Abstract
Asparagine-linked glycosylation is catalysed by oligosaccharyltransferase (OTase). In Trypanosoma brucei OTase activity is catalysed by single-subunit enzymes encoded by three paralogous genes of which TbSTT3B and TbSTT3C can complement a yeast Δstt3 mutant. The two enzymes have overlapping but distinct peptide acceptor specificities, with TbSTT3C displaying an enhanced ability to glycosylate sites flanked by acidic residues. TbSTT3A and TbSTT3B, but not TbSTT3C, are transcribed in the bloodstream and procyclic life cycle stages of T. brucei. Selective knockdown and analysis of parasite protein N-glycosylation showed that TbSTT3A selectively transfers biantennary Man5GlcNAc2 to specific glycosylation sites whereas TbSTT3B selectively transfers triantennary Man9GlcNAc2 to others. Analysis of T. brucei glycosylation site occupancy showed that TbSTT3A and TbSTT3B glycosylate sites in acidic to neutral and neutral to basic regions of polypeptide, respectively. This embodiment of distinct specificities in single-subunit OTases may have implications for recombinant glycoprotein engineering. TbSTT3A and TbSTT3B could be knocked down individually, but not collectively, in tissue culture. However, both were independently essential for parasite growth in mice, suggesting that inhibiting protein N-glycosylation could have therapeutic potential against trypanosomiasis.
Keywords: glycosylation, oligosaccharyltransferase, STT3, Trypanosoma
Introduction
The protozoan parasite Trypanosoma brucei is the causative agent of human African trypanosomiasis or sleeping sickness. The life cycle of T. brucei includes a bloodstream (trypomastigote) form in the mammalian host and a procyclic form in the midgut of the tsetse fly insect vector. In both forms, the cell surface of T. brucei is covered by a coat of glycosylphosphatidylinositol-anchored glycoproteins, the variant surface glycoproteins (VSGs) in the bloodstream (Cross, 1975) and the procyclins in the insect (Mowatt and Clayton, 1987; Richardson et al, 1988; Roditi et al, 1989). These proteins are N-glycosylated, a posttranslational modification that modulates folding, stability and activity of proteins that is essential in most eukaryotes (Varki, 1993; Sharon and Lis, 1995; Helenius and Aebi, 2004; Ohtsubo and Marth, 2006). N-glycosylation is catalysed by oligosaccharyltransferase (OTase) in the lumen of the endoplasmic reticulum (ER) (Helenius and Aebi, 2004) in which the transfer of oligosaccharide from a lipid-linked oligosaccharide (LLO) donor to selected asparagines in the sequons (N–x−S/T; x≠P) (Gavel and von Heijne, 1990) of translocated proteins occurs. Although OTase in higher eukaryotes is a multiprotein complex of up to eight subunits (Kelleher and Gilmore, 2006), the genome of T. brucei predicts that it lacks all except for the catalytic STT3 subunit, for which there are three complete paralogous genes (Berriman et al, 2005). Some of the additional subunits of the multiprotein complex OTases increase the diversity of efficiently glycosylated protein substrates. Ribophorin I (Ost1p) acts as a chaperone to enhance glycosylation of specific membrane proteins (Wilson and High, 2007), and the oxidoreductase activity of Ost3/6p increase the glycosylation efficiency of defined sequons (Schulz and Aebi, 2009; Schulz et al, 2009). In mammals, the presence of either STT3-A or STT3-B in the OTase complex also influences glycan substrate specificity and glycosylation rate, and both OTase isoforms act sequentially to maximise the efficiency of the N-glycosylation process (Kelleher et al, 2003; Ruiz-Canada et al, 2009). Although some organisms have extended the range of OTase activity by adding subunits to the core catalytic STT3, it is possible that T. brucei has instead duplicated and diversified a single-subunit OTase, as seems to be the case for the related kinetoplastid parasite Leishmania major (Nasab et al, 2008; Hese et al, 2009). This hypothesis is supported by the observation that, though in higher eukaryotes OTase has high specificity for the mature Glc3Man9GlcNAc2 glycan, which it transfers to many different glycosylation sequons, T. brucei selectively transfers Man5GlcNAc2 to one sequon and Man9GlcNAc2 to another in the VSG221 protein (Jones et al, 2005; Manthri et al, 2008). This suggests that a mechanism exists for glycan donor- and peptide acceptor-specific glycosylation in this organism, although the generality of that mechanism beyond VSG has not been tested.
It has been reported that the single-subunit STT3 OTases from T. cruzi and L. major can complement an STT3 deletion in yeast (Castro et al, 2006; Nasab et al, 2008; Hese et al, 2009). We considered, therefore, that yeast could be a useful ex vivo model system to investigate the potential substrate specificities of the T. brucei OTases and with which to compare the effects of individual TbSTT3 knockdowns in T. brucei itself.
Results
TbSTT3B and TbSTT3C complement a Δstt3 yeast mutant with different efficiencies and different acceptor sequon specificities
There are three putative intact STT3 genes in T. brucei (TbSTT3A, TbSTT3B and TbSTT3C). Although all three TbSTT3 proteins were efficiently expressed in wild-type yeast cells at similar levels (Supplementary Figure S1A), only expression of TbSTT3B and TbSTT3C, but not of TbSTT3A, could support yeast growth in the absence of endogenous STT3 (Figure 1A). The Δstt3+TbSTT3B cells grew almost as well as wild type at 23°C, but showed a mild temperature-sensitive growth phenotype at 37°C (Figure 1B); whereas Δstt3+TbSTT3C cells had a severe growth defect even at 23°C. Owing to the inability of TbSTT3A to support yeast Δstt3 growth, we determined its function in trypanosomes by an mRNA silencing approach (see below).
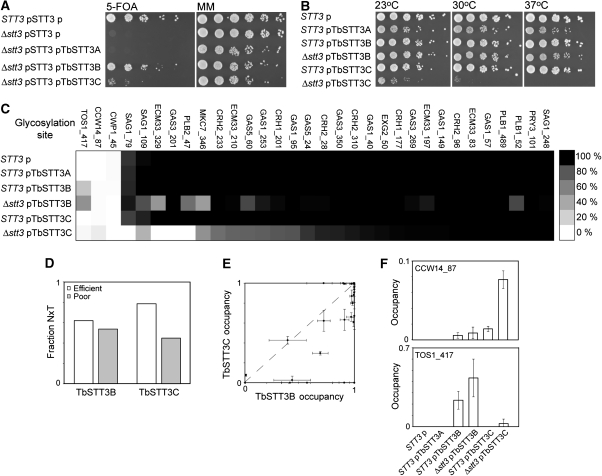
Figure 1.
Complementation of yeast Δstt3 by T. brucei OTases. (A) Yeast cells with (STT3) or without (Δstt3) genomic STT3, with the yeast STT3 gene encoded on a URA3 plasmid (pSTT3), and with various TbSTT3 genes (pTbSTT3A; pTbSTT3B; pTbSTT3C) or with empty vector (p) on a LEU2 plasmid were grown in serial dilution on minimal media (MM) or media containing 5-FOA for selection of ura-cells and (B) after selection were grown at different temperatures in serial dilution on MM. (C) Glycosylation occupancy of all sites measured for each strain, mapped from 100% occupancy, black, to 0% occupancy, white. Values are averages of three independent measurements. Data are shown in Tables SI and SII. (D) Proportion of threonine in the +2 position of efficiently and poorly glycosylated sequons. (E) Cross correlation of site-specific glycosylation efficiency by TbSTT3B and TbSTT3C. (F) Glycosylation of TOS1_417 and CCW14_87 sites by T. brucei STT3B and STT3C that are not used by yeast OTase. Error bars show range.
