Abstract
Fungal spores can account for large proportions of air particulate matter, and they may potentially influence the hydrological cycle and climate as nuclei for water droplets and ice crystals in clouds, fog, and precipitation. Moreover, some fungi are major pathogens and allergens. The diversity of airborne fungi is, however, not well-known. By DNA analysis we found pronounced differences in the relative abundance and seasonal cycles of various groups of fungi in coarse and fine particulate matter, with more plant pathogens in the coarse fraction and more human pathogens and allergens in the respirable fine particle fraction (<3 μm). Moreover, the ratio of Basidiomycota to Ascomycota was found to be much higher than previously assumed, which might also apply to the biosphere.
Keywords: atmospheric aerosol, bioaerosol, DNA analysis, fungal spores
Recent studies have shown that fungal spores and other biological particles can account for large proportions of aerosol particle mass in pristine rainforest air as well as in rural and urban environments (1–5). For example, fungal spores were found to account for up to 45% of coarse particle mass (>1 μm) in tropical rainforest air and up to 4–11% of fine particle mass (≤2.5 μm) in urban and rural air (3). Fungi have also been found in clouds, fog, and precipitation where these and other biological particles can act as nuclei for water droplets and ice crystals and can influence precipitation patterns and the Earth's energy budget (e.g., refs. 6–17). On average, the number and mass concentrations of fungal spores in continental boundary layer air are on the order of 103–104 m−3 and approximately 1 μg m−3, respectively, and the estimated global emissions of approximately 50 Tg yr−1 are among the largest sources of organic aerosol (1).
Some fungi are major pathogens or allergens for humans, animals, and plants, and air is the primary medium for their dispersal (18–20), but the diversity of fungi in air particulate matter is not well-known. The traditional cultivation, microscopy, and chemical tracer techniques applied in earlier studies were insufficient for a broad coverage of different fungal species, and recently reported first applications of molecular genetic techniques were very limited in scope and methodology as discussed below.
In this study, we investigated and characterized the diversity and frequency of occurrence of fungi in air particulate matter by DNA extraction and sequence analysis of the internal transcribed spacer region (ITS).
Results and Discussion
Over a period of 1 year, coarse and fine particle air filter samples were collected on glass fiber filters with a high-volume sampler and a nominal cut-off diameter of approximately 3 μm (Table S1). The average sampling time was 7 days, corresponding to a sampled air volume of approximately 3,000 m3, in central Europe. We investigated the diversity of fungi by DNA extraction, amplification (Table S2), and sequence analysis of the internal transcribed spacer region (ITS). Fungal DNA was found in all air samples. As shown in Table 1, the number of different species (species richness, S) detected in the fine particle fraction was not much lower than in the coarse fraction. The Shannon index (H′), Shannon evenness (E), and Simpson's index (D) values calculated from the frequency of occurrence of the different species were nearly the same for coarse and fine particles (Table 1). These diversity parameter values are similar to the values commonly obtained for fungi in soil and on plants as well as for bacteria in soil (21–23).
Table 1.
Diversity parameters for all fungi, Basidiomycota (BMC), Ascomycota (AMC), and Fungi Incertae Sedis (FIS) found in the investigated air samples (total, coarse, and fine PM)
| Parameter | Fungi |
BMC |
AMC |
FIS |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Coarse | Fine | Total | Coarse | Fine | Total | Coarse | Fine | Total | Coarse | Fine | |
| S | 368 | 227 | 203 | 238 | 160 | 124 | 124 | 62 | 78 | 4 | 3 | 1 |
| S* | 1136 | 713 | 647 | 658 | 448 | 377 | 500 | 327 | 267 | 9 | 5 | — |
| H′ | 5.2 | 4.8 | 4.7 | 4.9 | 4.5 | 4.2 | 4.1 | 3.3 | 3.7 | 1.3 | 1 | — |
| E | 0.88 | 0.88 | 0.88 | 0.88 | 0.89 | 0.87 | 0.84 | 0.80 | 0.85 | 0.96 | 0.94 | — |
| D | 0.01 | 0.015 | 0.018 | 0.015 | 0.017 | 0.027 | 0.033 | 0.066 | 0.054 | 0.1 | 0.167 | — |
Species richness (S measured, S* estimated), Shannon index (H′), Shannon Evenness (E), and Simpson's index (D).
The most frequent species (Cladosporium sp.) was detected in all air samples except 1. On the other hand, 70% of the detected species were found only in 1 sample (SI Text, and Table S3). The high proportion of species that were found only once and the limited number of investigated samples and DNA amplification products imply that the actual diversity of fungi in the sampled air masses was higher than detected. As shown in Table 1, estimates of the actual species richness (S*) based on the Chao-1 estimator approach (21, 24) are an approximate factor of 3 higher than the measured species richness values (S), suggesting that the total number of fungal species in the investigated air samples was >1,000.
