is still unclear whether lagging-strand telomere ends undergo a resection reaction similar to that of leading-strand ends. In human cells, in which it has been possible to physically distinguish leading- from lagging-strand ends, the evidence points to longer overhangs at lagging-strand ends (Chai et al, 2006). Nevertheless, it seems that the majority of ends terminate with a precise 3′ end sequence (Sfeir et al, 2005), suggesting that processing of both leading and lagging-strand ends share at least one common feature. In ciliates, the resection reaction, in addition to being much more constrained in magnitude, is very precisely regulated (Fan and Price, 1997; Jacob et al, 2001). Although we still lack information regarding the actual nucleases responsible for telomere end processing in any system, the MRX (MRN in mammalian cells) complex clearly has an important function that might explain why mutations in complex members lead to telomere shortening in yeast. Surprisingly, however, although the length of S phase overhangs is reduced in the absence of Mre11 (Larrivee et al, 2004), the exonuclease activity of Mre11 itself does not seem to be required for telomere length maintenance (Moreau et al, 1999; Lee et al, 2002). Perhaps multiple, partially redundant nuclease activities can carry out 5′ end resection at telomeres, as has recently been found to occur at DSBs (reviewed in Mimitou and Symington, 2009).
The CST complex and telomerase recruitment: final piece of the puzzle?
The pioneering genetic screens for telomere maintenance mutants carried out by Lundblad and co-workers (Lundblad and Szostak, 1989; Lendvay et al, 1996) revealed not only the three protein components of the telomerase holoenzyme (the catalytic subunit Est2 together with Est1 and Est3), but, in addition, an unusual (partial loss-of-function) allele of CDC13. Hartwell and co-workers originally identified CDC13 by way of a ts mutation (cdc13-1) that causes a G2/M cell-cycle arrest at the non-permissive temperature. They later showed that loss of Cdc13 function leads to massive, unregulated 5′ end resection of telomeres (Garvik et al, 1995), commonly referred to now as a telomere capping defect. The cdc13-2 allele, however, displays a very different behaviour, namely the ‘ever shorter telomere' (est) phenotype observed in telomerase mutants. In other words, cdc13-2 mutant protein seems to be proficient in telomere capping but unable to support telomerase action.
The characteristics of the cdc13-2 mutant, and the finding that Cdc13 protein binds specifically to TG-rich singled-stranded telomeric DNA in vitro and in vivo, immediately suggested that the protein might have a direct function at telomeres to either recruit or activate telomerase (or both). A series of remarkable hybrid-protein studies from Lundblad and co-workers, coupled with additional genetic analysis, lead to a more refined picture of the functional significance of the Cdc13–telomerase interaction. They began by showing that the expression of a Cdc13–Est1 fusion protein (Est1 interacts stably with the telomerase enzyme, Est2, and its RNA moiety, TLC1) leads to telomere elongation (Evans et al, 1999). Significantly, Cdc13–Est1 fusions containing either the cdc13-2 mutation or a telomerase-minus mutation in Est1 were perfectly capable of telomere maintenance. These results suggested that the defect conferred by cdc13-2 (or that of the est1-47 allele) related to telomerase recruitment or access to the telomere end, as it could be bypassed by covalently linking Cdc13 and Est1. Even more strikingly, they showed that the essential function of Est1 could be bypassed altogether by fusing Cdc13 directly to the telomerase catalytic subunit Est2. Together, these data supported a model in which a specific Cdc13–Est1 interaction serves to recruit telomerase to telomere ends. Further support for this notion came from the identification and characterization of a mutation in EST1 (est1-60) that suppresses the cdc13-2 Est phenotype (Pennock et al, 2001). Notably, cdc13-2 and est1-60 are reciprocal charge-swap mutations (Glu to Lys, and Lys to Glu, respectively), suggesting that they might define an important interaction site between these two proteins.
The ‘recruitment model' of Lundblad and co-workers found additional support from chromatin immunoprecipitation (ChIP) studies that began to assess the specific chromatin association of various telomeric proteins in live cells. Two studies showed that the cdc13-2 mutation abrogates Est2 binding either at a native telomere or at a TG-flanked DSB undergoing de novo telomere formation (Taggart et al, 2002; Bianchi et al, 2004), consistent with a defect in telomerase recruitment. The study examining binding at native telomeres (Taggart et al, 2002) showed that Cdc13, as well as both telomerase subunits examined (Est1 and Est2), display maximal association with telomeres during late S phase, consistent with the findings discussed above showing that telomerase action is restricted to this part of the cell cycle. This study also made the unexpected finding that Est2, but not Est1, is telomere associated in G1. Oddly, this association decreases as cells enter the cell cycle, only to peak again during late S phase. Subsequent work revealed that G1-specific Est2 association requires the Ku protein (Fisher et al, 2004), shown previously to associate with telomerase holoenzyme through an interaction with a stem-loop structure in the telomerase RNA, TLC1 (Peterson et al, 2001; Stellwagen et al, 2003). The significance of G1 binding of telomerase to telomeres, at a time when it does not carry out TG addition, is still unclear, but this pathway for telomerase recruitment seems not to be sufficient for telomere maintenance, although it does contribute to telomere elongation. In any event the Ku-dependent telomerase recruitment pathway contributes in part to efficient telomerase binding in S phase, because eliminating the Cdc13-dependent S phase-restricted pathway reduces telomerase binding by only about half, and complete elimination of binding requires inactivation of both pathways (Chan et al, 2008). Although a Cdc13-independent pathway for Est1 recruitment has been described that depends on the RPA (replication protein A) protein but does not affect Est2 telomere binding (Schramke et al, 2004), the above studies suggest that efficient telomerase recruitment in S phase correlates with Est1 binding at the telomere. Thus, it is tempting to speculate that the Est1-independent telomere association of telomerase might involve a form or location of the enzyme that is not permissive for telomere repeat addition. Conversely, Est1 may be responsible for delivering telomerase in a state with the potential for carrying out repeat synthesis, possibly due to positioning near the telomere terminus (Sabourin and Zakian, 2008). Interestingly, Est1 appears to be capable of binding to the telomere only when associated with telomerase (Chan et al, 2008).
One caveat associated with the ChIP assays used in these experiments is the uncertainty regarding precisely what the cross-linking reaction measures. For example, is the increase in Est1/Est2 binding observed in S phase the result of increased association with the telomere (recruitment), or instead an increase in the actual catalytic engagement of the telomerase enzyme (activation)? In the context of the telomere-healing assay, Cdc13-dependent cross-linking of telomerase was detected even when the enzyme itself was catalytically inactive, indicating that the ChIP assay, at least in this context, is measuring telomerase association with the telomere, rather than its catalytic action. Nevertheless, it is difficult to rule out the possibility that the Est1–Cdc13 interaction, in addition to promoting telomerase recruitment, may also facilitate telomerase action in some other manner. In this regard, it worth noting that a detailed genetic analysis of Est1 function uncovered evidence for additional roles of Est1 in the Ku-mediated pathway for telomerase action, as well as a possible telomerase activation function distinct from its recruitment interaction with Cdc13 (Evans and Lundblad, 2002). Furthermore, a function for Est1 in activating telomerase has been identified by in vitro experiments with Candida albicans proteins (Singh and Lue, 2003).