The sequences of TbSTT3B and TbSTT3C share 95% identity, whereas TbSTT3A share only 79% identity with them both (Supplementary Figure S2). To investigate whether the minor sequence differences between TbSTT3B and TbSTT3C influence peptide acceptor selection in yeast, we used a mass spectrometry-based method (Schulz and Aebi, 2009) to analyse the site-specific glycosylation occupancy of cell wall proteins in our ex vivo system. We analysed yeast cells with and without yeast STT3 and with different TbSTT3 genes (Figure 1C; Supplementary Tables SI and SII). Yeast cells with wild-type OTase and in addition a T. brucei OTase did not show a reduction in glycosylation at any normally used site, suggesting that expression of T. brucei OTases did not interfere with the normal yeast N-glycosylation machinery. In Δstt3+TbSTT3B cells, most sequons were glycosylated with comparable efficiency to wild-type yeast, whereas Δstt3 + TbSTT3C cells showed lower efficiency at more sites, correlating with the growth phenotypes of these cells (Figure 1A). Furthermore, TbSTT3B and TbSTT3C did indeed have different activities towards some glycosylation sequons, as some sites were more efficiently glycosylated by TbSTT3B and others by TbSTT3C (Figure 1C and D). As for other OTases (Kasturi et al, 1995; Petrescu et al, 2004) TbSTT3B and TbSTT3C showed a slight preference for threonine over serine in the +2 position of glycosylated sequons (Figure 1E). Significantly, cells expressing TbSTT3B and TbSTT3C partially glycosylated some sites never used by yeast OTase (Figure 1C and F) and the occupancy of these sites was increased in cells lacking yeast OTase activity. This effect is not solely a result of OTase dosage in cells overexpressing TbSTT3B or TbSTT3C, as the newly glycosylated site was different in each case (i.e. N417 of Tos1p by TbSTT3B and N87 of Ccw14p by TbSTT3C).
We also analysed the sequences surrounding the glycosylated sequons (±5 amino acids) and noticed a tendency towards efficient occupancy of acidic sequences by TbSTT3C. These data will be discussed in more detail later.
Only TbSTT3A and TbSTT3B transcripts are found in bloodstream and procyclic form T. brucei
We used semi-quantitative RT–PCR to assess the relative levels of the TbSTT3 mRNA transcripts in bloodstream and procyclic form trypanosomes (Supplementary Figure 1B). We found that the TbSTT3A and TbSTT3B genes were expressed in both life cycle stages, with TbSTT3A more highly expressed in the bloodstream form, whereas transcripts for the TbSTT3C gene were not detected in either life cycle stage of the parasite.
The TbSTT3A gene encodes a Man5GlcNAc2-specific OTase responsible for all paucimannose and complex N-glycans in T. brucei glycoproteins
We studied the roles of the TbSTT3 genes in trypanosomes using inducible RNA interference (RNAi) and glycoprotein structure analysis. Our principal reporter molecule is the VSG coat glycoprotein of the parasite that, in the variant 221 clone used in our studies, contains two N-glycosylation sites (see Figure 2A, inset): one at Asn263 occupied by small, biantennary, endoglycosidase-H (EndoH)-resistant paucimannose and complex structures, which originate from the transfer of Man5GlcNAc2 from the Man5GlcNAc2-PP-Dol precursor, and one at Asn428 occupied by conventional EndoH-sensitive triantennary oligomannose structures, which originate from the transfer of Man9GlcNAc2 from Man9GlcNAc2-PP-Dol (Jones et al, 2005; Manthri et al, 2008). Thus, analysis of VSG221 N-glycosylation allows us to simultaneously assess the effects of genetic or chemical perturbations on both mechanisms of protein N-glycosylation in this organism (Jones et al, 2005; Urbaniak et al, 2006; Manthri et al, 2008; Stokes et al 2008). Cell-surface VSG can be purified in a soluble form (sVSG221) after osmotic shock, a process that releases VSG from the parasite surface by the action of endogenous GPI-specific phospholipase C that cleaves the dimyristoylglycerol lipid component of the VSG GPI anchor (Cross, 1975, 1984; Cardoso de Almeida and Turner, 1983; Ferguson et al, 1985). This cell surface-derived sVSG221 is amenable to glycoform analysis as an intact glycoprotein by electrospray–mass spectrometry (ES-MS) and as Pronase glycopeptides by ES-MS and ES-MS/MS (Jones et al, 2005; Urbaniak et al, 2006; Manthri et al, 2008; Stokes et al 2008). In addition, the N-glycosylation status of both cell surface-derived sVSG221 and of newly synthesised (presumably ER-associated) VSG221, which is not released by osmotic shock (Ferguson et al 1986), can be assessed by EndoH and PNGaseF digestion and analysis by SDS–PAGE and Coomassie blue staining or anti-VSG221 western blot, respectively.
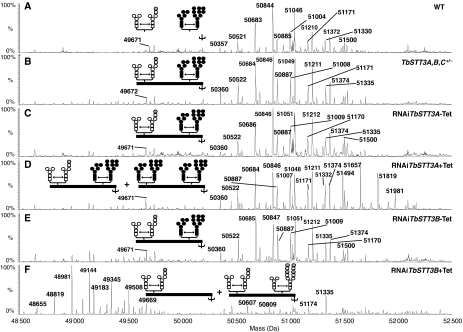
Figure 2.
Mass spectrometric analyses of intact mature sVSG221 glycoforms before and after selective knockdown of TbSTT3A and TbSTT3B expression. Samples of sVSG221 from (A) wild type cells, (B) TbSTT3A,B,C+/−, (C) TbSTT3A,B,C+/−-STT3ARNAi minus Tet, (D) TbSTT3A,B,C+/−-STT3ARNAi plus Tet, (E) TbSTT3A,B,C+/−-STT3BRNAi minus Tet and (F) TbSTT3A,B,C+/−-STT3BRNAi plus Tet were analysed by ES-MS. The spectra show the masses of the various glycoforms of the intact mature sVSG221 molecules. The inset cartoons represent our interpretation of those glycoform masses in terms of the ranges of N-glycans present at each of the two N-glycosylation sites. These assignments combine additional data from the ES-MS and ES-MS/MS analyses of Pronase glycopeptides from the same sVSG221 preparations (Supplementary Figure S3; Table SIV). In the inset cartoons, endo-H-resistant N-glycans are in white and endo-H-sensitive N-glycans are in black. Circles and squares (filled and open) represent mannose and N-acetylglucosmaine residues, respectively, and grey circles represent galactose residues.
The three intact TbSTT3 genes are arrayed in tandem on chromosome 5. One whole TbSTT3A,B,C array was replaced by homologous recombination with a puromycin acetyl transferase (PAC) selectable marker to obtain a bloodstream form TbSTT3A,B,C+/− mutant (Supplementary Figure S1C). This deletion did not affect the glycosylation profile of the VSG221 reporter glycoprotein (Figure 2A and B) but it provided us with a heterozygote cell line in which subsequent selective RNAi knockdowns of TbSTT3s would be maximised.
A 498 nt internal coding sequence was used to generate a bloodstream form cell line containing an inducible double-stranded RNA (dsRNA) construct targeting TbSTT3A in the TbSTT3A,B,C+/− heterozygote (Supplementary Figure S1E). Tetracycline induction substantially reduced TbSTT3A mRNA without affecting TbSTT3B mRNA (Figure 3A). The TbSTT3A knockdown did not have a major effect in the growth of the parasite in culture (Figure 3B) but it had a clear effect on the glycosylation of newly synthesised (ER-associated) VSG221. Thus, the majority of the ER-associated VSG221 in the TbSTT3A knockdown contained two EndoH-sensitive N-glycans, instead of one EndoH-sensitive and one EndoH-resistant N-glycan (Figure 3C). Analysis of cell surface-derived sVSG221 after 72 h of RNAi induction showed the presence of two distinct bands after EndoH digestion, suggesting this preparation contained two forms of sVSG221 with one and two EndoH-sensitive N-glycans, respectively, (Figure 3D). Further analysis of the cell surface-derived sVSG221 by ES-MS confirmed that two sets of overlapping glycoforms were present: one set identical to wild-type sVSG221 and another set with masses compatible with a modified sVSG221 with oligomannose structures at both N-glycosylation sites (Figure 2D; Supplementary Table SIII). This assignment was supported by subsequent Pronase digestion and ES-MS and ES-MS/MS identification of the sVSG221 glycopeptides (Supplementary Table SIV; Supplementary Figure S3A). The apparently greater proportion of modified versus wild-type VSG221 in the newly synthesised VSG221 versus mature cell surface-derived sVSG221 (compare Figure 3C and D) suggests that the small proportion of wild-type VSG221 present in the ER is enriched during ER exit and/or transit to the cell surface, which is consistent with the apparent importance of the correct glycosylation of the Asn263 site for VSG221 structure and folding (Blum et al, 1993; Izquierdo et al, 2009).