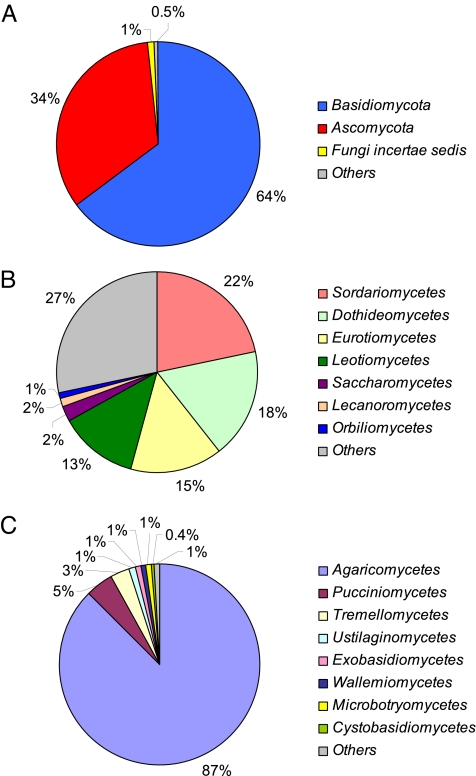
As illustrated in Fig. 1A, nearly all detected fungal species were Basidiomycota (BMC, club fungi, 64%) or Ascomycota (AMC, sac fungi, 34%), many of which actively eject their spores with aqueous jets or droplets (1). Only approximately 2% of the measured sequences were from Fungi Incertae Sedis (FIS, formerly called Zygomycota) or other fungi that could not be attributed to a phylum. This is consistent with the predominance of AMC and BMC in the biosphere, where the subkingdom of Dikarya (AMC plus BMC) accounts for 98% of the known species in the biological kingdom of Eumycota (fungi) (25). The AMC species found in the air samples were distributed over 4 major classes (Dothideomycetes, Eurotiomycetes, Leotiomycetes, and Sordariomycetes; Fig. 1B) and about one-third could not be attributed to any class. In contrast, most of the detected BMC species belonged to a single class, the Agaricomycetes (Fig. 1C). More information about the taxonomic classes and families of the detected fungi is available online (SI Text) (26).
Fig. 1.
Species richness of airborne fungi: relative proportions of different phyla (A), different classes of Ascomycota (B), and different classes of Basidiomycota (C).
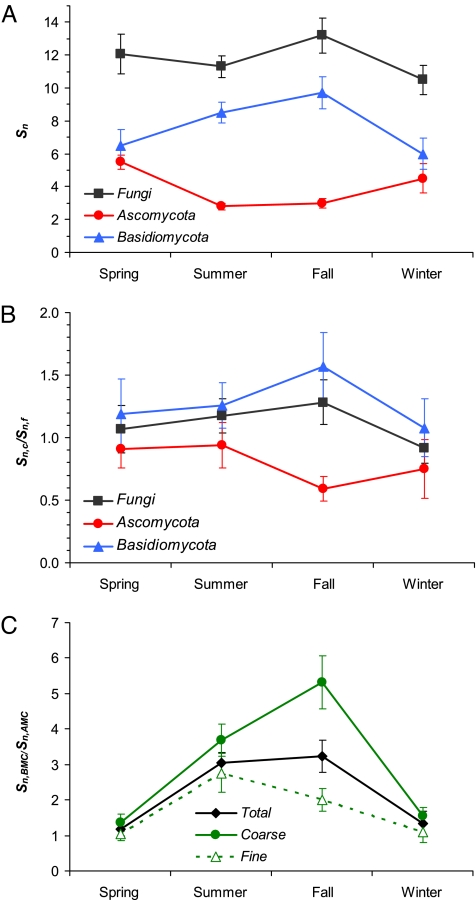
Fig. 2 shows seasonal variations in the normalized species richness (Sn: number of detected species divided by number of investigated samples), in the ratio of Sn between coarse and fine particles (Sn,c/Sn,f), and in the ratio of Sn between BMC and AMC (Sn,BMC/Sn,AMC). With regard to all fungal species, Sn and Sn,c/Sn,f exhibited little seasonal variability. AMC and BMC, however, exhibited pronouncedly different values and seasonal cycles of Sn and Sn,c/Sn,f. For AMC, Sn was highest in spring and lowest in summer and Sn,c/Sn,f was lowest in fall, whereas for BMC both Sn and Sn,c/Sn,f were highest in fall and lowest in winter. Throughout all seasons, Sn remained higher for BMC than for AMC (Fig. 2A) and Sn,c/Sn,f remained >1 for BMC but <1 for AMC (Fig. 2B). The ratio of BMC to AMC (Sn,BMC/Sn,AMC) was higher in summer and fall than in spring and winter, and it was higher for coarse than for fine particle samples (Fig. 2C). In other words, most BMC occurred in the coarse particle fraction during summer/fall, whereas most AMC were found in the fine particle fraction during winter/spring. This seems to reflect a predominance of BMC among the mushrooms that form fruiting bodies in summer and fall, and a high proportion of AMC among the molds, endophytes, and epiphytes spreading in winter and spring (27, 28). Statistical analyses (multiple linear regressions) indicate that ambient temperature and relative humidity are positively correlated with the relative proportion of BMC (R2 = 0.64, P value <0.001) and negatively correlated with the relative proportion of AMC (R2 = 0.67, P value <0.001).
Fig. 2.
Seasonal variations in the species richness normalized by the number of investigated air samples (Sn) (A), the ratio of normalized species richness between coarse and fine particles (Sn,c/Sn,f) (B), and the ratio of normalized species richness between Basidiomycota and Ascomycota (Sn,BMC/Sn,AMC) (C). Error bars illustrate inter-sample variability (relative standard error of the arithmetic mean of the species richness of individual samples).