As noted above, characterization of the cdc13-2 allele, and of Cdc13 fusion proteins (Evans and Lundblad, 1999; Pennock et al, 2001; Bianchi et al, 2004), identified an amino-terminal domain of the protein implicated in telomerase recruitment through an interaction with Est1. Tseng et al (2006) identified several potential Tel1 and Mec1 (ATR orthologue) phosphorylation sites (SQ motifs) within the ‘recruitment domain' (RD) of Cdc13 and showed that these sites are indeed targets, in vitro, of both kinases. Significantly, mutation of two of these sites to alanine causes an est phenotype, suggesting that these sites together define a regulated surface on the Cdc13 RD necessary for telomerase action. The most straightforward conclusion from these studies is that Tel1 phosphorylation of these SQ sites on Cdc13 is necessary to activate the RD for Est1 binding, though this model still awaits a direct experimental test. Consistent with this idea, though, cells deleted for TEL1, or for MRE11, which in the context of MRX is required for Tel1 recruitment to DNA ends, display a severe defect in both Est1 and Est2 telomere association during late S/G2 phase, but not in Cdc13 binding (Goudsouzian et al, 2006). The RD domain of Cdc13 has recently been shown also to be the target of phosphorylation by Cdk1 (Cdc28) that is necessary for efficient Est1 recruitment (Li et al, 2009; Tseng et al, 2009). Thus, multiple phosphorylation events, by the Tel1 and Cdk1 kinases, might be required to achieve adequate levels of Est1 and telomerase recruitment, in this manner coupling telomerase action to cell-cycle stage and telomere length (see below).
The role of Cdc13 in the telomerase pathway is clearly not limited to its interaction with Est1. In fact, genetic analysis has unveiled an inhibitory role of Cdc13 through the identification of several CDC13 alleles with increased telomere length (Grandin et al, 2000; Chandra et al, 2001). These effects appear to be mediated through direct interaction of Cdc13 with the Stn1 and Ten1 proteins and formation of the CST complex. Interestingly, the three CST proteins, each of which contains one or more oligosaccharide-oligonucleotide-binding (OB) folds, bear a strong structural resemblance to the three components of the RPA complex, which is itself involved in numerous DNA metabolism reactions (Gao et al, 2007). CST thus appears to be an RPA-like complex with telomere-specific functions. Grandin and Charbonneau first identified STN1 as a partial suppressor of the capping defect of cdc13-1 and later identified TEN1 as a suppressor of a similar defect in STN1-mutated cells (Grandin et al, 1997, 2001). However, it is clear that the role of CST is not limited to telomere protection and that both Stn1 and Ten1, like Cdc13, have a role in mediating repression of telomerase activity, as partial loss-of-function alleles of all three proteins cause telomere elongation. These findings lead the Lundblad and Charbonneau laboratories to propose models in which CST regulates telomere length by limiting telomerase access to the ends (Grandin et al, 2000; Chandra et al, 2001). The Lundblad laboratory, in particular, proposed that Cdc13 first acts positively in the telomerase pathway (by recruiting Est1) and then switches to an inhibitory role through interaction with Stn1 (Chandra et al, 2001). The repressive effect of CST appears to operate through an interaction between the C-terminal domain of Stn1 and the Cdc13 protein (Chandra et al, 2001; Puglisi et al, 2008). Thus, even though the Cdc13–Est1 interaction promotes telomere elongation, CST as a whole can inhibit telomere elongation. The current model is that CST might do so by modulating the lagging-strand replication machinery responsible for synthesis of the CA-strand. Indeed, Cdc13 also interacts with the Pol1 (catalytic) subunit of DNA polymerase α/primase (Qi and Zakian, 2000), whereas Stn1 displays physical and functional association with the Pol1 regulatory subunit Pol12 (Grossi et al, 2004; Petreaca et al, 2007; Puglisi et al, 2008; Gasparyan et al, 2009). As point mutations in both Pol1 and Pol12 affecting their interaction with Cdc13 and Stn1, respectively, cause telomere elongation (Qi and Zakian, 2000; Grossi et al, 2004; Puglisi et al, 2008), the CST–DNA polymerase α/primase interaction is presumed to inhibit at least one step in the telomerase pathway, possibly by modulating the length of the TG overhang available for Cdc13–Est1–telomerase action. This idea is consistent with the view, first proposed in ciliates, that synthesis of the CA-strand by lagging-strand replication is inhibitory for TG-strand synthesis (Fan and Price, 1997). Very recently it has been proposed that the switch in Cdc13-binding partners from Est1 to Stn1 is modulated by a specific Cdk1-dependent phosphorylation event on the telomerase RD of Cdc13 (Li et al, 2009). In addition, an elegant biochemical study has just uncovered a role, at least in vitro, for the Hsp82 chaperone in facilitating a transition between Cdc13/telomerase extendable and CST non-extendable complexes (DeZwaan et al, 2009). This work points to a role for the Cdc13 C-terminus in repressing telomerase through assembly of the CST complex, in a manner not solely dependent on CA-strand synthesis, and in addition identifies a function for the Cdc13 N-terminus in activating telomerase action independently of a recruitment function, in agreement with earlier genetic studies (Meier et al, 2001).
Putting the pieces together: how does telomere tract length modulate telomerase action?
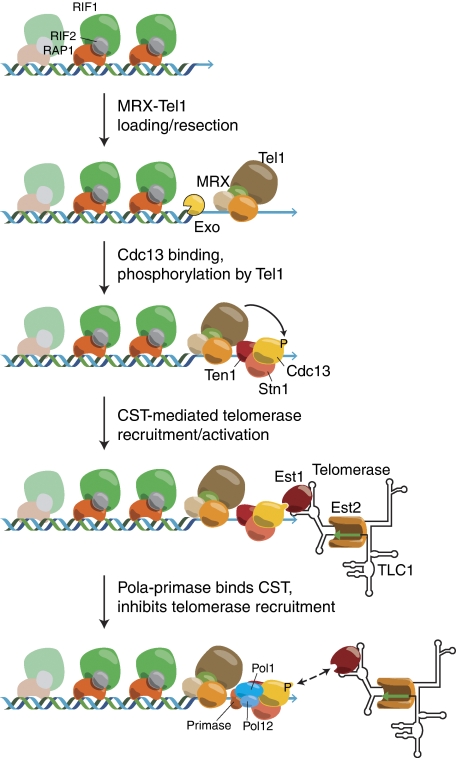
The constellation of data outlined above supports the following general working model of telomerase-dependent telomere elongation (Figure 2). First, replication fork passage sets the stage for a DNA end-processing reaction that leads to the generation (or extension) of a 3′ TG-rich single-stranded overhang on both leading- and lagging-strand ends. The Tel1 kinase and the MRX complex, which act in a common telomerase pathway for telomere maintenance (Ritchie and Petes, 2000), might promote this event, possibly through the recruitment and/or activation of multiple exonucleolytic activities. Genetic epistasis data suggest that some aspect of MRX/Tel1 function is directly inhibited in cis by the action of Rap1/Rif complexes bound to the duplex portion of the TG repeat tracts (Craven and Petes, 1999; Ray and Runge, 1999; Chan et al, 2001). Subsequent to end processing, the newly generated overhang promotes Cdc13 binding. (Whether this is a direct consequence of increased single-stranded binding sites for Cdc13 is still not clear (Tsukamoto et al, 2001).) Increased Cdc13 binding and/or a modification of telomere-bound Cdc13 (see below), in turn leads to telomerase holoenzyme recruitment, at least in part through the Cdc13–Est1 interaction, and subsequent TG-strand extension. Finally, a CST interaction with DNA polymerase α/primase complex leads to termination of telomerase action and promotes replication of the complementary CA-rich strand.