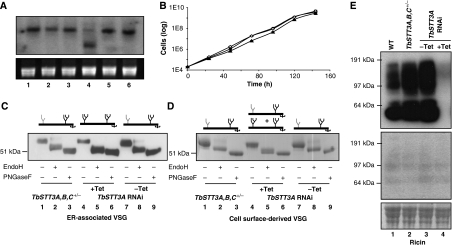
Figure 3.
The effects of RNAi silencing of STT3A on protein glycosylation in T. brucei. (A) Northern blot of total RNA extracted from the following cell lines: wild type (lane 1), TbSTT3A,B,C+/− (lane 2), TbSTT3A,B,C+/−-STT3ARNAi minus Tet (lane 3), TbSTT3A,B,C+/−-STT3ARNAi plus Tet (lane 4), TbSTT3A,B,C+/−-STT3BRNAi minus Tet (lane 5) and TbSTT3A,B,C+/−-STT3BRNAi plus Tet (lane 6) were probed with the STT3A probe (upper panel) and stained with ethidium bromide for rRNA as loading control (lower panel). (B) Growth of the following T. brucei cells at 37°C: wild type (closed circles), TbSTT3A,B,C+/−-STT3ARNAi minus Tet (open squares) and TbSTT3A,B,C+/−-STT3ARNAi plus Tet (closed triangles). (C) Cell ghosts containing ER-associated VSG221 and (D) cell surface-derived sVSG221 from the following cells: TbSTT3A,B,C+/− (lanes 1–3), TbSTT3A,B,C+/−-STT3ARNAi plus Tet (lanes 4–6) and TbSTT3A,B,C+/−-STT3ARNAi minus Tet (lanes 7–9) were digested or not, as indicated, with EndoH or PNGaseF were subjected SDS–PAGE and either western blot with anti-VSG221 antibody (C) or staining with Coomassie blue (D). The cartoons above the gels in panels (C) and (D) summarise the types of VSG221 present with respect to EndoH-resistant biantennary paucimannose and complex N-glycans (grey) and EndoH-sensitive triantennary oligomannose N-glycans (black). (E) Blots of total glycoprotein extracts of the following cells: wild type (lane 1), TbSTT3A,B,C+/− (lane 2), TbSTT3A,B,C+/−-STT3ARNAi minus Tet (lane 3) and TbSTT3A,B,C+/−-STT3ARNAi plus Tet (lane 4) were incubated with ricin without (upper panel) and with carbohydrate inhibitors (specificity control, middle panel) or stained with Pounceau red (loading control, lower panel).
These data suggest that TbSTT3A is responsible for the transfer of Man5GlcNAc2 to Asn263 of VSG221 and that, in its absence, Man9GlcNAc2 is transferred to that site instead via TbSTT3B. The model described in Manthri et al (2008) predicts that only sites that receive Man5GlcNAc2 can be processed to complex structures, because of the absence of Golgi α-mannosidase II gene in the T. brucei genome. Therefore, we also looked at the non-VSG glycoproteins of the parasite by solubilising parasite ghosts (after osmotic shock-mediated release of sVSG) and performing SDS–PAGE and ricin lectin blotting. The terminal galactose-specific lectin ricin detects T. brucei glycoproteins expressing complex N-linked oligosaccharides, including the giant poly-N-acetyllactosamine-containing N-glycans unique to this organism (Atrih et al, 2005). The striking reduction in ricin binding to the glycoproteins of the TbSTT3A knockdown cells (Figure 3E) is in complete agreement with the aforementioned model and suggests that it is general, rather than restricted to VSG N-glycosylation.
The TbSTT3B gene encodes a Man9GlcNAc2-specific OTase responsible for most or all oligomannose N-glycans in T. brucei glycoproteins
The same approach was taken to assess the function of TbSTT3B. A 467 nt internal coding sequence was used to create a bloodstream form TbSTT3A,B,C+/− heterozygote containing an inducible dsRNA construct targeting both TbSTT3B and TbSTT3C (Supplementary Figure S1E). As TbSTT3C mRNA is not present in bloodstream or procyclic form cells (Supplementary Figure S1B), tetracyline induction for 72 h leads to the selective knockdown of TbSTT3B mRNA (Figure 4A). As for TbSTT3A, the TbSTT3B knockdown did not substantially affect the growth of the parasite in culture (Figure 4B). Analysis of both newly synthesised, ER-associated, VSG221 and cell surface-derived sVSG221 after 72 h of TbSTT3B silencing showed the presence of two sets of VSG221 glycoforms (Figure 4C and D). The lower molecular weight band co-migrates with wild-type VSG221 treated with EndoH, suggesting it has only one occupied N-glycosylation site, whereas the higher molecular weight band has two EndoH-resistant N-glycans. Analysis of intact and Pronase digested cell surface-derived sVSG221 after 72 h of TbSTT3B knockdown by mass spectrometry (Figure 2F; Supplementary Tables SIII, SIV; Supplementary Figure S3B) confirmed the presence of a lower molecular weight group of sVSG221 glycoforms lacking glycans at Asn428 and a higher molecular weight group with Asn263 and Asn428 sites occupied by paucimannose and/or complex glycans, respectively.
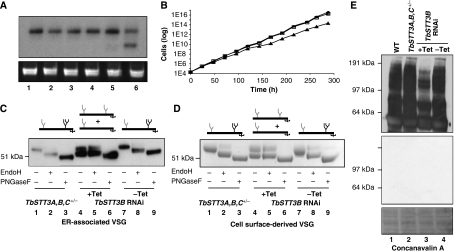
Figure 4.
The effects of RNAi silencing of STT3B on protein glycosylation in T. brucei. (A) Northern blot of total RNA extracted from the following cell lines: wild type (lane 1), TbSTT3A,B,C+/− (lane 2), TbSTT3A,B,C+/−-STT3ARNAi minus Tet (lane 3), TbSTT3A,B,C+/−-STT3ARNAi plus Tet (lane 4), TbSTT3A,B,C+/−-STT3BRNAi minus Tet (lane 5) and TbSTT3A,B,C+/−-STT3BRNAi plus Tet (lane 6) were probed with the STT3B probe (upper panel) and stained with ethidium bromide for rRNA as loading control (lower panel). (B) Growth of the following T. brucei cells at 37°C: wild type (closed circles), TbSTT3A,B,C+/−-STT3BRNAi minus Tet (open squares) and TbSTT3A,B,C+/−-STT3BRNAi plus Tet (closed triangles). (C) Cell ghosts containing ER-associated VSG221 and (D) cell surface-derived sVSG221 from the following cells: TbSTT3A,B,C+/− (lanes 1–3), TbSTT3A,B,C+/−-STT3BRNAi plus Tet (lanes 4–6) and TbSTT3A,B,C+/−-STT3BRNAi minus Tet (lanes 7–9) were digested or not, as indicated, with EndoH or PNGaseF were subjected SDS–PAGE and either western blot with anti-VSG221 antibody (C) or staining with Coomassie blue (D). The cartoons above the gels in panels (C) and (D) summarise the types of VSG221 present with respect to EndoH-resistant biantennary paucimannose and complex N-glycans (grey) and EndoH-sensitive triantennary oligomannose N-glycans (black). (E) Blots of total glycoprotein extracts of the following cells: wild type (lane 1), TbSTT3A,B,C+/− (lane 2), TbSTT3A,B,C+/−-STT3BRNAi plus Tet (lane 3) and TbSTT3A,B,C+/−-STT3BRNAi minus Tet (lane 4) were incubated with ConA without (upper panel) and with carbohydrate inhibitors (specificity control, middle panel) or stained with Pounceau red (loading control, lower panel).
These results are also consistent with our model for VSG221 N-glycosylation (Manthri et al, 2008), such that removal of TbSTT3B has no effect on the glycosylation of Asn263 but greatly reduces the efficiency of the glycosylation of the Asn428 site (which is normally served by the transfer of Man9GlcNAc2 via TbSTT3B). We confirmed the generality of the results seen with the VSG221 reporter by analysing the aforementioned non-VSG glycoprotein preparation by SDS–PAGE and concanavalin A (ConA) blotting (Figure 4E). ConA binds with highest affinity to oligomannose and hybrid glycans containing a Manα1-3(Manα1-6)Manα1-motif and less well to complex biantennary-type structures (Brewer and Bhattacharyya, 1986). Thus, the significant reduction in ConA binding to the glycoproteins from the TbSTT3B knockdown cells suggests that most or all of the conventional triantennary oligomannose N-linked glycans have disappeared in parallel with TbSTT3B knockdown.