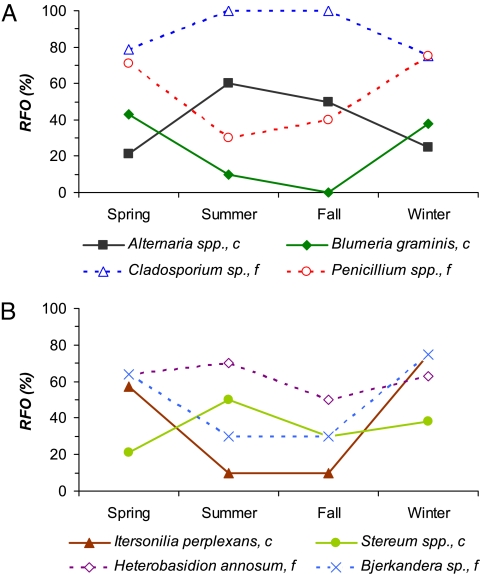
Fig. 3 illustrates that the relative frequency of occurrence (RFO) of individual species, that is, the proportion of samples in which these species were detected, is independent of the seasonal variability of AMC and BMC species richness. For example, the prominent allergenic AMC species Cladosporium sp. and Alternaria spp. occurred most frequently in summer and fall, whereas the mold Penicillium spp. and the plant pathogen Blumeria graminis (mildew) were mostly found in winter and spring. With regard to BMC, the seasonal cycle of Sn was not matched by the RFO of any frequently detected species. The fall maximum of BMC species richness is primarily due to species that were found only once and can be attributed to mushroom fruiting and enhanced activity of plant decomposers during that season (27).
Fig. 3.
Seasonal variations in the relative frequency of occurrence (RFO) of selected species of Ascomycota (A) and Basidiomycota (B) in coarse and fine air particulate matter, respectively (c, f).
Several allergens and pathogens were frequently found in both fine and coarse particle samples (e.g., Cladosporium sp., Alternaria spp., Penicillium spp., and Aspergillus spp.). Some plant pathogens, however, were found only or mostly in coarse particle samples (e.g., Blumeria graminis and Puccinia spp.), whereas some human pathogens were detected only in the fine particle samples (e.g., Candida tropicalis and Wallemia spp.). Overall, the species richness and cumulative frequency of occurrence of plant pathogens was found to be higher in the coarse particle fraction, whereas those of human pathogens and allergens were similar or higher in the fine particle fraction. Note that fine particles have longer residence times in the atmosphere (multiple days to weeks) and that they can reach the alveolar region of human lungs upon inhalation, whereas coarse particles are rapidly removed from the atmosphere (sedimentation, scavenging, and precipitation) and are deposited in the upper airways when inhaled. Thus, the scientific investigation and public discussion of climate and health effects are mostly focused on fine aerosol particles (11).
As discussed by Despres et al. (29), fungal DNA detected in aerosol filter samples is most likely to originate from spores which are known to resist environmental stress and survive atmospheric transport (30, 31), whereas DNA in fungal tissue fragments may be rapidly degraded by atmospheric photooxidants. Nevertheless, hyphae and tissue fragments can also be present in coarse and fine air particulate matter (32–34). Moreover, the disruption of fungal spores due to environmental stress may lead to the formation and sampling of smaller fragments, whereas the association of spores with each other or with other particulate matter may lead to the formation and sampling of larger particles. For example, unicellular fungi (like endophytic fungi) and plant pathogens (rust fungi, powdery mildew) are likely to be associated with plant fragments (35–37).
As outlined above, the species richness and the relative proportion of BMC were generally higher in coarse particle samples, while the species richness and the relative proportion of AMC were higher in fine particle samples. This is consistent with the following basic features of BMC and AMC (26, 38–40): BMC tend to have spores with relatively large aerodynamic diameters (≈5–10 μm), and many of them produce fruiting bodies that may fragment and release large tissue particles. In contrast, AMC are mostly single-celled (yeast) or filamentous (hyphal), and some of the most prominent AMC species like Cladosporium sp. and Penicillium spp. have spores with small aerodynamic diameters (≈2–5 μm). More information about the differences between coarse and fine particle samples and about fungal spore size is available online (SI Text and Table S3, Table S4, Table S5, Table S6, Table S7, and Table S8).
Earlier studies of airborne fungi have mostly not evaluated the diversity of AMC vs. BMC, or they have reported a predominance of AMC (e.g., refs. 41, 42). Most of these studies were based on cultivation techniques and have detected far fewer species (typically ≪100). Lugauskas et al. (43) reported a high number of different fungal species (≈430), but for this they had analyzed over 900 aerosol samples collected at multiple locations over several years, corresponding to a normalized species richness of only approximately 0.5 as opposed to ∼10 from our DNA-based investigations. Over 80% of all known fungi are not cultivable on synthetic media, and fast growing fungi can mask slower growing fungi (44). BMC are often out-competed by the more rapidly growing mold fungi (45), and some AMC like Cladosporium can actively inhibit the growth of others by production of inhibitory substances (46). Thus, cultivation-based studies of airborne fungi are likely biased against the detection of BMC (especially by the high abundance of Cladosporium).