Figure 2.
Proposed steps in telomere replication in the budding yeast S. cerevisiae. After passage of the DNA replication fork, the MRX (Mre11, Rad50, Xrs2) complex and the PI3K-related kinase Tel1 are recruited to the telomere ends. (Differences in leading and lagging-strand ends are omitted for simplicity). The MRX complex, Tel1 and Cdk1 (not indicated) control the exonucleolytic resection of the telomeric (CA-rich) 5′ end through as yet uncharacterized exonucleases and mechanisms. The 3′ single-strand overhang thus generated (or elongated) serves as a platform for binding of the CST (Cdc13–Stn1–Ten1) complex. Tel1 phosphorylates Cdc13 at multiple sites (Cdc13 is also phosphorylated by Cdk1). Phosphorylated Cdc13 recruits telomerase through an interaction with the Est1. Finally, an interaction between DNA polymerase α/primase complex and CST inhibits telomerase recruitment and promotes completion of lagging-strand synthesis (see text for details).
How might this series of events be regulated by TG tract length such that cells maintain a constant average telomere length? In principle, telomere length homeostasis results from a balance between two opposing reactions, incomplete replication/nucleolytic degradation, and telomerase-mediated elongation, either or both of which could be regulated by TG tract length. This first question was resolved by experiments in which site-specific recombination was used to shorten a single-telomere TG tract in yeast (Marcand et al, 1999). Significantly, the shortened telomere initially elongates at a relatively rapid rate (∼15 bp per generation), which decreases gradually as the end approaches the equilibrium length. However, in the absence of telomerase activity, telomere length decreases at a constant rate, independent of TG tract length, of ∼3 bp per generation. Taken together, these data strongly support the idea that telomere length is regulated solely through a progressive inhibition on telomerase action, which increases in a roughly linear manner with increased TG tract length.
This key finding then poses the following question: does telomere tract length influence the extent of telomerase action at an end (processivity), or instead the probability that an end will be reacted on by telomerase in a given cell cycle? To address this question, Lingner and co-workers developed an ingenious assay to measure telomere elongation events in individual cells during a single cell cycle, taking advantage of yeast mating to supply telomerase enzyme to a telomerase-minus cell carrying a uniquely marked telomere. Their findings (Teixeira et al, 2004) showed clearly that not all telomeres are elongated in every cell cycle. Instead, telomere elongation occurs preferentially at ends with the shortest TG tracts. The extent of elongation, though variable, does not depend on TG tract length. By extending this assay to cells expressing two distinguishable telomerase RNA template alleles, Chang et al (2007) were able to show that telomerase can undergo multiple rounds of productive association/dissociation with a given telomere in a single cell cycle. Interestingly, they found that telomerase activity does become measurably processive at extremely short (<125 bp) telomeres, in a transition that depends on the Tel1 protein (Arneric and Lingner, 2007). Finally, this analysis demonstrated that deletion of either RIF1 or RIF2 increases the frequency of elongation events, but not their extent (Teixeira et al, 2004). These experiments clearly suggest that TG tract length controls a switch between extendible and non-extendible telomere states in cis.
As short telomeres are also preferentially elongated by telomerase in mammalian cells (for details see Bianchi and Shore, 2008), it is possible that a related mechanism (probably different in its molecular details) exists in higher eukaryotes. Interestingly, in human cells telomeres are continuously extended irrespective of their length unless the amount of telomerase enzyme is sufficiently low, suggesting that low telomerase levels are necessary to favor the preferred elongation of shorter telomeres (Cristofari and Lingner, 2006). Although the effect of telomerase cellular concentration on the switch from non-extendable to extendable state at yeast telomeres has not been studied in detail, recent work (Mozdy et al, 2008) has shown that TLC1 template RNA levels limit telomere length and that over twenty genes affect TLC1 levels and average telomere length when mutated. This and related effects may explain, at least in part, the striking observation that nearly 300 genes (∼5% of the total genome), when deleted, lead to either an increased or decreased telomere length set-point in yeast (Askree et al, 2004; Gatbonton et al, 2006).
Given the likely steps involved in the telomerase pathway in yeast outlined above (see Figure 2), at least three plausible molecular mechanisms can be proposed to explain the switch at telomeres from extendable to non-extendable. The first model proposes that TG tract length directly influences the extent of the 5′ end resection reaction, such that short TG tracts are more extensively resected than longer ones. This increased TG-rich single-stranded DNA would then bind a larger number of Cdc13 molecules, thus increasing the probability of telomerase recruitment at the short TG tract telomeres. Support for this model comes from experiments in which Cdc13 binding at a DSB flanked by either 80 or 250 bp of TG repeat sequence was measured by a quantitative ChIP assay and found to be considerably reduced at the longer end (Negrini et al, 2007). A second (‘activation') model proposes that telomere tract length influences a step that stimulates the action of telomere-bound telomerase, evidence for which, as discussed above, comes from analysis of Est1 and Cdc13 (Meier et al, 2001; Evans and Lundblad, 2002; Singh and Lue, 2003; DeZwaan et al, 2009). Finally, a third ('recruitment') model posits that TG tract length regulates telomerase association with the telomere end. These three models make different predictions regarding the relative amounts of Cdc13, Est1, and Est2 protein bound at short versus long telomeres as cells traverse the S phase of the cell cycle. According to the resection model, one would expect to detect increased binding of all three proteins at a short telomere. The activation model predicts that one would find equal amounts of the three proteins at short and long telomeres (but see below), whereas the recruitment model predicts that Est2 binding should be increased at the short versus long ends. To directly address this issue, two different groups have recently adapted the site-specific recombination system for telomere shortening developed by Marcand et al (1999) for quantitative analysis of protein association by ChIP (Sabourin et al, 2007; Bianchi and Shore, 2007b). Both groups measured increased Est1 and Est2 binding at the shortened telomere compared with the un-shortened control, but equal amounts of Cdc13 at the long and short ends, consistent with the recruitment model. In addition these results suggest that the interaction between Cdc13 and Est1 might be controlled by telomere length. As discussed above, though, these conclusions are based on the assumption that the measured increase in the ChIP signal for Est1 and Est2 reflects an increase in association with the telomeric target site, rather than increased catalytic activity of a change in the precise molecular configuration of binding. For example, telomerase actively engaged in nucleotide addition may cross-link to telomeric DNA with a different efficiency than an enzyme bound to the telomere but not in the act of synthesis. Or, in a different scenario, Est1 might relocate telomerase within the telomeric complex to a more easily cross-linkable or antibody-accessible position. Although at this point, then, it can conservatively be argued that TG tract length effects what might be loosely defined as telomerase activation, the simplest interpretations of these results indicates that telomerase recruitment to telomere ends in yeast is regulated by telomere length. Distinguishing conclusively between these different possibilities appears to be beyond the reach of present ChIP techniques, which cannot monitor conformational or positional changes for proteins at a given chromatin site and are therefore unsuitable for assessing activation models.