TbSTT3A and TbSTT3B have distinct acceptor sequon specificities as well as oligosaccharide donor specificities
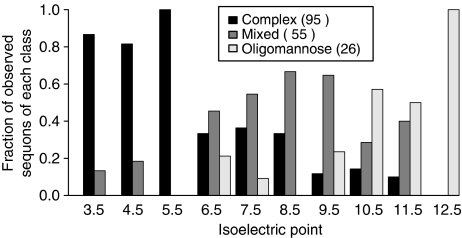
To establish if there are consensus sequences or patterns that control site-specific N-glycosylation by TbSTT3A or TbSTT3B, we developed a mass spectrometry approach to analyse wild-type bloodstream form T. brucei glycoproteins, summarised in ‘Materials and methods' and in Supplementary Figure S4. Briefly, the glycoproteins were captured from VSG-depleted trypanosome lysates by sequential affinity chromatography on immobilised ricin followed by immobilised ConA. The ricin- and ConA-binding fractions were eluted with appropriate sugars and individually processed by digestion with endoH followed by PNGaseF. Thus, endoH-sensitive glycopeptides appear 203 Da heavier by mass spectrometry because of the GlcNAc residue left attached to the relevant Asn residues by endoH whereas the remaining endoH-resistant, but PNGaseF-sensitive, sites appear 1 Da heavier by mass spectrometry because of the conversion of the relevant Asn residues to Asp by PNGaseF. Using this method, we noticed that many of the amino-acid sequences immediately adjacent to the sequons presenting complex or paucimannose N-glycans contained aspartic and/or glutamic acid residues, suggesting that charge might have a function in sequon recognition by TbSTT3A. To test this hypothesis, the isoelectric points (pIs) of the sequons themselves, plus five residues N- and C-terminal to them, were calculated. The exclusively EndoH-resistant sites, that is those modified by TbSTT3A, tend to be found within more acidic regions of protein sequence (mean±s.d. pI of 4.8±1.7) whereas the exclusively EndoH-sensitive sites, that is those modified by TbSTT3B, tend to be within neutral to basic regions (mean±s.d. pI of 9.5±2.2). The mixed sites, that is those that have been partly modified by TbSTT3A and partly by TbSTT3B, are relatively evenly spread between these pI extremes (Figure 5; Supplementary Tables SV and SVI). Therefore, the different polypeptide acceptor substrate specificities of TbSTT3A and TbSTTB seem to be, at least in part, related to the pIs of the peptide sequences immediately around the N-glycosylation sites.
Figure 5.
Distribution of the type of N-glycan occupancy related to the isoelectric point (pI) of the acceptor site peptide. The predicted pI values for each sequon ±5 amino acids were binned in unit intervals from 3 to 13. For each interval, bars are shown to indicate the observed proportions of sites modified with exclusively EndoH-resistant glycans (black), EndoH-sensitive glycans (white), both EndoH-resistant and EndoH-sensitive glycans (grey).
TbSTT3C expressed in yeast seems to have a similar preference for acidic acceptor sequences to TbSTT3A expressed in T. brucei
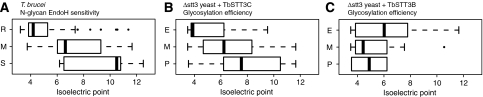
As mentioned earlier, we cannot make a direct comparison between the specificities of TbSTT3A and TbSTT3C because TbSTT3A does not complement Δstt3 yeast and TbSTTC is not expressed in trypanosomes. However, in T. brucei, sites receiving endoH-resistant paucimannose/complex glycans through TbSTT3A are relatively acidic and the same is true of sites exhibiting high glycosylation efficiency in Δstt3+TbSTT3C yeast cells (Supplementary Table S1). This is apparent from the data plotted in (Figure 6). Figure 6A shows the distributions of pIs of the acceptor sequons (±5 amino acids) for sites recognised principally by TbSTT3A (endoH-resistant sites), principally by TbSTT3B (endoH-sensitive sites) and by both (mixed sensitivity sites) in T. brucei. Figure 6B and C show the comparable distributions of pIs for sites that are efficiently (>80%), poorly (<20%) and moderately (20–80%) glycosylated in Δstt3+TbSTT3C and Δstt3+TbSTT3B yeast cells, respectively. These data illustrate the similar preferences of TbSTT3C and TbSTT3A for acidic sites, and the lack of any obvious preference of TbSTT3B when it is expressed in either Δstt3 yeast or in T. brucei.
Figure 6.
Correlations of N-glycan type in T. brucei and N-glycosylation efficiency in Δstt3 yeast with acceptor site pI. (A) Box plot (Tukey, 1977) of predicted pI distributions for the sequons ±5 amino acids occupied by endoH-resistant (R), mixed (M) or endoH-sensitive (S) N-glycans. (B, C) Box plots of predicted pI distributions for the sequons ± 5 amino acids exhibiting efficient (E), moderate (M) and poor (P) N-glycosylation in Δstt3 +TbSTT3C and TbSTT3B yeast cells, respectively. Note: Box plots are concise ways of illustrating a distribution of values. The black line shows the median, the box encompasses 50% of the data, the whiskers, 75% of the data and the outliers are shown as discrete points.
Both TbSTT3A and TbSTT3B catalysed N-glycosylation mechanisms are essential for infectivity in mice
Both N-glycosylation mechanisms are, individually, non-essential for parasite growth in culture (Figures 3B and 4B). However, we found that simultaneous knockdown of TbSTT3A and TbSTT3B halted the growth of bloodstream form trypanosomes in culture (Supplementary Figure S1F), showing that protein N-glycosylation per se is essential in culture.
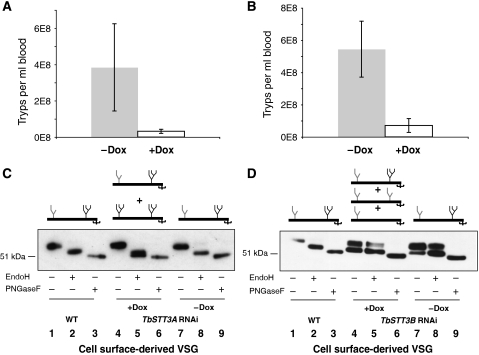
We next investigated whether the TbSTT3A/TbSTT3B redundancy seen in culture was also evident in animal infections. Although both RNAi cell lines grew well in doxycycline-free mice, the infections in the presence of RNAi-inducing doxycycline were very low in both cases (Figure 7A and B). Western blot analysis of the VSG221 from the few surviving cells showed that they were expressing significant amounts of wild-type N-glycosylated VSG221 (Figure 7C and D). Most likely these cells proliferated because they had lost some of the effects of the doxycycline-induced TbSTT3 knockdown, a well-known phenomenon in T. brucei RNAi experiments. These data suggest that both mechanisms of protein N-glycosylation are independently required for parasite growth in mice.
Figure 7.
Animal infectivity studies with STT3ARNAi and STT3BRNAi cell lines. (A) T. brucei TbSTT3A,B,C+/−-STT3ARNAi cells or (B) TbSTT3A,B,C+/−-STT3BRNAi cells present in the blood of mice dosed with (white bar) and without doxycycline (grey bar) for 2 days before and throughout the 3 day infection. (C) Purified sVSG221 of wild-type cells grown in culture (lanes 1–3, as a control), and TbSTT3A,B,C+/−-STT3ARNAi plus doxycycline (lanes 4–6) and TbSTT3A,B,C+/−-STT3ARNAi minus doxycycline (lanes 7–9) cells isolated from mouse blood, digested or not, as indicated, with EndoH or PNGaseF were subjected SDS–PAGE and western blot with anti-VSG221 antibody. (D) Purified sVSG221 of wild-type cells grown in culture (lanes 1–3, as a control), and TbSTT3A,B,C+/−-STT3BRNAi plus doxycycline (lanes 4–6), and TbSTT3A,B,C+/−-STT3BRNAi minus doxycycline (lanes 7–9) cells isolated from mouse blood, digested or not, as indicated, with EndoH or PNGaseF were subjected SDS–PAGE and western blot with anti-VSG221 antibody.