To our knowledge, only 3 studies have reported DNA analyses of fungi in atmospheric aerosol samples up to now. Boreson et al. (47) found 18 species in urban air (40% AMC, 6% BMC) and 31 species in desert air (65% AMC, 17% BMC), Despres et al. (29) found only 3 AMC and 1 BMC species in several dozens of filter samples of urban, rural, and high-alpine air, and Fierer et al. (48) measured over 1,500 fungal DNA sequences from 5 urban air samples and attributed 97% to AMC but did not specify the species richness. We have tested and applied multiple PCR primer pairs; these and other experimental details were identified as key elements for efficient amplification of DNA from AMC, BMC, and other fungi (see Materials and Methods). Consequently, we have been able to detect a wide range of species that are well-known from cultivation-based studies (e.g., Cladosporium spp., Penicillium spp., Alternaria spp., and Candida spp.) as well as non cultivable species (e.g., Blumeria graminis and Puccinia spp.) and previously unknown species as indicated by 136 operational taxonomic units (OTUs) with low NCBI similarity scores (<97%). Some of these might also stem from degraded DNA (29), but at least the 21 OTUs that were found in multiple samples are much more likely to represent unknown species. The other DNA-based studies of airborne fungi have neither found the expected high abundance of Cladosporium nor a high species richness of BMC, which we suppose to be due to limitations of the applied methods.
Ongoing investigations of aerosol samples from several locations around the world indicate that the species richness of fungi in continental air is generally higher for BMC than for AMC. In contrast, the number and proportion of all fungal species known from the biosphere are higher for AMC (≈60,000, ≈60–70%) than for BMC (25,000–30,000, ≈30–40%) (25, 49). Recently, however, Hunt et al. (50) compared culture- and DNA-based analyses of fungi in grassland soil samples and found that 54% of the fungal sequences were from BMC whereas no BMC species could be isolated by plate culturing. Thus, the high proportion of BMC in air as observed in our study (64%) may be explained by one or both of the following hypotheses:
BMC may be enriched in the atmosphere relative to the biosphere, because their proportion at the atmosphere-biosphere interface (i.e., on vegetation and soil surfaces) or their efficiency in dispersing spores may be enhanced relative to AMC (dry release vs. active wet discharge with aqueous jets or droplets) (1).
The ratio of BMC to AMC in the biosphere may have been underestimated in earlier studies for the reasons outlined above (bias of culture-based techniques, primer selection in DNA-based techniques). In this case, our atmospheric measurement results might reflect the biospheric proportions.
Clearly, our results will need to be complemented by further investigations to confirm or discard the above hypotheses. In any case, however, our data show that the diversity of airborne fungi—and in particular the species richness of BMC—is much higher than indicated by earlier studies.
As many fungal species in the biosphere are still unknown, the detection and characterization of fungi in atmospheric aerosol samples by DNA analysis can help to elucidate the regional and global spread and diversity of fungi. Moreover, DNA analyses also enables efficient and unambiguous identification of other primary biogenic aerosol particles and components [bacteria, animal, and plant fragments, etc., (29)].
Information about the diversity and abundance of airborne fungi and other bioaerosol particles is relevant for many areas of research such as biogeosciences, climate and ecology, human and veterinary medicine, industrial and environmental hygiene, agriculture, bioengineering, and security. Among the key perspectives for future investigations of air particulate matter by DNA analysis are the possible influence of different types of bioaerosols on the hydrological cycle (14, 16, 17), the spread of genetically modified organisms (51), and climate-related changes in the diversity and abundance of fungi and other organisms on local, regional, and global scales (1, 52).
Materials and Methods
Aerosol Sampling.
Aerosol samples were collected on glass fiber filters (Pall Corporation, Type A/A, 102-mm diameter) over 1 year in Mainz, Germany (130 m a.s.l., March 2006–April 2007). The sampling station was positioned on a mast at the top of the Max Planck Institute for Chemistry (MPIC, ≈5 m above the flat roof of the 3-story building) on the campus of the University of Mainz (49°59′31.36′′N 8°14′15.22′′E). The air masses sampled at MPIC represent a mix of urban and rural continental boundary layer air in central Europe. A high-volume dichotomous sampler [self-built based on Solomon et al. (53)] was used to separate and collect coarse and fine aerosol particles on a pair of glass fiber filters. The sampler was operated with a rotary vane pump (Becker VT 4.25) at a total flow rate of approximately 300 L min−1, corresponding to a nominal cut-off diameter of ≈3 μm. Coarse particles with aerodynamic diameters larger than the cut-off were collected through a virtual impactor operated in line with the inlet (≈30 L min−1), and fine particles with aerodynamic diameters smaller than the cut-off were collected from the main gas flow perpendicular to the inlet (≈270 L min−1).
The sampling period was generally 7 days, corresponding to a sampled air volume of approximately 3,000 m3. A few samples were collected over shorter periods (1–5 days, ≈400–2,000 m3). A listing of all investigated pairs of air filter samples (42 coarse and 42 fine particle samples) is given in Table S1. Before use, all glass fiber filters were decontaminated by baking at 500°C over night (29). Loaded filters were packed in aluminum foil (also prebaked at 500°C), and stored in a freezer at −80°C until DNA extraction. To detect possible contaminations from the sampler and sample handling, blank samples were taken at regular intervals (≈4 weeks). The filters were mounted in the sampler like for regular sampling, but the pump was turned on either not at all (“mounting blanks”) or for only 5 s (“start-up blank”).
DNA Extraction and Amplification.
Filter sample aliquots (⅛–¼) were extracted with a commercial soil extraction kit (LysingMatrixE, Fast DNA Spin Kit for Soil, MP Biomedicals) according to the supplier's instructions with the following modifications: 15-min centrifugation step after the lysis, partly additional 900 μL buffer and repeated beating and centrifugation. Both generated supernatants were combined for the further extraction process. Finally, the DNA was dissolved in 100 μL elution buffer. Decontaminated filters were included during the extractions as extraction blanks.
