Abstract
Many viruses use programmed –1 ribosomal frameshifting to express defined ratios of structural and enzymatic proteins. Pseudoknot structures in messenger RNAs stimulate frameshifting in upstream slippery sequences. The detailed molecular determinants of pseudoknot mechanical stability and frameshifting efficiency are not well understood. Here we use single-molecule unfolding studies by optical tweezers, and frameshifting assays to elucidate how mechanical stability of a pseudoknot and its frameshifting efficiency are regulated by tertiary stem-loop interactions. Mechanical unfolding of a model pseudoknot and mutants designed to dissect specific interactions reveals that mechanical stability depends strongly on triplex structures formed by stem-loop interactions. Combining single-molecule and mutational studies facilitates the identification of pseudoknot folding intermediates. Average unfolding forces of the pseudoknot and mutants ranging from 50 to 22 picoNewtons correlated with frameshifting efficiencies ranging from 53% to 0%. Formation of major-groove and minor-groove triplex structures enhances pseudoknot stem stability and torsional resistance, and may thereby stimulate frameshifting. Better understanding of the molecular determinants of frameshifting efficiency may facilitate the development of anti-virus therapeutics targeting frameshifting.
Keywords: optical tweezers, RNA triplexes, single-molecule, RNA folding
Messenger RNAs (mRNAs) designate proteins by sequences of codons consisting of 3 nucleotides each. The reading frame is defined by a start codon (AUG) and is usually maintained by ribosomes (with an error rate less than 3 × 10−5) (1). Programmed –1 ribosomal frameshifting (FS) has been found to express defined ratios of structural and enzymatic proteins in many viruses including HIV (2–6). Programmed FS has also been found during expression of cellular genes (6–8). Highly efficient FS at an mRNA slippery sequence from X XXY YYZ (0 frame) to XXX YYY Z (–1 frame) is often stimulated by a downstream pseudoknot structure (Fig. 1A). X can be any 3 identical nucleotides, Y can be either AAA or UUU, and Z is usually not G (6, 9–11). The slippery sequence and pseudoknot structure are typically separated by a single-stranded linker of 5–10 nucleotides. The natural high-efficiency FS stimulatory structure is often a hairpin (H)-type pseudoknot, which involves base pairing between nucleotides in a hairpin loop and nucleotides outside of the hairpin loop. It has been shown that tertiary minor-groove interactions between stem 1 and loop 2 (base triples) of pseudoknots are critical for programmed FS in certain viruses (12–16).
Fig. 1.
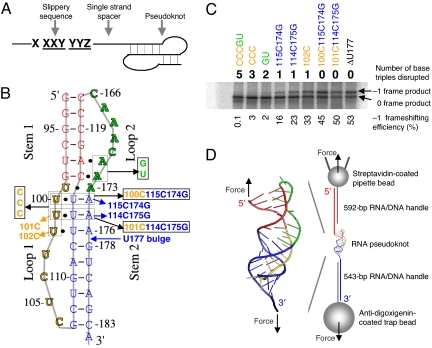
Minus-one ribosomal frameshifting (FS), pseudoknots, and experimental design. (A) Three components for programmed FS. (B) Pseudoknots used for bulk FS assay and single-molecule studies. All of the mutants were made based on ΔU177. The secondary structure of pseudoknot ΔU177 is also shown in Fig. 3C. In mutant CCCGU, all 5 base triples formed between stem 1 (shown in red) and loop 2 (shown in green) and between stem 2 (shown in blue) and loop 1 (shown in yellow) are disrupted. In TeloWT, the single nucleotide bulge U177 in stem 2 is added. (C) A typical SDS/PAGE result for the bulk FS assay. Shown on top and below the gel are the numbers of base triples disrupted and values of FS efficiency, respectively. (D) Experimental setup for mechanical (un)folding of RNA pseudoknots using optical tweezers. Pseudoknot ΔU177 is flanked by RNA/DNA hybrid handles, which are in turn attached to pipette bead and trap bead. The directions of applied force are shown with black arrows. Drawing is not to scale.