An additional important finding to emerge from these studies was the observation that Tel1 protein is significantly enriched at a shortened telomere (Sabourin et al, 2007; Bianchi and Shore, 2007b), a conclusion also supported by ChIP studies in cells lacking telomerase activity (Hector et al, 2007). As pointed out above, the Tel1 kinase is required for normal telomerase association with telomeres, is essential for telomerase action, together with the related Mec1 kinase (Ritchie et al, 1999), and may phosphorylate the Cdc13 RD in vivo to promote Est1 binding and thus telomerase recruitment (Tseng et al, 2006). This latter finding is particularly significant in light of the ChIP findings with Tel1, as it is very easy to imagine that increased Tel1 association with short telomeres will lead to increased Cdc13 phosphorylation, and in turn, increased telomerase recruitment. It should be noted that Cdk1-dependent phosphorylation of Cdc13 also promotes Est1 binding (Li et al, 2009). However, based on genetic data, this effect seems less important than that of Tel1, and it is unclear whether it is regulated by TG tract length.
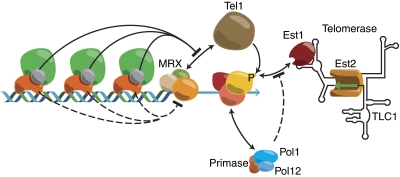
At this point, it seems necessary to understand why Tel1 recruitment (which, like at a DSB, depends on the C-terminus of Xrs2 (Sabourin et al, 2007)) is more efficient at short compared with longer TG tracts. Recent work sheds some light on this question. To begin with, Marcand and co-workers present evidence that Rif2 has a direct function in telomere capping, and suggest that this could result from a direct inhibition of MRX and/or Tel1 binding (Marcand et al, 2008). In a more recent report, Sugimoto and co-workers show, using a ‘telomere healing' (de novo formation) assay, that both Rif1 and Rif2 directly block Tel1 (but not MRX) association at longer TG tracts, with Rif2 showing a stronger effect (Hirano et al, 2009). Significantly, this study also provides biochemical evidence that Rif2 acts by competing with Tel1 for binding to a surface on the C-terminus of Xrs2. The molecular basis for Rif1 action remains unknown, and the authors are careful to point out that a Rap1-dependent but Rif-independent mechanism for telomere length control is likely to exist, consistent with earlier findings (Negrini et al, 2007; Marcand et al, 2008). A working model for TG tract length regulation of telomerase, based on the findings discussed above, is summarized in cartoon form in Figure 3.
Figure 3.
Summary of some of the proposed steps in TG tract length-dependent regulation of telomerase action. The Rap1–Rif complex generates a number-dependent negative regulatory signal that inhibits telomerase recruitment. Rif2 (grey sphere) competes with Tel1 for binding to the Xrs2 component of MRX, thus inhibiting Tel1 binding. Rap1 appears to block MRX–Tel1 binding through an uncharacterized mechanism. The molecular basis of Rif1 inhibition on telomerase action is unknown. Tel1 in turn phosphorylates Cdc13, creating a positive signal for telomerase recruitment through a direct interaction with its Est1 subunit. The DNA polymerase α/primase complex interacts with the CST complex, thus blocking telomerase recruitment. Regulation and timing of this step are not understood at present. Note that Est1, or other components of telomerase, may be activated through as yet unknown mechanisms, perhaps involving Tel1 phosphorylation (see text for discussion).
A direct effect of Tel1 on Cdc13, and in turn telomerase recruitment, appears not to be the only mechanism promoting elongation of short telomeres. In their Cre-LoxP-based telomere shortening experiments, Bianchi and Shore (2007a) made the unexpected finding that shortened telomeres, unlike normal-length controls, replicate early in S phase due to the earlier firing of subtelomeric DNA replication origins. They showed that this effect accelerates the elongation of a critically short telomere, perhaps providing an advantage to cells in the event of replication fork collapse at the telomere (Miller et al, 2006) or rapid telomere deletion (Li and Lustig, 1996). The mechanism by which telomere length feeds back to control nearby replication origins is not known at present, but seems unlikely to be fully explained by the effects of yeast Ku or SIR proteins on subtelomeric gene silencing and replication origin firing (Stevenson and Gottschling, 1999; Cosgrove et al, 2002). Intriguingly, a local effect of DSBs on the firing of nearby origins has recently been uncovered, which might be related to the phenomenon observed at shortened telomeres (Doksani et al, 2009). Although it is unclear why early replication accelerates telomere elongation, one possibility is that it increases the time that the telomere is available for telomerase binding, particularly during a period in early S phase when association of the inhibitor Rif1 is lowest (Bianchi and Shore, 2007a).
A number of loose ends
As suggested at various points above, several aspects of telomere replication, even in the well-studied yeast system, remain poorly understood. Thus, apart from the first hints of a mechanism by which Rif2 blocks Tel1 recruitment, we still lack a clear picture of how the Rap1–Rif ‘counting' factors directly act to decrease the probability of telomerase action. For example, it appears that MRX association at short telomeres is also increased relative to longer ones (Viscardi et al, 2007), yet the mechanism(s) leading to this difference are still unknown. Measurement of Mre11 binding adjacent to TG tracts of varying length suggests that long arrays of Rap1-binding sites exclude Mre11 through a Rif-independent mechanism either intrinsic to Rap1, or operating through an unidentified interacting partner (Negrini et al, 2007). This phenomenon may be related to the RAP1/TRF2-dependent inhibition of telomeric NHEJ observed in human cell extracts (Bae and Baumann, 2007). It is worth noting here that Levy and Blackburn showed that fusion of a heterologous oligomerization domain to Rap1 could bypass Rif function, at least to some extent, and restore telomere length regulation, suggesting that a tightly folded telomeric protein/DNA complex may be sufficient to impede MRX association and subsequent events in the telomerase pathway (Levy and Blackburn, 2004). Whether increased MRX association at short telomeres actually leads to their increased 5′ end resection is also not known. The fact that Cdc13 binding is not affected in telomere shortening experiments (Sabourin et al, 2007; Bianchi and Shore, 2007b) argues against increased resection, but an alternative hypothesis is that Cdc13 binding is controlled in some other manner by MRX (and/or Tel1), with increased telomeric single-stranded DNA being bound by RPA.
One intriguing feature of telomere replication emphasized throughout this review is the pervasive involvement of proteins that also have key functions in both the DNA damage checkpoint and DNA repair pathways provoked by DSBs (e.g. MRX complex, Tel1, and Ku proteins). The increased association of MRX and Tel1 at short telomeres, possibly to levels similar to those observed at DSBs, raises the question of whether short telomeres are in many ways recognized by the cell as DSBs. Do short telomeres activate a DNA checkpoint (G2/M arrest) response? Here the precise answer is still unclear. Viscardi et al (2007) report phosphorylation of the checkpoint kinase Rad53 (CHK2 in mammals) after telomere shortening, but neither this nor other reports (Sabourin et al, 2007; Bianchi and Shore, 2007b) indicate that this leads to any detectable cell-cycle arrest. Instead, one report (Michelson et al, 2005) suggests that an elongating telomere formed at a TG-flanked DSB actually exerts an ‘anti-checkpoint' effect on the non-TG-containing side of the break, though the origin of this checkpoint down-regulation has been questioned (Hirano and Sugimoto, 2007).