With the DNA extract from each of the filters listed in Table S1, several PCRs were performed to amplify DNA from fungi for sequence analysis. The 50-μL reaction mixture always contained the template DNA (0.5–2-μL sample extract), 1× PCR buffer, 0.2 mM each dNTP (Roth), 0.33 μM of each primer (Sigma-Aldrich), and 2.5 units of JumpStart REDTaq DNA polymerase (Sigma-Aldrich). A negative control was included in all PCR runs.
PCR was performed with the primer pairs listed in Table S2 and Fig. S1, respectively. The thermal profile (DNA Engine, Bio-Rad Laboratories) was as follows: initial denaturing at 94°C for 3 min; 35 cycles with denaturing at 94°C for 30 s, annealing at primer pair specific temperature for 30 s (Table S2), elongation at 72°C for 90 s, and a final extension step at 72°C for 5 min.
Fungal DNA was detected in all of the 84 investigated air filter samples, and 200 PCR products were obtained with different primer pairs as specified in Table S1 and Table S2. No DNA was detected in the mounting, start-up, extraction, and PCR blanks, indicating that no contamination occurred during sample handling and analysis in the laboratory.
PCR products were obtained with all primer combinations except for the Zygomycota-specific primer pair D (Table S1, Table S2, and Fig. S1). The primer pairs A and F amplified Basidiomycota (BMC) as well as Ascomycota (AMC) and the primer pairs that were designed to be specific for BMC (B) and AMC (C) indeed amplified only BMC and AMC, respectively. The sequences obtained with the primer pair E which should be specific for Chytridiomycota matched with members of the Pucciniomycetes (within the BMC) and Fungi incertae sedis (formerly Zygomycota).
Fungal DNA was also co-amplified with primer pairs (G–I) that were supposed to be specific for animal, insect, and archaea 18S or 16SrDNA (Table S1, Table S2, and Fig. S1). Table S1 gives information on obtained sequences resulting from co-amplification reactions in coarse particle filter extracts. The primer pairs G and H amplified BMC whereas the primer pair I amplified AMC. This primer behavior calls for reevaluation of the primer performance and requires attention before the primer pair is further used in samples that harbor material from different groups of organisms. The obtained fungal 18S sequences confirm knowledge from other studies that the ITS region is a better target region and 18S rDNA analysis often is not able to discriminate between species in environmental samples (54).
Cloning and Restriction Fragment Length Polymorphism.
Amplification products for sequencing were cloned using the TOPO TA Cloning Kit (Invitrogen) following the supplier's instructions. Colonies containing inserts were identified by blue-white selection and lysed in 20 μL water for 10 min at 95°C. The inserts of 12–24 colonies were amplified (“colony PCRs”) using 3 μL lysate in a 40-μL reaction. The PCR mixture always contained: 1× PCR Buffer, 2.5 mM MgCl2, 0.25 mM each dNTP (Roth), 0.25 μM of each primer (Sigma-Aldrich), 1.25 U TaqDNA Polymerase (Invitrogen). PCR was performed with the primer pair M13F-40 and M13R, and the thermal profile was as follows: initial denaturing at 94°C for 5 min; 40 cycles with 94°C for 30 s, annealing at 55°C for 1 min, elongation at 72°C for 1 min, and a final extension step at 72°C for 15 min.
Colony PCR was followed by a restriction fragment length polymorphism (RFLP) analysis to select as many as possible different clones for sequencing. Two microliters of the PCR-products were digested without further purification with 5 U of the enzymes TaqI, HinfI, or AluI (Fermentas). Restriction fragments were separated by gel electrophoresis in a 3% agarose gel stained with ethidium bromide and the images were documented with the Gel Doc XR system and analyzed with Quantity One software (Bio-Rad Laboratories). On the basis of the resulting restriction fragment patterns, representative colony PCR products with different numbers and sizes of fragments were selected for sequencing.
DNA Sequence Analysis, Taxonomic Attribution, and Statistical Parameters.
DNA sequences were determined with ABI Prism 377, 3100, and 3730 sequencers (Applied Biosystems) using BigDye-Terminator v3.1 chemistry at the DNA Core Facility of the Max Planck Institute for Plant Breeding Research, Cologne. For comparison with known sequences, databank queries using the Basic Local Alignment Search Tool (BLAST) were performed via the website of the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/). Each sequence was identified to the lowest taxonomic rank common to the top BLAST hits (up to ≈100 database sequences with highest similarity and total scores). For 17 of the 1,513 sequences determined in this study, the ITS1 and ITS2 regions matched in different genera. These 17 sequences were assumed to be chimeric results of PCR recombination and were excluded from further analysis.
For each aerosol filter sample, sequences that produced the same BLAST results were pairwise aligned using the program BioEdit (BioEdit 7.05; http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and the similarity between them was calculated. Sequences with similarity scores greater than or equal to 97% were clustered into an operational taxonomic unit (OTU). The different OTUs can be regarded as different fungal species, because the sequence variability due to intraspecific ITS divergence is usually <3% (55). PCR errors can also introduce some sequence variability (56). The approximate 100 PCR cycles typically applied in our study (amplification, colony, and sequencing PCRs) are, however, unlikely to cause substitutions of more than 1% of the base pairs in the measured sequences, because the total error rates of Taq polymerase are typically in the range of 10−5-10−4 errors per base pair (57, 58) When sequences obtained by co-amplification with nonfungal primer pairs (18S; G, H, I; Table S2) yielded the same best match results as ITS sequences from the same sample, they were attributed to the same OTU, because they are likely to have the same origin (see Fig. S1).