All of the secondary and tertiary structures in coding regions of mRNA have to be unfolded for translation. With the mRNA slippery sequence at the aminoacyl (A) and peptidyl (P) sites of the ribosome, the downstream pseudoknot structure is believed to be in contact with the helicase domain of the ribosome and provides mechanical resistance to ribosomal translocation (17–23). The detailed molecular determinants of pseudoknot mechanical stability and FS efficiency are not well understood.
Here we used an H-type pseudoknot derived from human telomerase RNA (ΔU177), which has a well-defined structure with extensive major-groove and minor-groove stem-loop interactions (24) as a model system to investigate the correlation among structure, mechanical stability, and FS efficiency. Mutants were designed to dissect the contributions of specific interactions of the major-groove base triples on mechanical stability and FS efficiency. Furthermore, the structural features of the folding intermediate states were mapped by combining single-molecule unfolding using optical tweezers and mutational studies.
Results
Bulk FS Assay In Vitro.
The high-resolution structure of pseudoknot ΔU177 with unusual 3 consecutive major-groove base triples (Fig. 1 B and D) (24) allows for the elucidation of the contributions of major-groove interactions to FS. The FS efficiency ranges from 53% for ΔU177 to essentially 0% for mutant CCCGU with all 5 base triples disrupted (Fig. 1C). Disruption of either 2 base triples in the minor groove (mutant GU) or 3 base triples in the major groove (mutant CCC) results in FS efficiency lower than 5%. Upon disruption of only 1 major-groove base triple, mutant 101C and mutant 102C have FS efficiencies of 11% and 33%, respectively. Upon substituting a major-groove U·A-U with a C+·G-C base triple (Fig. S1), the FS efficiencies of mutant 101C114C175G and mutant 100C115C174G remain above 40%. Clearly, all of the major-groove and minor-groove base triples are required for high-efficiency (>40%) FS.
Single-Molecule Mechanical Unfolding and Folding Using Optical Tweezers.
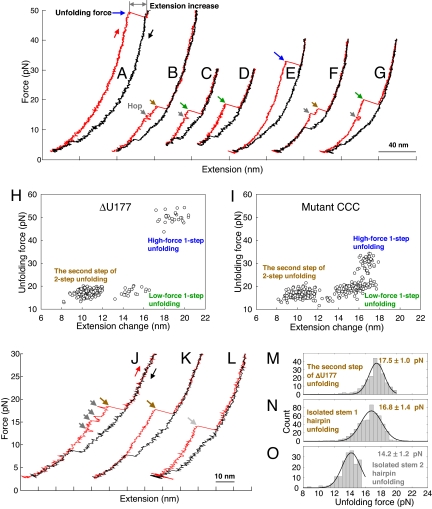
The pseudoknots (Fig. 1B) were also used to probe the effect of base triples on mechanical stability using optical tweezers (Fig. 1D, see Methods) (25, 26). The folding transitions are typically observed below 10 picoNewtons (pN) presumably due to the slow folding rate (Fig. 2 A–G). Three classes of ΔU177 unfolding reactions were observed: (1) 1-step unfolding at approximately 50 pN with extension increase of 19 nm (Fig. 2A); (2) 1-step unfolding at approximately 18 pN with extension increase of 15 nm (Fig. 2 C and D); and (3) 2-step unfolding with the second step unfolding force at approximately 18 pN with extension increase of 11 nm (Fig. 2 B and J). Three classes of unfolding reactions were also observed for mutant CCC (Fig. 2 E–G) and other pseudoknots (Fig. S2). The extension increase values for the 1-step reactions are well predicted by the Worm-Like-Chain model (27, 28) for the unfolding of the native pseudoknot. Presence of 3 classes of unfolding reactions is evident in the plots of unfolding force-versus-extension change (Fig. 2 H and I, and Fig. S3).
Fig. 2.