The inhibitory roles of the CST and DNA polymerase α/primase complexes, clearly linked through the Cdc13–Pol1 and Stn1–Pol12 interactions, are still not well understood, but may involve direct interference with the Cdc13–Est1 interaction (Chandra et al, 2001; Puglisi et al, 2008; Li et al, 2009). As neither Cdc13 (Sabourin et al, 2007; Bianchi and Shore, 2007b) nor Stn1 (Puglisi et al, 2008) binding are affected by TG tract length, a telomere-length dependent inhibitory effect exerted by Stn1 through its interaction with Cdc13 and Pol12 would seem likely to involve a length-dependent modification of the protein or its interacting partner(s), but this remains to be tested (Li et al, 2009). Alternatively, it is possible that the CST complex exerts a sort of ‘default' repressive action on telomerase activity that is not modulated by repeat array length. It also remains to be seen if and how the inhibitory effect of Stn1 on telomerase action is mechanistically linked to the completion of lagging-strand synthesis at telomeres. These open questions underscore the importance of understanding the precise temporal order of events associated with telomere replication, a question that should be accessible through careful ChIP analysis in synchronized cell cultures. One such very recent analysis (Moser et al, 2009) indicates that leading- and lagging-strand replication at telomeres in the fission yeast Schizosaccharomyces pombe are temporally uncoupled, with the latter considerably delayed. Studies in ciliates showing a physical association between telomerase and the lagging-strand machinery (Ray et al, 2002), and the appearance of Stn1 homologues in all eukaryotes examined (Gao et al, 2007; Martin et al, 2007), suggest that the yeast studies may be of much more general significance. Future work along these lines should lead to a more detailed understanding of how conventional and telomerase-based replication are coupled and how the interactions between the respective molecular machineries influence telomere length regulation.
Finally, recent work has unexpectedly revealed efficient recruitment of the telomerase machinery to non-telomeric DSBs (Oza et al, 2009). Although these breaks are at some frequency healed by telomerase-dependent de novo telomere formation (Myung et al, 2001b; Stellwagen et al, 2003), the amount of Cdc13 and Est2 present at these DSBs might seem very high compared with the efficiency at which these ends are healed by telomere addition. Together, these data imply that powerful mechanisms must exist to limit telomerase action at DSBs, which would in most cases be a very dangerous way to repair the DNA damage (Pennaneach et al, 2006). On the other hand, these mechanisms seem to be inactive or ineffective at DSBs containing even only short stretches of TG repeat sequence (Grossi et al, 2001; Hirano and Sugimoto, 2007; Negrini et al, 2007). Part of this inhibitory function appears to be carried out by Pif1, a helicase that can remove telomerase from its substrate both in vitro and in vivo (Boule et al, 2005), and is required to repress telomere formation at accidental DSBs (Myung et al, 2001a). However, regulation of telomerase at DSBs is likely to involve other factors, and elucidating how elements of the DNA damage response pathway influence the destiny of telomerase recruited at telomere ends versus DSBs remains an area of exciting challenges.
Telomere length regulation from fission yeast to humans
Although several components involved in budding yeast telomere replication are conserved, in one form or another, a brief examination of the related proteins in either fission yeast or mammals immediately reveals the remarkable evolutionary flexibility of telomere biology (Palm and de Lange, 2008). Telomere length regulation in these systems, a detailed discussion of which is beyond the scope of this review, is less well understood. We will, thus, limit ourselves to a short outline of some of the key features and open questions. A more in depth treatment of these topics can be found elsewhere (Gilson and Geli, 2007; Verdun and Karlseder, 2007; Bianchi and Shore, 2008; Palm and de Lange, 2008).
In fission yeast, the telomere duplex DNA-binding protein is called Taz1, a single-Myb domain containing protein similar to mammalian TRF1 and TRF2. Taz1 recruits both Rap1 and Rif1 orthologues to fission yeast telomeres, in which these proteins negatively regulate telomerase activity (reviewed in Rog and Cooper, 2008). In this system, Rif1 interacts with the telomere independently of Rap1, which instead recruits a novel telomeric protein called Poz1 (Miyoshi et al, 2008). Poz1 in turn interacts with Tpz1, the fission yeast orthologue of mammalian TPP1 (see below), which binds to both Pot1 (protection of telomeres 1; Baumann and Cech, 2001), a single-stranded DNA-binding OB-fold protein, and Ccq1, a protein involved in telomerase recruitment (Miyoshi et al, 2008; Tomita and Cooper, 2008). One current model for telomere length regulation in fission yeast (Miyoshi et al, 2008) posits a switch between telomerase inactive and active forms actuated by a transfer of the Poz1–Tpz1–Pot1–Ccq1 complex from Rap1 to the 3′ single-stranded overhang. Precisely how this switch would be regulated, presumably as a function of TG repeat tract length, is still unclear.
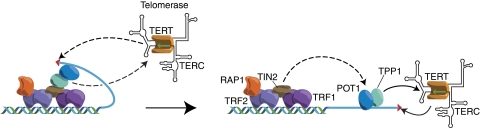
In mammals, telomeric duplex DNA is bound by two Myb domain protein homodimers, TRF1 and TRF2. In these systems, the Rap1 orthologue is recruited by a direct interaction with TRF2, whereas a novel protein, TIN2, interacts with both TRF1 and TRF2 and provides the key bridge to the TPP1–POT1 telomere end-binding complex. Together, this complex of six proteins is referred to as the ‘shelterin' complex, to denote its key role in both telomere protection and telomerase-based telomere maintenance (de Lange, 2005). It is unclear whether a telomere-length dependent switch in telomerase regulation similar to the one postulated above for fission yeast also operates in mammals, but this is of course a possibility given the similarities between the telomeric complexes in the two systems. Indeed, a mechanism to relay information regarding repeat-array length to the overhang that is the substrate for telomerase has been proposed in human cells, based on the dual ability of POT1 to indirectly interact with double-stranded repeat binding factors and also to directly bind the single-stranded overhang (Loayza and De Lange, 2003; Wang et al, 2007; Xin et al, 2007; see Figure 4). Consistent with these ideas, TPP1 interacts with telomerase and has been proposed to recruit the enzyme to chromosome ends (Xin et al, 2007). Again, the details of how this process might be regulated are still unknown, and the involvement of additional proteins is likely, perhaps including human Est1.
Figure 4.
Possible mechanisms of telomerase regulation by the shelterin complex in mammalian cells. The formation of a t-loop by the shelterin complex (not indicated here) is thought to sequester the telomere 3′ terminus from telomerase. Opening of the t-loop will generate a structure (left) that may still provide only limited access to telomerase, perhaps due to the TPP1–TIN2 interaction. In this case, a transition to a more open structure (right) might be required to allow efficient recruitment of telomerase by POT1–TPP1. See main text for more details and a description of related events thought to occur at telomeres in the fission yeast S. pombe.
Interestingly, the human MRX complex (MRN, where NBS1 is the human orthologue of Xrs2) and Ku86 both have roles in the telomerase pathway (O'Connor et al, 2004), just as in the yeast systems, but their mechanism of action might differ considerably in the mammalian system. MRN, which binds telomeres in late S/G2 phase of the cell cycle (Verdun et al, 2005; Verdun and Karlseder, 2006), recruits ATM (the mammalian Tel1 orthologue) to telomeres. One of ATM functions is phosphorylation of TRF1, which leads to its dissociation from telomeres (Wu et al, 2007), a curious twist on the situation in budding yeast in which the Rap1–Rif complex instead acts to inhibit telomere binding of Tel1 (ATM) (Hirano et al, 2009). An attractive notion, still untested, is that MRN–ATM association with telomeres is modulated by telomere repeat tract length, with ATM then acting on (several?) telomeric proteins to positively influence telomerase action. Regarding the apparent special role of TPP1 in fission yeast and mammalian telomere replication, we note that both S. cerevisiae and C. albicans Est3 proteins have recently been proposed to be structural homologues of TPP1 (Lee et al, 2008; Yu et al, 2008). Despite this structural similarity and a possible shared role in telomerase stimulation, the yeast Est3 proteins might have evolved at least partly distinct telomeric functions (e.g. with regard to telomere capping), or at least use different interacting partners. These findings point to how evolution has used a basic toolbox of telomeric proteins to derive different functional modules, and emphasize the importance of studying telomere biology in different organisms.