Table S3 gives an overview of the 368 detected OTUs with corresponding characteristic NCBI sequence accession numbers, taxonomic class and species names, and frequencies of occurrence, that is, numbers of air samples and coarse or fine particle filters, respectively, in which they were detected. Note that some OTUs exhibited high similarity (98–100%) with multiple NCBI sequences attributed to different orders, families, genera, or species, which may be due to database entries with incorrect/inconsistent taxonomic information or high interspecific ITS similarity (e.g., BMC2: Bjerkandera/Thanatephorus; BMC3: Trametes/Tricholoma/Phellinus sp.). About one-third of the OTUs exhibited less than 97% similarity with NCBI sequences (<90% for 37 OTUs, 90–96% for 99 OTUs) and may represent species that are unknown or at least not listed in the NCBI database. Alternatively, the measured DNA may have been partially degraded in the air or upon sampling and analysis (59–62). This is, however, not very likely for fungal spores that are known to resist environmental stress and survive atmospheric transport (29). Moreover, 21 OTUs with low best match similarity scores were found in more than 1 sample, which appears unlikely for damaged DNA. Thus, we assume that most if not all OTUs represent different fungal species, and that ≈30% of these have not yet been characterized by DNA analysis and listing in the NCBI database.
The detection and apparent frequency of occurrence of different species can also be affected by technical factors like extraction efficiency of DNA, varying rDNA copy number in the species, primer matching and performance, amplification efficiency of the target region, and cloning success. As discussed above, the high proportion of fungal species that were detected only once indicates, that a higher number of samples and clones would have to be investigated for a more complete coverage of species diversity. Thus, the detected and reported numbers and frequencies of occurrence of species, families, and classes should be regarded as a lower limit for the actual diversity and frequency of occurrence of airborne fungi.
To characterize and compare the diversity of fungal species (OTUs) in the investigated air masses, we have calculated the parameters defined in Table S4 for the whole data set and for different subsets (phyla, seasons, etc.). The results are summarized in Table S5, Table S6, and Table S7 and additional information is given in SI Text. Statistical analyses were performed with the software package SPSS (release 16.0.1, SPSS Inc.). t tests were performed to confirm statistical significance of discussed differences between seasons, AMC/BMC and coarse/fine particle samples (P values <0.05). Multiple linear regressions were performed with species richness values (running mean of 5 consecutive samples) and local air quality data (ZIMEN Luftmessnetz, Rheinland Pfalz, Station Mainz-Mombach: temperature, relative humidity, wind speed and direction, atmospheric pressure, ozone concentration, and rainfall averaged over the sampling periods).
Nucleotide Sequence Accession Numbers.
The sequences from the 368 OTUs of the present study have been deposited in the GenBank database under following accession numbers: FJ820489–FJ820856 (Table S9).
Supplementary Material
Acknowledgments.
We thank J. Cimbal and I. Germann for technical assistance; M.O. Andreae, W. Elbert, H. Paulsen, and D. Begerow for helpful discussions and support; U. Kampe for providing air quality data; and the Max Planck Institute for Plant Breeding Research for DNA sequencing. This work was supported by the Max Planck Society, German Research Foundation Grant DE1161/2-1, and Landesexzellenzcluster Geocycles Contribution No. 494.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition footnote: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ820489–FJ820856).
This article contains supporting information online at www.pnas.org/cgi/content/full/0811003106/DCSupplemental.
References
- 1.Elbert W, Taylor PE, Andreae MO, Pöschl U. Contribution of fungi to primary biogenic aerosols in the atmosphere: Wet and dry discharged spores, carbohydrates, and inorganic ions. Atmos Chem Phys. 2007;7:4569–4588. [Google Scholar]
- 2.Bauer H, et al. Significant contributions of fungal spores to the organic carbon and to the aerosol mass balance of the urban atmospheric aerosol. Atmos Environ. 2008;42:588–593. [Google Scholar]
- 3.Womiloju TO, Miller JD, Mayer PM, Brook JR. Methods to determine the biological composition of particulate matter collected from outdoor air. Atmos Environ. 2003;37:4335–4344. [Google Scholar]
- 4.Held A, et al. Aerosol size distributions measured in urban, rural and high-alpine air with an electrical low pressure impactor (ELPI) Atmos Environ. 2008;42:8502–8512. [Google Scholar]
- 5.Hock N, et al. Rural continental aerosol properties and processes observed during the Hohenpeissenberg Aerosol Characterization Experiment (HAZE2002) Atmos Chem Phys. 2008;8:603–623. [Google Scholar]
- 6.Jaenicke R. Abundance of cellular material and proteins in the atmosphere. Science. 2005;308:73–73. doi: 10.1126/science.1106335. [DOI] [PubMed] [Google Scholar]