Representative force-extension curves and summary of mechanical unfolding results. Red and black curves are for pulling and relaxing, respectively, with waiting time at approximately 3 pN for 0 or 10 s. (A–D) Representative force-extension curves of ΔU177. (E–G) Representative force-extension curves of mutant CCC (with 3 major-groove U·A-U base triples disrupted, see Fig. 1B). (H) Summary of unfolding force-versus-extension change of ΔU177. (I) Summary of unfolding force-versus-extension change of mutant CCC. (J) A force-extension curve of ΔU177 folding intermediate structure with low-force 2-step unfolding. (K) A force-extension curve of isolated stem 1 hairpin derived from ΔU177 (see Fig. S4 A). (L) A force-extension curve of isolated stem 2 hairpin derived from ΔU177. (M) Unfolding force histogram of the second step of low-force 2-step unfolding of the folding intermediate structure of pseudoknot ΔU177. (N) Unfolding force histogram of isolated stem 1 hairpin derived from ΔU177 (see Fig. S4 B). (O) Unfolding force histogram of isolated stem 2 hairpin derived from ΔU177.
Stem 1 Forms Before Stem 2.
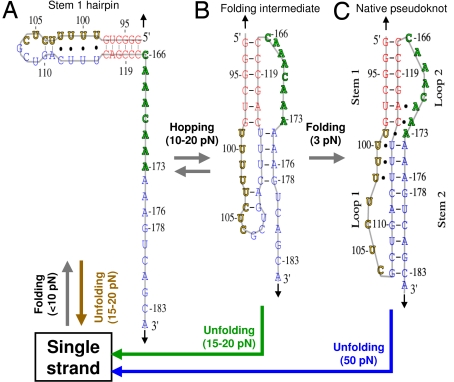
Equilibrium thermal unfolding/folding studies by UV absorbance-versus-temperature melting experiments, by slowly varying temperature (0.3–0.5 °C/minute from 5–98 °C), have suggested that stem 1 forms first followed by formation of stem 2 (24). To test if stem 1 forms before stem 2 in the mechanical folding of ΔU177, we made isolated stem 1 and stem 2 hairpins, using extended RNA/DNA handles to hybridize the 3′ and 5′ parts of ΔU177, respectively (Fig. S4). The unfolding force comparison (Fig. 2 J–O) clearly shows that the second step of ΔU177 unfolding (17.5 ± 1.0 pN) is similar to that of isolated stem 1 hairpin unfolding (16.8 ± 1.4 pN), not the isolated stem 2 hairpin unfolding (14.2 ± 1.2 pN). Thus, the second step of low-force 2-step unfolding corresponds to the disruption of stem 1 hairpin, that is, stem 1 is fully formed in the folding intermediate structure of ΔU177 (Fig. 3B) and other pseudoknots (see Fig. S5). The first step of low-force 2-step unfolding corresponds to the disruption of partially formed stem 2 (see below).
Fig. 3.
Possible folding intermediate structures of pseudoknot ΔU177. The positions and directions of applied force are indicated with black arrows. The unfolding transitions are indicated with the same colors as those in Fig. 2. (A) Stem 1 hairpin. In a force-ramp experiment, stem 1 hairpin typically folds from single strand when force is below 10 pN. Stem 1 hairpin unfolds to single strand at 15–20 pN. (B) A folding intermediate with stem 2 partially formed. In a force-ramp experiment, the folding intermediate rapidly forms after stem 1 hairpin is formed when force is below 10 pN. Upon increasing force to between 10 and 20 pN, the folding intermediate is in equilibrium with stem 1 hairpin and hopping between the 2 structures were observed. The folding intermediate may unfold apparently in 1 step or 2 steps at 15–20 pN. (C) Native pseudoknot. Folding transition from the folding intermediate to native pseudoknot is slow even at 3 pN as indicated by subsequent unfolding trajectories (see Fig. 2). Native pseudoknot unfolds to single strand in 1 step at approximately 50 pN.
Pseudoknots with Mutations in Loop 1 and Loop 2.