Discussion
In common with the related kinetoplastids T. cruzi and L. major (Ivens et al, 2005), and with the unrelated protist Giardia lamblia (Morrison et al, 2007), the genome of T. brucei lacks identifiable homologues of all known OTase subunits except for the catalytic subunit, STT3, for which it has three intact genes, TbSTT3A,B and C (Berriman et al, 2005; Kelleher and Gilmore 2006). The goal of this work was to elucidate the specificities and roles of the three TbSTT3 proteins in T. brucei.
Although we cannot formally exclude the possibility that heterologous expression of TbSTT3s in yeast might affect their specificities, or that STT3 protein levels necessarily correlate directly with enzymatic activity, the heterologous complementation of Stt3p mutant yeast strains with STT3 proteins from protozoan organisms seems, thus far, to be a good experimental system to study STT3 function and specificity (Shams-Eldin et al, 2005; Castro et al, 2006; Nasab et al, 2008; Hese et al, 2009). Studies on the four STT3 genes of L.major showed that three of them complement the yeast Δstt3 mutant with different efficiencies, that they can function without association with other yeast OTase subunits (Nasab et al, 2008; Hese et al, 2009) and that they have overlapping but distinct peptide substrate specificities (Nasab et al, 2008; Hese et al, 2009). Similar ex vivo studies carried out here showed that TbSTT3B and TbSTT3C are functional single-subunit OTases able to rescue the yeast ΔStt3 mutant. Furthermore, the glycosylation site occupancy data highlighted the different polypeptide acceptor specificities of TbSTT3B and TbSTT3C and, in particular, a preference for TbSTT3C (which rescued the yeast Δstt3 mutant at the expense of a severe growth phenotype) for acceptor sequons in relatively acidic local environments. Despite these successes, the ex vivo approach provided no information on TbSTT3A because its acceptor and/or donor specificity is/are incompatible with the growth of Saccharomyces cerevisiae. To address this issue, we studied the expression of the TbSTT3 genes in the bloodstream and procyclic form T. brucei and the functions of the two expressed genes (TbSTT3A and TbSTT3B) in bloodstream form trypanosomes using inducible RNAi knockdown in bloodstream form parasites.
TbSTT3A is more highly expressed in bloodstream form than procyclic form T. brucei and its selective knockdown produced a profound glycosylation phenotype in bloodstream form parasites that show that TbSTT3A is largely or solely responsible for the transfer of the biantennary Man5GlcNAc2 precursor during the biosynthesis of glycoproteins in the ER of T. brucei. The data also confirmed that the processing of biantennary Man5GlcNAc2 attached to protein is the only route to complex N-glycans in this organism (Manthri et al, 2008). In contrast, TbSTT3B is highly expressed in both life cycle stages and its selective knockdown in bloodstream form parasites showed that TbSTT3B is largely or solely responsible for the transfer of the triantennary Man9GlcNAc2 precursor during the biosynthesis of glycoproteins in the ER of T. brucei and confirmed that the processing of triantennary Man9GlcNAc2 attached to protein is the only route to oligomannose N-glycans in this organism (Manthri et al, 2008). As noted earlier (Manthri et al, 2008), the strict demarcation between these different routes to complex and oligomannose N-glycans seems to be due to the absence of the Golgi mannosidase II activity that permits the conversion of triantennary Man5GlcNAc2 to Man3GlcNAc2 in other eukaryotes.
This clear selectivity for different LLO donors by TbSTT3A and TbSTT3B is novel. Mammalian and yeast OTase complexes, which operate in organisms with full repertoires of the ALG genes responsible for LLO biosynthesis, are selective for Glc3Man9GlcNAc2-PP-Dol whereas protists and fungi that lack groups of ALG genes (Samuelson et al, 2005) display either little specificity for donor LLO structure or are tuned to selectively transfer the end LLO product of their modified dolichol cycles (Kelleher et al, 2007). In the case of T. brucei, the end product of LLO synthesis is Man9GlcNAc2-PP-Dol and TbSTT3B does indeed have a preference for this donor whereas TbSTT3A has specificity for the first topologically competent LLO intermediate Man5GlcNAc2-PP-Dol.
The two OTases also differ substantially in apparent peptide acceptor substrate specificity, with TbSTT3A showing unusual selectivity for glycosylation sequons flanked by acidic residues. TbSTT3C also showed a similar preference for acidic sites; however, it is difficult to compare TbSTT3A and TbSTT3C because the two could not be studied in the same organism. Nevertheless, it is tempting to speculate that TbSTT3C (which is more closely related to TbSTT3B) might have hybrid properties, perhaps sharing the features of the acidic peptide acceptor specificity of TbSTT3A with the more conventional Man9GlcNAc2-PP-Dol LLO donor specificity of TbSTT3B. It may be that TbSTT3C is expressed at specific parasite life cycle stage(s) when the organism might require oligomannose structures attached to relatively acidic glycosylation sites. In this respect, it is worth noting that the cell-surface BARP glycoproteins, which are specifically expressed in the epimastigote form of the parasite in the tsetse salivary glands (Urwyler et al, 2007), present one or two relatively acidic N-glycosylation sites per molecule.
Analysis of VSG glycosylation in the TbSTT3A RNAi knockdown shows that, in the absence of TbSTT3A, TbSTT3B is able to transfer Man9GlcNAc2 to the acidic Asn263 site rather efficiently, such that most of the VSG in the ER contains Man9GlcNAc2 at both Asn263 and Asn428 (Figure 4C). This is consistent with our model that TbSTT3A rapidly modifies acidic sites, possibly in a co-translational fashion, whereas TbSTT3B subsequently glycosylates any remaining sites, regardless of pI. In other words, TbSTT3A has distinct acceptor peptide substrate specificity, whereas TbSTT3B has very broad specificity. The latter is also borne out by the yeast data in Figure 1. This helps to explain why in the procyclic form of the parasite, which expresses predominantly TbSTT3B, the relatively acidic N-glycosylation sites of the procyclins are occupied by triantennary oligomannose Man5GlcNAc2 glycans (Treumann et al, 1997) and why the lysosomal glycoprotein p67 contains complex N-glycans in the bloodstream form of the parasite but exclusively oligomannose structures in the procyclic form (Kelley et al, 1995.
It is interesting to note that, though in the TbSTT3A knockdown the ER resident VSG is predominantly modified with oligomannose glycans at both Asn263 and Asn428, the mature cell-surface VSG is made up of these mutant VSG glycoforms plus wild-type VSG glycoforms (Figures 3D and 4D). Presumably, the incomplete knockdown of TbSTT3A mRNA allows some wild-type VSG to be synthesised and this is enriched during the process of ER exit and/or transit to the cell surface. It is possible that incorrect glycosylation of the Asn263 site might destabilise VSG221 and lead to its degradation. This site seems to be particularly important in VSG221 because the wild-type paucimannose and small complex N-glycans at Asn263 occupy the same space as a short α-helix of peptide in other VSG variants (Blum et al, 1993) and because this site seems to interact selectively with the calreticulin/UDP-glucose:glycoprotein glucosyltransferase protein folding/quality control system (Izquierdo et al, 2009).
Analysis of the VSG molecules in the TbSTT3B RNAi knockdown shows that, in the absence of TbSTT3B, TbSTT3A barely modifies Asn428 at all (Figures 3F, 4C and D) and, when it does, it transfers exclusively Man5GlcNAc2, leading to the synthesis of complex glycans at this site. The underglycosylation of Asn428 in the TbSTT3B knockdown is more severe than that seen in the TbALG3 null mutant, which has normal levels of TbSTT3A and TbSTT3B but can only make Man5GlcNAc2-PP-Dol (Manthri et al, 2008). This indicates that TbSTT3B that normally uses Man9GlcNAc2-PP-Dol can in its absence use Man5GlcNAc2-PP-Dol, albeit relatively inefficiently, to glycosylate Asn428.