- 7.Christner BC, et al. Ubiquity of biological ice nucleators in snowfall. Science. 2008;319:1214. doi: 10.1126/science.1149757. [DOI] [PubMed] [Google Scholar]
- 8.Bauer H, et al. The contribution of bacteria and fungal spores to the organic carbon content of cloud water, precipitation and aerosols. Atmos Res. 2002;64:109–119. [Google Scholar]
- 9.Pouleur S, Richard C, Martin JG, Antoun H. Ice nucleation activity in Fusarium acuminatum and Fusarium avenaceum. Appl Environ Microbiol. 1992;58:2960–2964. doi: 10.1128/aem.58.9.2960-2964.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreae MO, Crutzen PJ. Atmospheric aerosols: Biogeochemical sources and role in atmospheric chemistry. Science. 1997;276:1052–1058. [Google Scholar]
- 11.Pöschl Atmospheric aerosols: Composition, transformation, climate, and health effects. Angew Chem Int Ed. 2005;44:7520–7540. doi: 10.1002/anie.200501122. [DOI] [PubMed] [Google Scholar]
- 12.Andreae MO, Rosenfeld D. Aerosol-cloud-precipitation interactions. Part 1. The nature and sources of cloud-active aerosols. Earth Sci Rev. 2008;89:13–41. [Google Scholar]
- 13.Rosenfeld D, et al. Flood or drought: How do aerosols affect precipitation? Science. 2008;321:1309–1313. doi: 10.1126/science.1160606. [DOI] [PubMed] [Google Scholar]
- 14.Möhler O, DeMott PJ, Vali G, Levin Z. Microbiology and atmospheric processes: The role of biological particles in cloud physics. Biogeosciences. 2007;4:1059–1071. [Google Scholar]
- 15.Deguillaume L, et al. Microbiology and atmospheric processes: Chemical interactions of primary biological aerosols. Biogeosciences. 2008;5:1073–1084. [Google Scholar]
- 16.Prenni AJ, et al. Relative roles of biogenic emissions and Saharan dust as ice nuclei in the Amazon basin. Nature Geoscience. 2009 doi: 10.1038/ngeo517. [Google Scholar]
- 17.Pratt KA, et al. In situ detection of biological particles in cloud ice-crystals. Nature Geoscience. 2009 doi: 10.1038/ngeo521. [Google Scholar]
- 18.Kurup VP, Shen H-D, Banerjee B. Respiratory fungal allergy. Microbes Infec. 2000;2:1101–1110. doi: 10.1016/s1286-4579(00)01264-8. [DOI] [PubMed] [Google Scholar]
- 19.Adhikari A, Sen MM, Gupta-Bhattacharya S, Chanda S. Airborne viable, non-viable, and allergenic fungi in a rural agricultural area of India: A 2-year study at five outdoor sampling stations. Sci Total Environ. 2004;326:123–141. doi: 10.1016/j.scitotenv.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Brown JKM, Hovmøller MS. Epidemiology—Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science. 2002;297:537–541. doi: 10.1126/science.1072678. [DOI] [PubMed] [Google Scholar]
- 21.Hill TCJ, Walsh KA, Harris JA, Moffett BF. Using ecological diversity measures with bacterial communities. FEMS Microbiol Ecol. 2003;43:1–11. doi: 10.1111/j.1574-6941.2003.tb01040.x. [DOI] [PubMed] [Google Scholar]
- 22.Richard F, Moreau PA, Selosse MA, Gardes M. Diversity and fruiting patterns of ectomycorrhizal and saprobic fungi in an old-growth Mediterranean forest dominated by Quercus ilex L. Can J Bot. 2004;82:1711–1729. [Google Scholar]
- 23.Maria GL, Sridhar KR. Richness and diversity of filamentous fungi on woody litter of mangroves along the west coast of India. Curr Sci. 2002;83:1573–1580. [Google Scholar]
- 24.Chao A. Nonparametric-estimation of the number of classes in a population. Scand J Stat. 1984;11:265–270. [Google Scholar]
- 25.James TY, et al. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 26.Hibbett DS, et al. A higher-level phylogenetic classification of the Fungi. Mycol Res. 2007;111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Levetin E. Studies on airborne basidiospores. Aerobiologia. 1990;6:177–180. [Google Scholar]
- 28.Liang-Dong Guo G-RHYW. Seasonal and tissue age influences on endophytic fungi of Pinus tabulaeformis (Pinaceae) in the Dongling Mountains, Beijing. J Integr Plant Biol. 2008;50:997–1003. doi: 10.1111/j.1744-7909.2008.00394.x. [DOI] [PubMed] [Google Scholar]
- 29.Després VR, et al. Characterization of primary biogenic aerosol particles in urban, rural, and high-alpine air by DNA sequence and restriction fragment analysis of ribosomal RNA genes. Biogeosciences. 2007;4:1127–1141. [Google Scholar]
- 30.Griffin DW. Terrestrial microorganisms at an altitude of 20,000 m in Earth's atmosphere. Aerobiologia. 2004;20:135–140. [Google Scholar]
- 31.Griffin DW, Kellogg CA. Dust storms and their impact on ocean and human health: Dust in Earth's atmosphere. Ecohealth. 2004;1:284–295. [Google Scholar]
- 32.Gorny RL, et al. Fungal fragments as indoor air biocontaminants. Appl Environ Microbiol. 2002;68:3522–3531. doi: 10.1128/AEM.68.7.3522-3531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Green BJ, et al. Airborne fungal fragments and allergenicity. Med Mycol. 2006;44:245–S255. doi: 10.1080/13693780600776308. [DOI] [PubMed] [Google Scholar]
- 34.Marshall WA. Seasonality in Antarctic airborne fungal spores. Appl Environ Microbiol. 1997;63:2240–2245. doi: 10.1128/aem.63.6.2240-2245.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnold AE. Understanding the diversity of foliar endophytic fungi: Progress, challenges, and frontier's. Fungal Biol Rev. 2007;21:51–66. [Google Scholar]