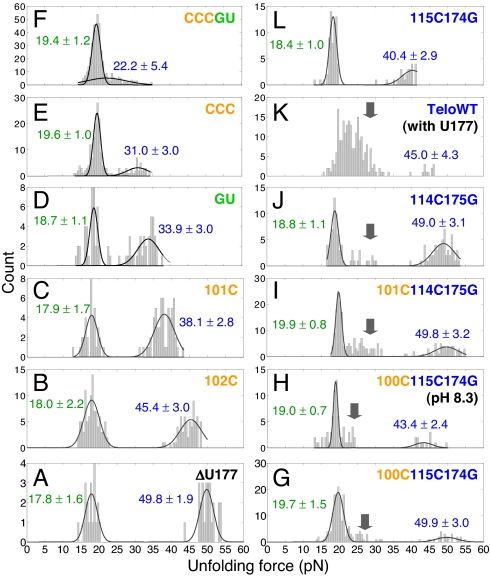
Fitting the unfolding forces of most of the pseudoknots with 2 Gaussian distributions (Fig. 4) suggests that base triples significantly affect the higher unfolding forces, but the lower unfolding forces are relatively independent of the mutations. Thus, the lower unfolding forces correspond to folding intermediate structures with no base triples formed and the higher unfolding forces correspond to native pseudoknots.
Fig. 4.
Unfolding force histograms for 1-step unfolding reactions. Values shown are peak values of Gaussian distributions with standard deviations. Mutations have significant effect on the native pseudoknots (higher unfolding forces, shown in blue) but not the folding intermediate structures (lower unfolding forces, shown in green). With the entire 5 base triples disrupted, mutant CCCGU still has 2 clusters of unfolding forces (F). Folding intermediate structures appear for pseudoknots with mutations in stem 2 (see arrows). Multiple folding intermediates may exist for TeloWT (K). All of the unfolding forces were measured at pH 7.3 except for mutant 100C115C174G, which was also measured at pH 8.3 (see H).
With the entire 5 base triples disrupted, mutant CCCGU still has 2 clusters of unfolding forces for 1-step unfolding corresponding to a folding intermediate structure (19.4 ± 1.2 pN) and native structure (22.2 ± 5.4 pN), respectively (Fig. 4F). Thus, in the folding intermediate structure stem 1 is fully formed, stem 2 is only partially formed, and base triples are not formed. Fig. 3B shows a possible folding intermediate structure with molecular extension similar to the native pseudoknot structure, which was proposed previously by modeling studies in the absence of force (29, 30), although it was not resolved by bulk thermal unfolding/folding experiments (24, 31). Presumably, the low-force apparent 1-step and 2-step unfolding trajectories correspond to the same folding intermediate structure, with partially formed stem 2 and no base triples formed.
The fact that the unfolding force distribution of ΔU177 is not a combination of those of mutant CCC and mutant GU (Fig. 4 A, D, and E) suggests that major-groove and minor-groove base triples form cooperatively. Slow kinetics of native pseudoknot folding from the folding intermediates is due to 2 intrinsically slow transitions. First, full formation of stem 2 in the native pseudoknot involves disruption of the preformed Watson-Crick base pairs (U102-A111 and C104-G110) and loop structures in stem 1 hairpin (Fig. 3). Second, the kinetics for the formation of non-Watson-Crick U99·A173 pair and triplex structures in native pseudoknot may be intrinsically slow. Slow kinetics of pseudoknot formation involving disruption of preformed structure in stem 1 hairpin (32) and intramolecular triplex formation (33) was previously observed in bulk experiments. In addition, the applied force parallel to the helical axis may restrict the conformational changes such as rotation required for formation of stem 2, non-Watson-Crick U99·A173 pair, and triplex structures (Figs. 1D and 3).
Pseudoknots with Mutations in Stem 2.