In summary, the available data show that (i) TbSTT3A has strict specificity for the Man5GlcNAc2-PP-Dol LLO donor and is highly selective for acidic peptide acceptor sites. (ii) TbSTT3B is selective, but not specific, for the Man9GlcNAc2-PP-Dol LLO donor and has broad specificity for peptide acceptor sites. (iii) TbSTT3C can use Glc3Man9GlcNAc2-PP-Dol donor and has more selectivity for acidic peptide acceptors than TbSTT3B. Although we conclude that local pI around the glycosylation sequon correlates with sequon usage by the different TbSTT3s, we acknowledge that other factors, such as position in the polypeptide and, thus, local secondary and tertiary structure are also likely to have significant functions in differential N-glycosylation in T. brucei. These aspects are currently under investigation.
Some subunits of the multiprotein complex OTase of higher eukaryotes tune protein- or site-specific N-glycosylation activity (Wilson and High, 2007; Ruiz-Canada et al, 2009; Schulz et al, 2009). This additional control or regulation of OTase activity possibly arises from an evolutionary advantage in some organisms for increased diversity of glycosylation sites. Although higher eukaryotes may have achieved this by the addition of proteins to the OTase complex, the data presented here support the hypothesis that gene duplication and diversification of STT3 activity and specificity have occurred in T. brucei. Regardless of the reason, the dual N-glycosylation mechanism used by T. brucei produces an extraordinarily high N-glycosylation efficiency, with 96% of all sites occupied as compared to ∼50–60% for other eukaryotes (Petrescu et al, 2004). It is remarkable that minor differences in amino-acid sequence, especially between TbSTT3B and TbSTT3C, lead to substantial alterations in their polypeptide acceptor substrate specificities (Supplementary Figure S2). The WWDYG motif and the surrounding amino-acid sequences are totally conserved in the three proteins, whereas the residues in and near the DxxK motif (Igura et al, 2008) present more variability. The novel LLO donor and acceptor peptide specificities of the T. brucei STT3 proteins certainly raise possibilities for their usage in glycoprotein engineering in heterologous systems.
The exact reason(s) why T. brucei has evolved parallel and selective routes to oligomannose and complex glycan synthesis is not clear. However, it could have occurred to compensate for T. brucei's apparent loss of Golgi mannosidase II genes (which disallows complex glycan formation) and/or to provide a ‘fast-track' to paucimannose/complex glycan synthesis from Man5GlcNAc2-PP-Dol. The latter might provide a selective advantage when one considers that about 50% of the N-glycosylation sites across all of its highly abundant VSG molecules are paucimannose or complex (Mehlert et al, 1998). Thus, fast tracking would minimise the drain on Dol-P-Man donors (necessary for converting Man5GlcNAc2-PP-Dol to Man9GlcNAc2-Dol) that are also required for GPI anchor biosynthesis, which is prodigious in this organism because it makes and turns over a 10-fold excess of GPI precursors (Ralton et al, 1993). Thus, overall, one might speculate that the unusual physiology of this protozoan parasite, which must synthesise an entire contiguous coat of protective surface glycoprotein to survive, may have provided the evolutionary pressure for N-glycosylation acceptor sites and OTase specificities to co-evolve.
Finally, it is surprising that the separate knockdown of TbSTT3A and TbSTT3B, with consequent disappearance of most paucimannose/complex or oligomannose N-glycans, respectively, from all glycoproteins in the parasite can be tolerated in culture, with only a marginal growth phenotype in both cases. On the other hand, both STT3 activities are independently essential in a mouse infection model. This result emphasises that gene functionality should be tested in vivo, that is in an animal model of infection, before making final conclusions about gene essentiality. The essentiality of both TbSTT3A and TbSTT3B in vivo and their novel specificities (particularly that of TbSTT3A) may offer potential therapeutic targets for future exploitation.
Materials and methods
TbSTT3 plasmid constructs
Each of the three TbSTT3 ORFs were amplified from genomic DNA by PCR and cloned into a pGEM vector; and into a pBAD Escherichia coli expression vector, introducing a C-terminal myc-His6 tag; and then into a pRS425_GPD_CYC1 yeast expression vector (pTbSTT3A, pTbSTT3B and pTbSTT3C).
Yeast manipulation
Yeast strains used were Y24390 (EUROSCARF, Frankfurt, Germany), YBS1 (Mat@ his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 Δstt3∷kanMX4 pSTT3), YBS2 (Mat@ his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 Δstt3∷kanMX4 pTbSTT3B) and YBS3 (Mat@ his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 Δstt3∷kanMX4 pTbSTT3C) (Nasab et al, 2008). Cells were grown at permissive temperature in YPD; for maintaining plasmids, in minimal medium; for selection of kanMX4 carrying strains with 100 μg/ml G418; or for selection of ura-cells with 5-fluoroorotic acid (5-FOA) (Boeke et al, 1987) and 1 M sorbitol. Haploid cells with genomic deletion of STT3 and expressing a TbSTT3 were created essentially as described (Castro et al, 2006), but using pSTT3 (Zufferey et al, 1995) containing the yeast STT3 locus in a URA3 YEp352 vector. Addition of 5-FOA to the media allowed for selection of cells that had lost the URA plasmid encoding the wild-type yeast STT3. These cells could only survive if expression of a plasmid-borne T. brucei STT3 paralogue complemented the lack of yeast STT3.
Site-specific glycosylation occupancy determination
Proteins covalently linked to the cell wall polysaccharide matrix were prepared from yeast cells, detected with liquid chromatography–electrospray ionisation–tandem mass spectrometry (LC-ES-MS/MS), and analysed as described earlier (Schulz and Aebi, 2009). Glycosylation sites with more than 95% occupancy were defined as being ‘efficiently' glycosylated.
Cultivation of trypanosomes
Bloodstream form T. brucei genetically modified to express T7 polymerase and the tetracycline repressor protein were cultivated in HMI-9 medium as described in Wirtz et al (1999).
Generation of constructs and transfomation of bloodstream form T. brucei
530-bp 5′- and 406-bp 3′-UTR sequences were PCR amplified from genomic DNA using HiFi Taq Platinum with 5′-atgCGGCCGcgaacggatggcagtcatttgc-3′ and 5′-gtttaaacttacggaccgtcaagcttgtacgtataaggagggg-3′ and 5′-gacggtccgtaagtttaaacggatccgttatgctgttgttaggg-3′ and 5′-ataagtaaGCGGCCGCaatacgcaacctccacaagc-3′ as forward and reverse primers, respectively. The two PCR products were used together in a further PCR reaction to yield a product containing the 5′-UTR linked to the 3′-UTR by a short HindIII and BamHI cloning site (underlined) and SalI and NotI restriction sites at each end (capital letters). The PCR product was cloned into the SalI and NotI sites of the pGEM-5Zf(+) vector (Promega) and the PAC drug resistance gene, with exogenous T7 promoter and terminator sequences, was introduced into the targeting vector through the HindIII and BamHI sites. For RNAi internal coding sequences were PCR amplified from genomic DNA using HIFI Taq Platinum with 5′-cgcGGATCCccctctttgggggtgc-3′ and 5′-ccgCTCGAGattgggtagatcagtcacg -3′ (BamHI and XhoI sites, respectively, in capital letters) for Tb927.5.890 (TbSTT3A), and 5′-cgcGGATCCgatgatttctttggttacc -3′ and 5′-ccgCTCGAGctcagaatatatccggaa -3′ (BamHI and XhoI sites, respectively, in capital letters) for Tb927.5.900 (TbSTT3B). The 498 nt (TbSTT3A) and 467 nt (TbSTT3A) internal coding sequences were cloned into p2T7TA (Alibu et al, 2005), cut with BamHI and XhoI, to generate p2T7-STT3A and p2T7-STT3B. These plasmids were then digested with NotI, and purified for transfection. Finally, for RNAi of TbSTT3A/B/C a 493 nt internal coding sequence was PCR amplified from genomic DNA using HIFI Taq Platinum with 5′-cgcGGATCCgtagccattcatcgtgtgc -3′ and 5′-ccgCTCGAGggtacgcaaatcgcaagg -3′ (BamHI and XhoI sites, respectively, in capital letters) and cloned into p2T7TA, to generate p2T7-TbSTT3All. These plasmids were then digested with NotI, precipitated and washed in 70% ethanol, dissolved in sterile distilled water, and used for T. brucei transformation (Wirtz et al, 1999).
Northern blotting
Total RNA for Northern blots was prepared using Qiagen RNeasy Midi kits. Samples of RNA (5 μg) were run on formaldehyde–agarose gels and transferred to Hybond N nylon membrane (Amersham Biosciences) for hybridisation with [α-32P] dCTP-labeled T. brucei Tb927.5.890 or Tb927.5.900 probe (Amersham Rediprime II).