Telomeric single-stranded DNA can form alternative DNA structures that may have important functions in both telomere capping and length regulation. In many different organisms, telomeres can adopt a so-called ‘t-loop' conformation (Griffith et al, 1999), in which the 3′ single-stranded overhang is base paired to internal repeats in the T2AG3 array, presumably hiding the chromosome end from the DNA damage checkpoint machinery, but perhaps also blocking telomerase access. TRF2, by promoting positive supercoiling, can induce t-loop formation in vitro (Amiard et al, 2007). Dramatic telomere shortening events can occur through homologous recombination at t-loop junctions (Wang et al, 2004), but it still remains to be seen whether t-loops have a function in homeostatic regulation of the telomerase pathway of length regulation. In addition to the t-loop, G-rich telomere overhangs can form so-called G-quadruplex structures, which have been extensively studied in vitro. The best evidence for the existence of telomeric G-quadruplex structures in vivo, and for their role in telomere replication, comes from studies in ciliates (Paeschke et al, 2008). Understanding the possible role of these structures in telomere length regulation, or DNA metabolism in general, remains a difficult task (Johnson et al, 2008).
Studies on the maturation of human telomerase are beginning to reveal new factors that have a function in telomerase assembly (reviewed in Collins, 2006). The human telomerase holoenzyme minimally contains the catalytic subunit (TERT), the telomerase RNA (TERC), and an RNA ‘H/ACA box'-binding protein called dyskerin (Cohen et al, 2007). In an exciting recent development, Artandi and co-workers have isolated a novel protein, TCAB1 (telomerase and Cajal body protein 1), based on its interaction with dyskerin, which associates with Cajal bodies and which might be important for delivering telomerase to telomeres (Venteicher et al, 2009). A role for Cajal bodies in allowing the association of telomerase with telomeres in S phase has recently been recognized (Jady et al, 2006; Tomlinson et al, 2006), and the interaction of telomerase with Cajal bodies appears to be important for telomerase action at chromosome ends (Cristofari et al, 2007). Significantly, TCAB1, which associates with active telomerase, is required for telomerase association both with Cajal bodies and with telomeres, and for telomere repeat synthesis at chromosome ends. Thus, the emerging view is that the mammalian telomerase enzyme has hijacked a Cajal body pathway for spliceosomal RNA maturation as part of a mechanism to deliver the telomerase RNP in an active form to telomeres. Why Cajal bodies might function as ‘delivery stations' to allow efficient recruitment of telomerase to telomeres is presently unclear, and other possibilities (e.g. that Cajal bodies facilitate assembly of an ‘active' enzyme capable of functioning in the context of native DNA ends) cannot at present be ruled out (Venteicher and Artandi, 2009).
Finally, though telomeres have been long thought to be transcriptionally silent chromosome domains, it is now clear that telomere repeats are indeed transcribed, in organisms as diverse as yeast and human (Azzalin et al, 2007; Luke et al, 2008; Schoeftner and Blasco, 2008). The potential role of telomeric RNA, dubbed ‘TERRA', as well as that of telomeric chromatin structure in homeostatic regulation of the telomerase pathway are both exciting open questions, and the reader is referred to the accompanying reviews (Bühler and Gasser, 2009; Luke and Lingner, 2009; Schoeftner and Blasco, 2009) for detailed discussions of these topics.
Concluding remarks
In little over 20 years since the discovery of the telomerase enzyme, considerable progress has been made in unravelling the molecular mechanisms underlying the control of this unique mode of chromosome end replication. The general picture that has emerged is one in which the protein complexes assembled on telomere repeat sequences have a dual function, on the one hand assisting the recruitment and activation of telomerase enzyme at telomeres, yet on the other hand collaborating in what seem to be multiple modes of negative feedback on telomerase action that serve to maintain telomere repeat length homeostasis. One striking and recurrent observation from this work is the connection between telomerase-based replication and the DNA damage response, both at the level of the proteins involved and the enzymatic and structural transitions of the telomeric DNA itself. In fact, telomeres in an extendable state bear a striking resemblance to DNA DSBs, and we still lack a clear understanding of how the cell can so faithfully channel these two related structures towards the appropriate ‘repair' pathways. Furthermore, regulation of telomerase action has multiple connections to conventional DNA replication, both at the level of the DNA polymerases themselves and control of DNA replication initiation at telomere-proximal origins, all of which need to be more clearly understood. These and other questions constitute significant challenges for the field in the coming years. Considering the recent and unexpected discovery of telomeric RNA, we should anticipate additional surprises as a diverse set of telomere length regulatory mechanisms come into better focus.
Acknowledgments
Work in the Shore laboratory is supported by grants from the Swiss National Fund (grant 31003A116716), from the Swiss National Fund ‘Frontiers in Genetics' National Centre, and from the Canton of Geneva. Work in the Bianchi laboratory is funded by the Medical Research Council. We apologize to those whose work could not be cited or discussed due to space limitations.