Upon substituting the U·A-U with a C+·G-C base triple (Fig. S1), the native structures of mutant 100C115C174G and mutant 101C114C175G have unfolding forces of 49.9 pN and 49.8 pN, respectively, close to that of the native structure of ΔU177 (Fig. 4 A, G, and I). A protonated C+·G-C base triple with 2 hydrogen bonds in the Hoogsteen C+·G pair is thermodynamically more stable than an unprotonated C·G-C base triple with 1 hydrogen bond in the Hoogsteen C·G pair. Remarkably, we found the native pseudoknot unfolding force for mutant 100C115C174G decreases from 49.9 to 43.4 pN upon increasing pH from 7.3 to 8.3 (Fig. 4 G and H). The pKa of residue C100 of mutant 100C115C174G was measured to be 7.8 (24). Instead, the relatively pH independent lower unfolding forces suggest that the major-groove triplex is not formed in the folding intermediate of mutant 100C115C174G.
An additional cluster of unfolding forces ranging between 20 and 35 pN appears for the pseudoknots with mutations in stem 2 (labeled with arrows in Fig. 4 G–K), with the exception of mutant 115C174G. The cluster of unfolding forces may correspond to a folding intermediate structure with complete formation of stem 1 and stem 2, but without base triples formed. Hopping between the folding intermediate structures and stem 1 hairpin at 10–20 pN becomes less frequent by destabilizing stem 1 hairpin and/or stabilizing stem 2 (Fig. 2 A–G and J and Figs. S2, S5, and S6). Thus, a U-A to C-G mutation destabilizes the preformed stem 1 hairpin and stabilizes stem 2 thereby facilitating complete formation of stem 2 in the folding intermediate structure.
Recent NMR studies of TeloWT (with U177 bulge) (Fig. 1B) revealed that all of the base triples are formed in the presence of the U177 bulge, and that the Watson-Crick A-U pairs in stem 2 become less stable than the Hoogsteen U·A pairs (31). Consistently, TeloWT has a broad distribution of unfolding forces (between 15 and 35 pN) for the low-force 1-step unfolding (Fig. 4K) probably due to the formation of multiple folding intermediate structures. Limited unfolding trajectories (Fig. 4K and Table S1) were obtained for the native structure of TeloWT (45.0 ± 4.3 pN) because it rarely formed after it was first mechanically unfolded even with waiting times of 30 s at approximately 3 pN. The results further suggest that the lower unfolding forces correspond to the folding intermediate structures.
Native Pseudoknot Mechanical Stability and –1 Frameshifting Efficiency.
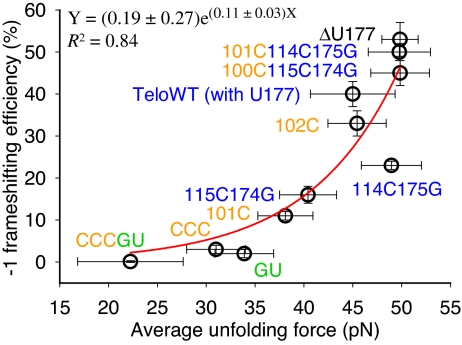
A single exponential function fits well the FS efficiency-versus-average unfolding force of native pseudoknots (Fig. 5). Extrapolation of the single exponential function indicates that the FS efficiency is 0.2% with an unfolding force of 0 pN, and 100% FS corresponds to an average unfolding force of 57 pN. Previous mechanical unfolding studies of RNA using optical tweezers, including a pseudoknot with lengthened 12-bp stem 1 and 29-nucleotide (nt) loop 2 compared to ΔU177 (25), have shown that the highest unfolding force is below 60 pN (21, 22, 34). Thus, it is reasonable to interpret the fitting function only for forces below 60 pN. It is likely that ribosomes would be stalled by a pseudoknot with an unfolding force of 60 pN or higher, resulting in abortive translation.
Fig. 5.
A single exponential function fits well (R2 = 0.84) the correlation between FS efficiency and average unfolding force. Error bars for unfolding force are standard deviations from multiple measurements. Error bars for FS efficiency are standard errors.
Relatively weak correlation was observed between FS efficiency and native pseudoknot unfolding free energy (Fig. S7).