- 36.Eichmann R, Hückelhoven R. Accommodation of powdery mildew fungi in intact plant cells. J Plant Physiol. 2008;165:5–18. doi: 10.1016/j.jplph.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 37.Hata K, Atari R, Sone K. Isolation of endophytic fungi from leaves of Pasania edulis and their within-leaf distributions. Mycoscience. 2002;43:369–373. [Google Scholar]
- 38.Taylor JW, Spatafora J, Berbee M. Ascomycota. Sac Fungi. 2006. http://tolweb.org/Ascomycota/20521/2006.10.09 in The Tree of Life Web Project, http://tolweb.org/
- 39.Hibbett DS. After the gold rush, or before the flood? Evolutionary morphology of mushroom-forming fungi (Agaricomycetes) in the early 21st century. Mycol Res. 2007;111:1001–1018. doi: 10.1016/j.mycres.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Swann E, Hibbett DS. Basidiomycota. The Club Fungi. 2007. http://tolweb.org/Basidiomycota/20520/2007.04.20 in The Tree of Life Web Project, http://tolweb.org/
- 41.Lee T, et al. Culturability and concentration of indoor and outdoor airborne fungi in six single-family homes. Atmos Environ. 2006;40:2902–2910. doi: 10.1016/j.atmosenv.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adhikari A, et al. Correlation of ambient inhalable bioaerosols with particulate matter and ozone: A two-year study. Environ Pollut. 2006;140:16–28. doi: 10.1016/j.envpol.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Lugauskas A, Sveistyte L, Ulevicius V. Concentration and species diversity of airborne fungi near busy streets in Lithuanian urban areas. Ann Agric Environ Med. 2003;10:233–239. [PubMed] [Google Scholar]
- 44.Bridge P, Spooner B. Soil fungi: Diversity and detection. Plant Soil. 2001;232:147–154. [Google Scholar]
- 45.Thorn RG. Soil fungi. In: Summer ME, editor. Handbook of Soil Sci. Boca Raton: CRC Press; 1999. pp. C22–C37. [Google Scholar]
- 46.Scott PM, Vanwalbe. W, Maclean WM. Cladosporin, a new antifungal metabolite from Cladosporium cladosporioides. J Antibiot. 1971;24:747–755. doi: 10.7164/antibiotics.24.747. [DOI] [PubMed] [Google Scholar]
- 47.Boreson J, Dillner AM, Peccia J. Correlating bioaerosol load with PM2.5 and PM10cf concentrations: A comparison between natural desert and urban-fringe aerosols. Atmos Environ. 2004;38:6029–6041. [Google Scholar]
- 48.Fierer N, et al. Short-term temporal variability in airborne bacterial and fungal populations. Appl Environ Microbiol. 2008;74:200–207. doi: 10.1128/AEM.01467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Meeus T, Renaud F. Parasites within the new phylogeny of eukaryotes. Trends Parasitol. 2002;18:247–251. doi: 10.1016/s1471-4922(02)02269-9. [DOI] [PubMed] [Google Scholar]
- 50.Hunt J, Boddy L, Randerson PF, Rogers HJ. An evaluation of 18S rDNA approaches for the study of fungal diversity in grassland soils. Microbial Ecol. 2004;47:385–395. doi: 10.1007/s00248-003-2018-3. [DOI] [PubMed] [Google Scholar]
- 51.Wang C, St Leger RJ. A scorpion neurotoxin increases the potency of a fungal insecticide. Nat Biotech. 2007;25:1455–1456. doi: 10.1038/nbt1357. [DOI] [PubMed] [Google Scholar]
- 52.Klironomos JN, et al. Increased levels of airborne fungal spores in response to Populus tremuloides grown under elevated atmospheric CO2. Can J Bot. 1997;75:1670–1673. [Google Scholar]
- 53.Solomon PA, Moyers JL, Fletcher RA. High-volume dichotomous virtual impactor for the fractionation and collection of particles according to aerodynamic size. Aerosol Sci Tech. 1983;2:455–464. [Google Scholar]
- 54.Reddy SR, Pindi PK, Reddy SM. Molecular methods for research on arbuscular mycorrhizal fungi in India: problems and prospects. Curr Sci. 2005;89:1699–1709. [Google Scholar]
- 55.O'Brien HE, et al. Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol. 2005;71:5544–5550. doi: 10.1128/AEM.71.9.5544-5550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.v. Wintzingerode F, Göbel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: Pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 57.Keohavong P, Thilly WG. Fidelity of DNA-Polymerases in DNA Amplification. Proc Natl Acad Sci USA. 1989;86:9253–9257. doi: 10.1073/pnas.86.23.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ling LL, Keohavong P, Dias C, Thilly WG. Optimization of the polymerase chain reaction with regard to fidelity: Modified T7, Taq, and vent DNA polymerases. PCR Methods Appl. 1991;1:63–69. doi: 10.1101/gr.1.1.63. [DOI] [PubMed] [Google Scholar]
- 59.Höss M, et al. DNA damage and DNA sequence retrieval from ancient tissues. Nucleic Acid Res. 1996;24:1304–1307. doi: 10.1093/nar/24.7.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 61.Pääbo S. Ancient DNA: Extraction, characterization, molecular cloning, and enzymatic amplification. Proc Natl Acad Sci USA. 1989;86:1939–1943. doi: 10.1073/pnas.86.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sinha RP, Hader DP. UV-induced DNA damage and repair: A review. Photochem Photobiol Sci. 2002;1:225–236. doi: 10.1039/b201230h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.