References
- Adams AK, Holm C (1996) Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol Cell Biol 16: 4614–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, Paoletti J, Gilson E, Giraud-Panis MJ (2007) A topological mechanism for TRF2-enhanced strand invasion. Nat Struct Mol Biol 14: 147–154 [DOI] [PubMed] [Google Scholar]
- Arneric M, Lingner J (2007) Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep 8: 1080–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Kupiec M (2005) Cell cycle-dependent regulation of double-strand break repair: a role for the CDK. Cell Cycle 4: 259–261 [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Bae NS, Baumann P (2007) A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell 26: 323–334 [DOI] [PubMed] [Google Scholar]
- Baumann P, Cech TR (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya MK, Lustig AJ (2006) Telomere dynamics in genome stability. Trends Biochem Sci 31: 114–122 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Negrini S, Shore D (2004) Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol Cell 16: 139–146 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Shore D (2007a) Early replication of short telomeres in budding yeast. Cell 128: 1051–1062 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Shore D (2007b) Increased association of telomerase with short telomeres in yeast. Genes Dev 21: 1726–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Shore D (2008) How telomerase reaches its end: mechanism of telomerase regulation by the telomeric complex. Mol Cell 31: 153–165 [DOI] [PubMed] [Google Scholar]
- Blackburn EH (2006) A history of telomere biology. In Telomeres, de Lange T, Lundblad V, Blackburn E (eds), 2nd edn, pp 551–563. Cold Spring Harbor, New York: Cold Spring Harbor Press [Google Scholar]
- Blackburn EH, Gall JG (1978) A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol 120: 33–53 [DOI] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91: 25–34 [DOI] [PubMed] [Google Scholar]
- Boule JB, Vega LR, Zakian VA (2005) The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 438: 57–61 [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP (1996) Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res 24: 4639–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler M, Gasser S (2009) Silent chromatin at middles and ends: lessons from yeasts. EMBO J (e-pub ahead of print 23 July 2009; doi:10.1038/emboj.2009.185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson MJ, Hartwell L (1985) CDC17: an essential gene that prevents telomere elongation in yeast. Cell 42: 249–257 [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Reddel RR (2008) Telomere uncapping and alternative lengthening of telomeres. Mech Ageing Dev 129: 99–108 [DOI] [PubMed] [Google Scholar]
- Chai W, Du Q, Shay JW, Wright WE (2006) Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell 21: 427–435 [DOI] [PubMed] [Google Scholar]
- Chan A, Boule JB, Zakian VA (2008) Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet 4: e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Chang J, Prescott J, Blackburn EH (2001) Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr Biol 11: 1240–1250 [DOI] [PubMed] [Google Scholar]
- Chandra A, Hughes TR, Nugent CI, Lundblad V (2001) Cdc13 both positively and negatively regulates telomere replication. Genes Dev 15: 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Arneric M, Lingner J (2007) Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev 21: 2485–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR (2007) Protein composition of catalytically active human telomerase from immortal cells. Science 315: 1850–1853 [DOI] [PubMed] [Google Scholar]
- Collins K (2006) The biogenesis and regulation of telomerase holoenzymes. Nat Rev Mol Cell Biol 7: 484–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad MN, Wright JH, Wolf AJ, Zakian VA (1990) RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell 63: 739–750 [DOI] [PubMed] [Google Scholar]
- Cosgrove AJ, Nieduszynski CA, Donaldson AD (2002) Ku complex controls the replication time of DNA in telomere regions. Genes Dev 16: 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RJ, Petes TD (1999) Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae. Genetics 152: 1531–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J (2007) Human telomerase RNA accumulation in cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell 27: 882–889 [DOI] [PubMed] [Google Scholar]
- Cristofari G, Lingner J (2006) Telomere length homeostasis requires that telomerase levels are limiting. EMBO J 25: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- DeZwaan DC, Toogun OA, Echtenkamp FJ, Freeman BC (2009) The Hsp82 molecular chaperone promotes a switch between unextendable and extendable telomere states. Nat Struct Mol Biol 16: 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE (1999) Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99: 723–733 [DOI] [PubMed] [Google Scholar]
- Dionne I, Wellinger RJ (1998) Processing of telomeric DNA ends requires the passage of a replication fork. Nucleic Acids Res 26: 5365–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, Bermejo R, Fiorani S, Haber JE, Foiani M (2009) Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell 137: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SK, Bertuch AA, Lundblad V (1999) Telomeres and telomerase: at the end, it all comes together. Trends Cell Biol 9: 329–331 [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V (1999) Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120 [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V (2002) The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics 162: 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Price CM (1997) Coordinate regulation of G- and C strand length during new telomere synthesis. Mol Biol Cell 8: 2145–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher TS, Taggart AK, Zakian VA (2004) Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol 11: 1198–1205 [DOI] [PubMed] [Google Scholar]
- Frank CJ, Hyde M, Greider CW (2006) Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol Cell 24: 423–432 [DOI] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V (2007) RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 14: 208–214 [DOI] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparyan HJ, Xu L, Petreaca RC, Rex AE, Small VY, Bhogal NS, Julius JA, Warsi TH, Bachant J, Aparicio OM, Nugent CI (2009) Yeast telomere capping protein Stn1 overrides DNA replication control through the S phase checkpoint. Proc Natl Acad Sci USA 106: 2206–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Geli V (2007) How telomeres are replicated. Nat Rev Mol Cell Biol 8: 825–838 [DOI] [PubMed] [Google Scholar]
- Goudsouzian LK, Tuzon CT, Zakian VA (2006) S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol Cell 24: 603–610 [DOI] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M (2000) Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol Cell Biol 20: 8397–8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M (2001) Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J 20: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Reed SI, Charbonneau M (1997) Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev 11: 512–527 [DOI] [PubMed] [Google Scholar]
- Gravel S, Larrivee M, Labrecque P, Wellinger RJ (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science 280: 741–744 [DOI] [PubMed] [Google Scholar]
- Greenwell PW, Kronmal SL, Porter SE, Gassenhuber J, Obermaier B, Petes TD (1995) TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82: 823–829 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH (1987) The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 51: 887–898 [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Grossi S, Bianchi A, Damay P, Shore D (2001) Telomere formation by rap1p binding site arrays reveals end-specific length regulation requirements and active telomeric recombination. Mol Cell Biol 21: 8117–8128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D (2004) Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev 18: 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CFJ, Sussel L, Shore D (1992) A RAP1-interacting protein involved in silencing and telomere length regulation. Genes Dev 6: 801–814 [DOI] [PubMed] [Google Scholar]
- Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW (2007) Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell 27: 851–858 [DOI] [PubMed] [Google Scholar]
- Hirano Y, Fukunaga K, Sugimoto K (2009) Rif1 and rif2 inhibit localization of tel1 to DNA ends. Mol Cell 33: 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Sugimoto K (2007) Cdc13 telomere capping decreases Mec1 association but does not affect Tel1 association with DNA ends. Mol Biol Cell 18: 2026–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob NK, Skopp R, Price CM (2001) G-overhang dynamics at Tetrahymena telomeres. EMBO J 20: 4299–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Richard P, Bertrand E, Kiss T (2006) Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell 17: 944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Smith JS, Kozak ML, Johnson FB (2008) In vivo veritas: using yeast to probe the biological functions of G-quadruplexes. Biochimie 90: 1250–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G, Boakye KA, Lustig AJ (1992) C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol 12: 5159–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Gotta M, Gorham HC, Pryde FE, Louis EJ, Gasser SM (1998) Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr Biol 8: 653–656 [DOI] [PubMed] [Google Scholar]
- Larrivee M, LeBel C, Wellinger RJ (2004) The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev 18: 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Mandell EK, Tucey TM, Morris DK, Lundblad V (2008) The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat Struct Mol Biol 15: 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Bressan DA, Petrini JH, Haber JE (2002) Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair (Amst) 1: 27–40 [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V (1996) Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144: 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Blackburn EH (2004) Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol Cell Biol 24: 10857–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Lustig AJ (1996) A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev 10: 1310–1326 [DOI] [PubMed] [Google Scholar]
- Li S, Makovets S, Matsuguchi T, Blethrow JD, Shokat KM, Blackburn EH (2009) Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell 136: 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cooper JP, Cech TR (1995) Telomerase and DNA: no longer a lagging strand problem? Science 269: 1533–1534 [DOI] [PubMed] [Google Scholar]
- Loayza D, De Lange T (2003) POT1 as a terminal transducer of TRF1 telomere length control. Nature 424: 1013–1018 [DOI] [PubMed] [Google Scholar]