Discussion
Frameshifting may be determined by multiple factors including thermodynamic stability and length of stem and loop, torsional resistance, and specific 3-dimensional structures. For example, higher unfolding forces and FS efficiency were observed for the infectious bronchitis virus (IBV) pseudoknot with an 11-bp stem 1 compared with a 10-bp stem 1 (22). Reverse correlation was observed between the rate of mechanical unfolding and FS efficiency in a specific narrow range of forces for similar pseudoknots derived from IBV, but with a much shorter loop 2 (8 nt vs. 32 nt) and varying compositions of G-C versus A-U pairs and length of stem 1 (21). Bulk FS assays revealed that a specific interaction between stem 1 and loop 2 is required for high-efficiency FS for the IBV pseudoknots with a shorter loop 2 (15). Similarly, minor-groove (stem 1-loop 2) triplexes were revealed by a crystal structure and shown to be critical for high-efficiency FS in the beet western yellows virus (BWYV) (13, 14). Our results suggest that formation of both major-groove (loop 1-stem 2) and minor-groove (stem 1-loop 2) triplex structures enhance mechanical stability of the stems thereby stimulating FS.
For the pseudoknots studied here with short stems (6-bp stem 1 and 9-bp stem 2), initial breaking of several base pairs in stem 1 and/or stem 2 may result in cooperative unfolding of the pseudoknot structures with applied mechanical stretching force >20 pN. Initiation of pseudoknot unfolding by disruption of terminal base pairs in stem 1 may mimic the directional 5′ to 3′ helicase activity of ribosomes (35, 36). The triplex structures may enhance local stability of the terminal stem 1 base pairs and thus, increase the FS efficiency.
Unwinding of stem 1 requires rotation of the rest of the structure; formation of stable stem 2 with extensive loop 1-stem 2 interactions (Fig. 1D) prevents this rotation. The torsional resistance of the pseudoknot is enhanced, thereby stimulating frameshifting (19). Indeed, we found that destabilization of major-groove triplex structures formed between loop 1 and stem 2 significantly decreases FS efficiency, presumably due to lower global torsional resistance of the pseudoknot. Initiation of cooperative pseudoknot unfolding by disruption of stem 2 by force may mimic the helicase activity of ribosomes.
Triplex structures in a pseudoknot may also have specific interactions with ribosomes and thus facilitate high-efficiency FS (12, 13). Structural and functional studies have revealed that tertiary interactions such as minor-groove triplexes and a base quadruplex in several viral mRNA pseudoknots are important (12–16), but may not be sufficient (37) in stimulating high-efficiency FS. Single-molecule studies may reveal the correlation between tertiary structure and programmed FS efficiency for these viral pseudoknots. FS efficiency may also be affected by folding kinetics of native pseudoknots, with multiple ribosomes (polysomes) translating 1 mRNA. It was shown that increasing the distance between elongating ribosomes allowed pseudoknot folding, resulting in a 2-fold increase in FS efficiency (38). Combined single-molecule mechanical unfolding/folding and bulk FS assays will provide further insight into reading-frame regulation (e.g., +1 and –1 frameshifting, and stop codon readthrough) by cis-acting mRNA pseudoknot structures and provide benchmarks for modeling of pseudoknot unfolding by mechanical force and by ribosomes.
Methods
Bulk FS Assay.
The plasmid p2luc developed by Gesteland, Atkins, and coworkers (39) was used as the reporter system for studying FS. Synthetic DNAs containing the slippery sequence (T TTA AAC), single-stranded linker (GGGTT), and the pseudoknot sequences were cloned into p2luc, and the FS elements containing reporter was then transcribed into mRNA by T7 RNA polymerase. The transcribed RNA was then capped in the 5′-end using mMESSAGE mMACHINE kit (Ambion). A commercially available rabbit reticulocyte lysate (Promega) was used for in vitro bulk FS assays following the recommendation of manufacturer with an incubation time of 90 min at 30 °C. Each reaction contains 250 ng capped mRNA and 2.5 μL reticulocyte lysate. With a 0 frame stop codon (UGA, residues 109–111) located at the 5′ side of stem 2 of the pseudoknot, a protein product of 37.4 kDa is released. If FS occurs, translation continues and terminates at a downstream –1 frame stop codon with a fusion protein product of 40.1 kDa. A shorter –1 frame fusion protein (40.1 kDa instead of 100 kDa) was designed to minimize ribosome fall-off events. The translated protein products labeled with [35S]methionine were separated by 12% SDS/PAGE. The efficiency of FS was calculated by the ratio of –1 frame product to both –1 and 0 frame products with correction for the difference in methionine content of the proteins.