Small-scale soluble-form VSG isolation
Soluble-form VSG (sVSG) was isolated from 100 ml cultures containing ∼2 × 108 bloodstream form T. brucei as described in Cross (1984).
ES-MS analysis of intact and Pronase digested VSG
Samples of the sVSG preparations were diluted to ∼0.07 μg/μl in 50% acetonitrile, 1% formic acid, loaded into nanotips (Micromass type F), and analysed by positive ion ES-MS on a Q-Star XL instrument (Applied Biosystems). Data were collected and processed using the Bayesian protein reconstruction algorithm of Analyst software. Samples of sVSG were digested with Pronase and the resulting glycopeptides enriched and analysed by ES-MS and ES-MS/MS as described in Manthri et al (2008).
ER–VSG SDS–PAGE and western blotting
sVSG-depleted T. brucei ghosts (Bohme and Cross, 2002), equivalent to ∼2 × 105 cells were subjected to SDS–PAGE and transferred to polyvinylidene difluoride Hybond-P membranes (Amersham Biosciences). The membranes were incubated for 1 h with anti-VSG221 rabbit polyclonal antibody diluted 1:4000 in PBS and then 1 h with anti-rabbit conjugated to horseradish peroxidase diluted 1:10000 with the same buffer. Membranes were developed with ECL (Amersham Biosciences) according to the manufacturer's instructions.
Lectin blotting of T. brucei cell extracts
Bloodstream form T. brucei cells were washed twice in trypanosome dilution buffer, solubilised with 2% SDS and 4 M urea, subjected to SDS–PAGE and transferred to nitrocellulose membranes (Amersham Biosciences). The membranes were incubated for 1 h with biotin-conjugated ricin (Vector Labs) diluted 1/3000, with or without inhibitors (10 mg/ml galactose and 10 mg/ml lactose) or 1 h with ConA (Sigma) diluted 1/3000, with or without inhibitor (0.5 M α-methyl mannose). Then, membranes were incubated for 1 h with extravidin-HRP diluted 1/10000, and developed with ECL (Amersham Biosciences) according to the manufacturer's instructions.
Mouse infection studies
The RNAi inducible cells were subcultured and grown without selection drugs for 48 h with and without 1 μg/ml tetracycline. The parasites were then introduced into groups of five mice (dosed with and without doxycycline, respectively) by intraperitoneal injection of 3 × 105 parasites. The plus doxycycline group of animals were dosed with doxycycline in the drinking water (0.2 mg/ml in a 5% sucrose solution) for 1 week before infection and until the experiment was terminated. Infections were assessed by tail bleeding and cell counting.
Extraction of ricin and ConA-binding glycoproteins
Bloodstream form T. brucei, were isolated from infected rats, purified over DEAE-cellulose and subjected to hypotonic lysis to release cytosolic components as well as the majority of the VSG coat as sVSG (Cross, 1984). The cell ghost pellets (1011 cell equivalents) were solubilised in 50 ml of 8 M urea, 3% SDS, 50 mM Tris–HCl, pH 6.8. The SDS/urea extract was diluted 50 times in buffer A (50 mM Tris–HCl, pH 6.8, 400 mM NaCl, and 0.8% Triton X-100, 0.1 mM TLCK, 1 μg/ml leupeptin and 0.1% sodium azide). Ricin-coupled agarose (Vector Laboratories, 4-ml packed volume) was added to the suspension, rotated gently overnight at 4°C, recovered by gentle centrifugation and packed into a 5 cm × 1 cm column. The column was washed with 5 volumes of buffer A and bound material was eluted with 30 mg/ml lactose and 30 mg/ml galactose in 4-fold diluted buffer A. ConA-coupled Agarose (Vector Laboratories) was added to the recovered supernatant from the earlier pull-down with Ricin–Agarose adjusted earlier to 150 mM NaCl, 5 mM CaCl2, 5 mM MnCl2 and 1 mM MgCl2, and recovered by gentle centrifugation and packed into a 5 cm × 1 cm column. Bound material was eluted with 0.5 M methyl-alpha-D-mannopyranoside in 4-fold diluted Buffer A.
Sequential endoglycosidase digestion of eluted glycoproteins and mass spectrometry analysis
Aliquots of the eluted Ricin-bound and ConA-bound fractions were independently subjected to EndoH followed by PNGaseF digestion (Roche Applied Science), subjected to SDS–PAGE and the gel lanes were then divided into 10 independent slices. Proteins in each slice were excised, reduced and alkylated, by the addition of 10 mM DTT and then 50 mM iodoacetamide. The samples were digested with 25 μl of 12.5 μg/ml modified trypsin (Roche Applied Science) in 20 mM ammonium bicarbonate. An aliquot of the tryptic digest (recovered in 1% formic acid in water) was analysed by LC-MS on an LTQ-Orbitrap mass spectrometer (ThermoElectron) coupled to a Dionex 3000 nano-LC system. The Orbitrap was set to analyse the survey scans at 60 000 resolution and the top five ions in each duty cycle selected for MS/MS in the LTQ linear ion trap. The raw files were processed to generate a Mascot generic file using the program Raw2msm and searched against an in-house curated T. brucei database using the Mascot search engine v.2.2 (Matrix Science) run on an in-house server using the following criteria: peptide tolerance=6 p.p.m., trypsin as the enzyme and carboxyamidomethylation of cysteine as a fixed modification. Variable modifications were oxidation of methionine, N-acetylglucosamine modification of Asn and deamidation of Asn to Asp.
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
We thank Doug Lamont, FingerPrints Proteomics Facility (supported by the Wellcome Trust Strategic Award 083481) and Angela Mehlert and Daniel Turnock for advice and assistance. LI and MLSG were supported by a Wellcome Trust Programme Grant (085622) to MAJF. JR was supported by an MRC Programme Grant to MAJF and GJB. JBP was supported by the European Network of Excellence ENFIN (contract LSHG-CT-2005-518254) to GJB. BLS and MA were supported by the Swiss National Science Foundation, grant 3100A0-105541 to MA, and the GlycoInit initiative of ETH, Zurich, and thank the Functional Genomics Center Zurich for their input.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alibu VP, Storm L, Haile S, Clayton C, Horn D (2005) A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Mol Biochem Parasitol 139: 75–82 [DOI] [PubMed] [Google Scholar]
- Atrih A, Richardson JM, Prescott AR, Ferguson MA (2005) Trypanosoma brucei glycoproteins contain novel giant poly-N-acetyllactosamine carbohydrate chains. J Biol Chem 280: 865–871 [DOI] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ et al. (2005) The genome of the African trypanosome Trypanosoma brucei. Science 309: 416–422 [DOI] [PubMed] [Google Scholar]
- Blum ML, Down JA, Gurnett AM, Carrington M, Turner MJ, Wiley DC (1993) A structural motif in the variant surface glycoproteins of Trypanosoma brucei. Nature 362: 603–609 [DOI] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol 154: 164–175 [DOI] [PubMed] [Google Scholar]
- Bohme U, Cross GA (2002) Mutational analysis of the variant surface glycoprotein GPI-anchor signal sequence in Trypanosoma brucei. J Cell Sci 115: 805–816 [DOI] [PubMed] [Google Scholar]
- Brewer CF, Bhattacharyya L (1986) Specificity of concanavalin A binding to asparagine-linked glycopeptides. A nuclear magnetic relaxation dispersion study. J Biol Chem 261: 7306–7310 [PubMed] [Google Scholar]
- Cardoso de Almeida ML, Turner MJ (1983) The membrane form of variant surface glycoproteins of Trypanosoma brucei. Nature 302: 349–352 [DOI] [PubMed] [Google Scholar]
- Castro O, Movsichoff F, Parodi AJ (2006) Preferential transfer of the complete glycan is determined by the oligosaccharyltransferase complex and not by the catalytic subunit. Proc Natl Acad Sci USA 103: 14756–14760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross GA (1975) Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology 71: 393–417 [DOI] [PubMed] [Google Scholar]
- Cross GA (1984) Release and purification of Trypanosoma brucei variant surface glycoprotein. J Cell Biochem 24: 79–90 [DOI] [PubMed] [Google Scholar]
- Ferguson MAJ, Halder K, Cross GAM (1985) Trypanosoma brucei variant surface glycoprotein has a sn-1,2-dimyristylglycerol membrane anchor at its COOH-terminus. J Biol Chem 260: 4963–4968 [PubMed] [Google Scholar]