- Luke B, Lingner J (2009) TERRA—telomeric repeat containing RNA. EMBO J (e-pub ahead of print 23 July 2009; doi:10.1038/emboj.2009.166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J (2008) The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32: 465–477 [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643 [DOI] [PubMed] [Google Scholar]
- Lustig AJ (2003) Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat Rev Genet 4: 916–923 [DOI] [PubMed] [Google Scholar]
- Lustig AJ, Kurtz S, Shore D (1990) Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250: 549–553 [DOI] [PubMed] [Google Scholar]
- Lustig AJ, Petes TD (1986) Identification of yeast mutants with altered telomere structure. Proc Natl Acad Sci USA 83: 1398–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D (2009) Taming the tiger by the tail: modulation of DNA damage responses by telomeres. EMBO J (e-pub ahead of print 23 July 2009; doi:10.1038/emboj.2009.176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Gilson E (1999) Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J 18: 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Mann C, Gilson E (2000) Cell cycle restriction of telomere elongation. Curr Biol 10: 487–490 [DOI] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D (1997) A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990 [DOI] [PubMed] [Google Scholar]
- Marcand S, Pardo B, Gratias A, Cahun S, Callebaut I (2008) Multiple pathways inhibit NHEJ at telomeres. Genes Dev 22: 1153–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V, Du LL, Rozenzhak S, Russell P (2007) Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci USA 104: 14038–14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier B, Driller L, Jaklin S, Feldmann HM (2001) New function of CDC13 in positive telomere length regulation. Mol Cell Biol 21: 4233–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson RJ, Rosenstein S, Weinert T (2005) A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes Dev 19: 2546–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP (2006) Semi-conservative DNA replication through telomeres requires Taz1. Nature 440: 824–828 [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2009) Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci 34: 264–272 [DOI] [PubMed] [Google Scholar]
- Mishra K, Shore D (1999) Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr Biol 9: 1123–1126 [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F (2008) Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320: 1341–1344 [DOI] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS (1999) The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Subramanian L, Chang YT, Noguchi C, Noguchi E, Nakamura TM (2009) Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J 28: 810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW, Claus TE, Szostak JW (1988) Characterization of two telomeric DNA processing reactions in Saccharomyces cerevisiae. Mol Cell Biol 8: 4642–4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD (2001a) Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076 [DOI] [PubMed] [Google Scholar]
- Myung K, Datta A, Kolodner RD (2001b) Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104: 397–408 [DOI] [PubMed] [Google Scholar]
- Negrini S, Ribaud V, Bianchi A, Shore D (2007) DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev 21: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, Moore JK, Haber JE, Lundblad V (1998) Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol 8: 657–660 [DOI] [PubMed] [Google Scholar]
- O'Connor MS, Safari A, Liu D, Qin J, Songyang Z (2004) The human Rap1 protein complex and modulation of telomere length. J Biol Chem 279: 28585–28591 [DOI] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL (2009) Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev 23: 912–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ (2008) Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat Struct Mol Biol 15: 598–604 [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Pennaneach V, Putnam CD, Kolodner RD (2006) Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol Microbiol 59: 1357–1368 [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V (2001) Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387–396 [DOI] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE (2001) The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet 27: 64–67 [DOI] [PubMed] [Google Scholar]
- Petreaca RC, Chiu H-C, Nugent CI (2007) The role of Stn1p in Saccharomyces cerevisiae telomere capping can be separated from its interaction with Cdc13p. Genetics 177: 1459–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polotnianka RM, Li J, Lustig AJ (1998) The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol 8: 831–834 [DOI] [PubMed] [Google Scholar]
- Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D (2008) Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J 27: 2328–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA (2000) The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev 14: 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL (2001) Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Ray A, Runge KW (1999) Varying the number of telomere-bound proteins does not alter telomere length in tel1Delta cells. Proc Natl Acad Sci USA 96: 15044–15049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Karamysheva Z, Wang L, Shippen DE, Price CM (2002) Interactions between telomerase and primase physically link the telomere and chromosome replication machinery. Mol Cell Biol 22: 5859–5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie KB, Mallory JC, Petes TD (1999) Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol 19: 6065–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie KB, Petes TD (2000) The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O, Cooper JP (2008) Telomeres in drag: dressing as DNA damage to engage telomerase. Curr Opin Genet Dev 18: 212–220 [DOI] [PubMed] [Google Scholar]
- Sabourin M, Tuzon CT, Zakian VA (2007) Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell 27: 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin M, Zakian VA (2008) ATM-like kinases and regulation of telomerase: lessons from yeast and mammals. Trends Cell Biol 18: 337–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA (2008) Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10: 228–236 [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA (2009) A ‘higher order' of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J, (e-pub ahead of print 23 July 2009; doi:10.1038/emboj.2009.197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramke V, Luciano P, Brevet V, Guillot S, Corda Y, Longhese MP, Gilson E, Geli V (2004) RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat Genet 36: 46–54 [DOI] [PubMed] [Google Scholar]
- Sfeir AJ, Chai W, Shay JW, Wright WE (2005) Telomere-end processing the terminal nucleotides of human chromosomes. Mol Cell 18: 131–138 [DOI] [PubMed] [Google Scholar]
- Shore D (1994) RAP1: a protean regulator in yeast. Trends in Genetics 10: 408–412 [DOI] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE (1994) TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409 [DOI] [PubMed] [Google Scholar]
- Singh SM, Lue NF (2003) Ever shorter telomere 1 (EST1)-dependent reverse transcription by Candida telomerase in vitro: evidence in support of an activating function. Proc Natl Acad Sci USA 100: 5718–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE (2003) Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev 17: 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JB, Gottschling DE (1999) Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev 13: 146–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Shore D (1991) Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc Natl Acad Sci USA 88: 7749–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Blackburn EH (1982) Cloning yeast telomeres on linear plasmid vectors. Cell 29: 245–255 [DOI] [PubMed] [Google Scholar]
- Taggart AK, Teng SC, Zakian VA (2002) Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Tomita K, Cooper JP (2008) Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev 22: 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP (2006) Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell 17: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SF, Lin JJ, Teng SC (2006) The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res 34: 6327–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SF, Shen ZJ, Tsai HJ, Lin YH, Teng SC (2009) Rapid Cdc13 turnover and telomere length homeostasis are controlled by Cdk1-mediated phosphorylation of Cdc13. Nucleic Acids Res 37: 3602–3611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Taggart AK, Zakian VA (2001) The role of the Mre11-Rad50-Xrs2 complex in telomerase- mediated lengthening of Saccharomyces cerevisiae telomeres. Curr Biol 11: 1328–1335 [DOI] [PubMed] [Google Scholar]
- van Steensel B, de Lange T (1997) Control of telomere length by the human telomeric protein TRF1. Nature 385: 740–743 [DOI] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE (2009) A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323: 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Artandi SE (2009) TCAB1: driving telomerase to Cajal bodies. Cell Cycle 8: 1329–1331 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Crabbe L, Haggblom C, Karlseder J (2005) Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell 20: 551–561 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J (2006) The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 127: 709–720 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J (2007) Replication and protection of telomeres. Nature 447: 924–931 [DOI] [PubMed] [Google Scholar]
- Viscardi V, Bonetti D, Cartagena-Lirola H, Lucchini G, Longhese MP (2007) MRX-dependent DNA damage response to short telomeres. Mol Biol Cell 18: 3047–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M (2007) The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510 [DOI] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T (2004) Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119: 355–368 [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Ethier K, Labrecque P, Zakian VA (1996) Evidence for a new step in telomere maintenance. Cell 85: 423–433 [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA (1993) Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72: 51–60 [DOI] [PubMed] [Google Scholar]
- Wotton D, Shore D (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev 11: 748–760 [DOI] [PubMed] [Google Scholar]
- Wu Y, Xiao S, Zhu XD (2007) MRE11-RAD50-NBS1 and ATM function as co-mediators of TRF1 in telomere length control. Nat Struct Mol Biol 14: 832–840 [DOI] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z (2007) TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 445: 559–562 [DOI] [PubMed] [Google Scholar]
- Yu EY, Wang F, Lei M, Lue NF (2008) A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol 15: 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]