Mechanical Unfolding and Folding Using Optical Tweezers.
The RNA molecules used for single-molecule studies were made by PCR amplification of the same plasmids used for FS assays followed by in vitro transcription by T7 polymerase. The constructs for single-molecule experiments were made, as described previously (28), by annealing the RNA with 2 PCR products to form 5′ and 3′ RNA/DNA handles with 592 and 543 base pairs, respectively. Two-nucleotide single strand linkers were used to separate the pseudoknot and RNA/DNA handles. The terminal ends of the RNA/DNA handles were labeled with biotin or digoxigenin to form attachment with streptavidin- and antidigoxigenin-coated polystyrene beads, respectively. The antidigoxigenin-coated bead was trapped in an optical trap and the streptavidin-coated bead was held on a micro pipette by suction (26). The pipette was connected to a piezoelectric stage (MCL). In a force-ramp experiment, the force was changed by approximately 10 pN/s (with an approximately constant stiffness of 0.1 pN/nm) by moving the stage at 100 nm/s with data acquisition rate at 100 Hz. In force-jump and force-drop experiments, the force was maintained by an electronic force feedback. All of the single-molecule experiments were done at 200 mM NaCl, 10 mM Tris-HCl, 0.1 mM EDTA, pH 7.3 or 8.3, and 22 ± 1 °C.
Supplementary Material
Acknowledgments.
We thank Dr. J.-D. Wen for discussions and for the MatLab codes for data analysis, and Dr. S.B. Smith for help and advice on the optical tweezers instruments. This work was supported by Grant GM10840 from the National Institutes of Health (I.T.), Grant NSC 95–2311-B-005–013 from the National Science Council of Taiwan (K.-Y.C.), and a Howard Hughes Medicial Institute grant to C.B.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905046106/DCSupplemental.
References
- 1.Atkins JF, Elseviers D, Gorini L. Low activity of β-galactosidase in frameshift mutants of Escherichia coli. Proc Natl Acad Sci USA. 1972;69:1192–1195. doi: 10.1073/pnas.69.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacks T, Varmus HE. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 3.Jacks T, et al. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 4.Baril M, Dulude D, Gendron K, Lemay G, Brakier-Gingras L. Efficiency of a programmed –1 ribosomal frameshift in the different subtypes of the human immunodeficiency virus type 1 group M. RNA. 2003;9:1246–1253. doi: 10.1261/rna.5113603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plant EP, Dinman JD. The role of programmed -1 ribosomal frameshifting in coronavirus propagation. Front Biosci. 2008;13:4873–4881. doi: 10.2741/3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farabaugh PJ. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manktelow E, Shigemoto K, Brierley I. Characterization of the frameshift signal of Edr, a mammalian example of programmed -1 ribosomal frameshifting. Nucleic Acids Res. 2005;33:1553–1563. doi: 10.1093/nar/gki299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wills NM, Moore B, Hammer A, Gesteland RF, Atkins JF. A functional -1 ribosomal frameshift signal in the human paraneoplastic Ma3 gene. J Biol Chem. 2006;281:7082–7088. doi: 10.1074/jbc.M511629200. [DOI] [PubMed] [Google Scholar]
- 9.ten Dam EB, Pleij CW, Bosch L. RNA pseudoknots: Translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinman JD, Icho T, Wickner RB. A -1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brierley I, Jenner AJ, Inglis SC. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen LX, Tinoco I., Jr The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J Mol Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- 13.Su L, Chen L, Egli M, Berger JM, Rich A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat Struct Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YG, Su L, Maas S, O'Neill A, Rich A. Specific mutations in a viral RNA pseudoknot drastically change ribosomal frameshifting efficiency. Proc Natl Acad Sci USA. 1999;96:14234–14239. doi: 10.1073/pnas.96.25.14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liphardt J, Napthine S, Kontos H, Brierley I. Evidence for an RNA pseudoknot loop-helix interaction essential for efficient -1 ribosomal frameshifting. J Mol Biol. 1999;288:321–335. doi: 10.1006/jmbi.1999.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nixon PL, et al. Solution structure of a luteoviral P1–P2 frameshifting mRNA pseudoknot. J Mol Biol. 2002;322:621–633. doi: 10.1016/s0022-2836(02)00779-9. [DOI] [PubMed] [Google Scholar]