- Ferguson MAJ, Duszenko M, Lamont GS, Overath P, Cross GA (1986) Biosynthesis of Trypanosoma brucei variant surface glycoproteins. N-glycosylation and addition of a phosphatidylinositol membrane anchor. J Biol Chem 261: 356–362 [PubMed] [Google Scholar]
- Gavel Y, von Heijne G (1990) Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng 3: 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Aebi M (2004) Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem 73: 1019–1049 [DOI] [PubMed] [Google Scholar]
- Hese K, Otto C, Routier FH, Lehle L (2009) The yeast oligosaccharyltransferase complex can be replaced by STT3 from Leishmania major. Glycobiology 19: 160–171 [DOI] [PubMed] [Google Scholar]
- Igura M, Maita N, Kamishikiryo J, Yamada M, Obita T, Maenaka K, Kohda D (2008) Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J 27: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G et al. (2005) The genome of the kinetoplastid parasite, Leishmania major. Science 309: 436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo L, Atrih A, Rodrigues JA, Jones DC, Ferguson MA (2009) Trypanosoma brucei UDP-glucose: glycoprotein glucosyltransferase has unusual substrate specificity and protects the parasite from stress. Eukaryot Cell 8: 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DC, Mehlert A, Guther ML, Ferguson MA (2005) Deletion of the glucosidase II gene in Trypanosoma brucei reveals novel N-glycosylation mechanisms in the biosynthesis of variant surface glycoprotein. J Biol Chem 280: 35929–35942 [DOI] [PubMed] [Google Scholar]
- Kasturi L, Eshleman JR, Wunner WH, Shakin-Eshleman SH (1995) The hydroxy amino acid in an Asn-X-Ser/Thr sequon can influence N-linked core glycosylation efficiency and the level of expression of a cell surface glycoprotein. J Biol Chem 270: 14756–14761 [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Banerjee S, Cura AJ, Samuelson J, Gilmore R (2007) Dolichol-linked oligosaccharide selection by the oligosaccharyltransferase in protist and fungal organisms. J Cell Biol 177: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R (2006) An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16: 47R–62R [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Karaoglu D, Mandon EC, Gilmore R (2003) Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Mol Cell 12: 101–111 [DOI] [PubMed] [Google Scholar]
- Kelley RJ, Brickman MJ, Balber AE (1995) Processing and transport of a lysosomal membrane glycoprotein is developmentally regulated in African trypanosomes. Mol Biochem Parasitol 74: 167–178 [DOI] [PubMed] [Google Scholar]
- Manthri S, Guther ML, Izquierdo L, Acosta-Serrano A, Ferguson MA (2008) Deletion of the TbALG3 gene demonstrates site-specific N-glycosylation and N-glycan processing in Trypanosoma brucei. Glycobiology 18: 367–383 [DOI] [PubMed] [Google Scholar]
- Mehlert A, Zitzmann N, Richardson JM, Treumann A, Ferguson MA (1998) The glycosylation of the variant surface glycoproteins and procyclic acidic repetitive proteins of Trypanosoma brucei. Mol Biochem Parasitol 91: 145–152 [DOI] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, Davids BJ, Dawson SC, Elmendorf HG, Hehl AB, Holder ME, Huse SM, Kim UU, Lasek-Nesselquist E, Manning G, Nigam A et al. (2007) Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317: 1921–1926 [DOI] [PubMed] [Google Scholar]
- Mowatt MR, Clayton CE (1987) Developmental regulation of a novel repetitive protein of Trypanosoma brucei. Mol Cell Biol 7: 2838–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasab FP, Schulz BL, Gamarro F, Parodi AJ, Aebi M (2008) All in one: Leishmania major STT3 proteins substitute for the whole oligosaccharyltransferase complex in Saccharomyces cerevisiae. Mol Biol Cell 19: 3758–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo K, Marth JD (2006) Glycosylation in cellular mechanisms of health and disease. Cell 126: 855–867 [DOI] [PubMed] [Google Scholar]
- Petrescu AJ, Milac AL, Petrescu SM, Dwek RA, Wormald MR (2004) Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology 14: 103–114 [DOI] [PubMed] [Google Scholar]
- Ralton JE, Milne KG, Guther ML, Field RA, Ferguson MA (1993) The mechanism of inhibition of glycosylphosphatidylinositol anchor biosynthesis in Trypanosoma brucei by mannosamine. J Biol Chem 268: 24183–24189 [PubMed] [Google Scholar]
- Richardson JP, Beecroft RP, Tolson DL, Liu MK, Pearson TW (1988) Procyclin: an unusual immunodominant glycoprotein surface antigen from the procyclic stage of African trypanosomes. Mol Biochem Parasitol 31: 203–216 [DOI] [PubMed] [Google Scholar]
- Roditi I, Schwarz H, Pearson TW, Beecroft RP, Liu MK, Richardson JP, Buhring HJ, Pleiss J, Bulow R, Williams RO et al. (1989) Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J Cell Biol 108: 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Canada C, Kelleher DJ, Gilmore R (2009) Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136: 272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson J, Banerjee S, Magnelli P, Cui J, Kelleher DJ, Gilmore R, Robbins PW (2005) The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc Natl Acad Sci USA 102: 1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz BL, Aebi M (2009) Analysis of glycosylation site occupancy reveals a role for Ost3p and Ost6p in site-specific N-glycosylation efficiency. Mol Cell Proteomics 8: 357–364 [DOI] [PubMed] [Google Scholar]
- Schulz BL, Stirnimann CU, Grimshaw JPA, Brozzo MS, Fritsch F, Mohorko E, Capitani G, Glockshuber R, Grütter MG, Aebi M (2009) Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc Natl Acad Sci USA 106: 11061–11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shams-Eldin H, Blaschke T, Anhlan D, Niehus S, Muller J, Azzouz N, Schwarz RT (2005) High-level expression of the Toxoplasma gondii STT3 gene is required for suppression of the yeast STT3 gene mutation. Mol Biochem Parasitol 143: 6–11 [DOI] [PubMed] [Google Scholar]
- Sharon N, Lis H (1995) Lectins--proteins with a sweet tooth: functions in cell recognition. Essays Biochem 30: 59–75 [PubMed] [Google Scholar]
- Stokes MJ, Güther MLS, Turnock DC, Martin KL, Alphey MS, Ferguson MAJ (2008) The synthesis of UDP-N-acetylglucosamine is essential for bloodstream form Trypanosoma brucei in vitro and in vivo and UDP-N-acetylglucosamine starvation reveals a hierarchy in parasite protein glycosylation. J Biol Chem 283: 16147–16161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treumann A, Zitzmann N, Hulsmeier A, Prescott AR, Almond A, Sheehan J, Ferguson MA (1997) Structural characterisation of two forms of procyclic acidic repetitive protein expressed by procyclic forms of Trypanosoma brucei. J Mol Biol 269: 529–547 [DOI] [PubMed] [Google Scholar]
- Tukey JW (1977) Exploratory Data Analysis. Reading, MA, London: Addison-Wesley [Google Scholar]
- Urbaniak MD, Turnock DC, Ferguson MAJ (2006) Characterisation of galactose starvation in a bloodstream form Trypanosoma brucei UDP-Glucose 4′-epimerase conditional null mutant. Eukaryotic Cell 5: 1906–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwyler S, Studer E, Renggli CK, Roditi I (2007) A family of stage-specific alanine-rich proteins on the surface of epimastigote forms of Trypanosoma brucei. Mol Microbiol 63: 218–228 [DOI] [PubMed] [Google Scholar]
- Varki A (1993) Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3: 97–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CM, High S (2007) Ribophorin I acts as a substrate-specific facilitator of N-glycosylation. J Cell Sci 120: 648–657 [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 99: 89–101 [DOI] [PubMed] [Google Scholar]
- Zufferey R, Knauer R, Burda P, Stagljar I, te Heesen S, Lehle L, Aebi M (1995) STT3, a highly conserved protein required for yeast oligosaccharyl transferase activity in vivo. EMBO J 14: 4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File