- 17.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 18.Plant EP, et al. The 9-Å solution: How mRNA pseudoknots promote efficient programmed -1 ribosomal frameshifting. RNA. 2003;9:168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plant EP, Dinman JD. Torsional restraint: A new twist on frameshifting pseudoknots. Nucleic Acids Res. 2005;33:1825–1833. doi: 10.1093/nar/gki329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green L, Kim CH, Bustamante C, Tinoco I., Jr Characterization of the mechanical unfolding of RNA pseudoknots. J Mol Biol. 2008;375:511–528. doi: 10.1016/j.jmb.2007.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen TM, Reihani SN, Oddershede LB, Sorensen MA. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc Natl Acad Sci USA. 2007;104:5830–5835. doi: 10.1073/pnas.0608668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao S, Chen SJ. Predicting ribosomal frameshifting efficiency. Phys Biol. 2008;5:16002. doi: 10.1088/1478-3975/5/1/016002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 25.Chen G, Wen J-D, Tinoco I., Jr Single-molecule mechanical unfolding and folding of a pseudoknot in human telomerase RNA. RNA. 2007;13:2175–2188. doi: 10.1261/rna.676707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith SB, Cui Y, Bustamante C. Optical-trap force transducer that operates by direct measurement of light momentum. Methods Enzymol. 2003;361:134–162. doi: 10.1016/s0076-6879(03)61009-8. [DOI] [PubMed] [Google Scholar]
- 27.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 28.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 29.Yingling YG, Shapiro BA. Dynamic behavior of the telomerase RNA hairpin structure and its relationship to dyskeratosis congenita. J Mol Biol. 2005;348:27–42. doi: 10.1016/j.jmb.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 30.Cao S, Chen SJ. Biphasic folding kinetics of RNA pseudoknots and telomerase RNA activity. J Mol Biol. 2007;367:909–924. doi: 10.1016/j.jmb.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim NK, et al. Solution structure and dynamics of the wild-type pseudoknot of human telomerase RNA. J Mol Biol. 2008;384:1249–1261. doi: 10.1016/j.jmb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyatt JR, Puglisi JD, Tinoco I., Jr RNA pseudoknots: Stability and loop size requirements. J Mol Biol. 1990;214:455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- 33.Roberts RW, Crothers DM. Kinetic discrimination in the folding of intramolecular triple helices. J Mol Biol. 1996;260:135–146. doi: 10.1006/jmbi.1996.0388. [DOI] [PubMed] [Google Scholar]
- 34.Li PTX, Bustamante C, Tinoco I., Jr Unusual mechanical stability of a minimal RNA kissing complex. Proc Natl Acad Sci USA. 2006;103:15847–15852. doi: 10.1073/pnas.0607202103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 36.Wen J-D, et al. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cornish PV, Stammler SN, Giedroc DP. The global structures of a wild-type and poorly functional plant luteoviral mRNA pseudoknot are essentially identical. RNA. 2006;12:1959–1969. doi: 10.1261/rna.199006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopinski JD, Dinman JD, Bruenn JA. Kinetics of ribosomal pausing during programmed -1 translational frameshifting. Mol Cell Biol. 2000;20:1095–1103. doi: 10.1128/mcb.20.4.1095-1103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.