Abstract
Most plastid proteins are encoded by the nuclear genome, and consequently, need to be transported into plastids across multiple envelope membranes. In diverse organisms possessing secondary plastids, nuclear-encoded plastid precursor proteins (preproteins) commonly have an N-terminal extension that consists of an endoplasmic reticulum (ER)-targeting signal peptide and a transit peptide-like sequence (TPL). This bipartite targeting peptide is believed to be necessary for targeting the preproteins into the secondary plastids. Here, we newly demonstrate the function of the bipartite targeting peptides of an algal group, chlorarachniophytes, and characterize the functional domains of the TPL in the precursor of a plastid protein, ATP synthase delta subunit (AtpD), using a GFP as a reporter molecule. We show that the C-terminal portion of the TPL is important for targeting the AtpD preprotein from the ER into the chlorarachniophyte plastids, and several positively charged amino acids in the TPL are also necessary for transporting the preprotein across the 2 innermost plastid membranes. Compared with other groups with secondary plastids, the TPL functional domains of the chlorarachniophytes are unique, which might be caused by independent acquisition of their plastids.
Keywords: ATP synthase delta subunit, secondary endosymbiosis, transit peptides, bipartite targeting peptide, periplastidal compartment
Plastids evolved either from the primary or the secondary endosymbioses (1, 2). Three major photosynthetic eukaryote groups [the green plants (land plants and green algae), red algae, and glaucophytes] evolved from a common ancestor that acquired plastids by a single primary endosymbiosis between a cyanobacterium and a nonphotosynthetic eukaryote. These plastids, which are surrounded by 2 envelope membranes, are called the “primary plastids” (3, 4). In subsequent secondary endosymbioses, different photosynthetic organisms with primary plastids (e.g., green and red algae) were engulfed and retained by 3 or more nonphotosynthetic eukaryotes of different lineages; they evolved into so-called “secondary plastids,” which are surrounded by 3 or 4 membranes (2, 5). Diverse algal groups, including chlorarachniophytes, cryptophytes, dinoflagellates, euglenophytes, haptophytes, heterokonts, and a nonphotosynthetic parasitic group, apicomplexans, harbor the secondary plastids (2).
In these endosymbiotic processes, numerous genes have transferred from the endosymbionts to the nuclear genomes of the host (6, 7), and the proteins expressed from these genes need to be sent back into the plastids (i.e., the former endosymbionts) across multiple plastid membranes (8–10). To be targeted correctly into the plastids, the precursors of nuclear-encoded plastid proteins generally possess plastid-targeting presequences (11). The acquisitions of the targeting sequences are thought to be essential factors for the integration of endosymbionts into host cells, to end up as organelles. The nuclear-encoded precursor proteins (preproteins) for the primary plastids have an N-terminal plastid-targeting sequence called a transit peptide (TP), and they are posttranslationally transported from the cytoplasm into the plastids (11–13). However, the preproteins for diverse secondary plastids commonly have N-terminal bipartite plastid-targeting sequences, each of which consists of a signal peptide (SP) and a TP-like (TPL) sequence (11). The SP is known, in several groups with secondary plastids, to be responsible for the cotranslational transport of preproteins into the endoplasmic reticulum (ER), and the TPL is believed to be necessary for targeting the preproteins into the plastid stroma across multiple envelope membranes (8–11).
Among the eukaryotic groups possessing secondary plastids, the primary sequences of the N-terminal bipartite plastid-targeting peptides are not conserved (11), which makes it difficult to characterize the common sequence motifs or structural elements essential to their function. Recent in vivo, in vitro, and in silico studies have revealed the functional domains of TPLs in several groups. Euglenophytes and dinoflagellates have secondary plastids surrounded by 3 smooth membranes. Many TPLs of those algal groups contain a remarkable hydrophobic region that anchors the preproteins to the ER-derived vesicular membrane during vesicular transportation from the ER into the plastids via Golgi bodies (14, 15). The plastids of heterokonts and cryptophytes are surrounded by 4 membranes; the outermost membrane is continuous with the ER. Their TPLs possess a conserved aromatic amino acid (such as phenylalanine) at the N terminus, and it has been demonstrated that this aromatic amino acid is necessary in allowing preproteins to pass through the 2 innermost plastid membranes (16–19). Apicomplexans have nonphotosynthetic plastids (apicoplasts) surrounded by 4 smooth membranes. It has been shown that several positively charged amino acids and a possible Hsp70 binding site in the TPLs are both important for targeting preproteins from the ER into the apicoplasts (20–22). These studies have indicated that, although “TPL” is commonly present in those eukaryotic groups, its function in the preprotein transport process seems differ among them. However, our knowledge of the TPL function is limited to a few groups. To obtain the “whole picture” of the diversity and evolution of plastid-targeting peptides in photosynthetic eukaryotes, we need to study other algal groups with secondary plastids that have not yet been sufficiently studied.
One such algal group is the chlorarachniophytes, a marine unicellular algal group that has acquired plastids via a secondary endosymbiosis between a green alga and a colorless cercozoan protist (23, 24). Each chlorarachniophyte plastid is bounded by 4 smooth membranes and contains a highly reduced nucleus, referred to as the nucleomorph, of the green algal endosymbiont in the periplastidal compartment (PPC), the space between the second and third plastid membranes (25, 26). The combination of these features is unique in secondary plastids, providing incentive to study plastid targeting in this algal group. Previous in silico analyses predicted that the nuclear-encoded plastid preproteins of chlorarachniophytes have N-terminal bipartite targeting peptides containing a SP and a TPL (27, 28). However, the TPL sequences are not well conserved among different preproteins and poorly characterized by those analyses, so detailed in vivo analyses are required to reveal the TPL functional domains in the chlorarachniophytes.
In this article, we demonstrate the function of bipartite targeting peptides in chlorarachniophytes, using a transient transformation system. We used one particular plastid-targeted preprotein [ATP synthase delta subunit protein of a chlorarachniophyte Bigelowiella natans (BnAtpD)] as a model; we then characterized the functional domains of its TPL, using GFP as a reporter molecule. We demonstrate that the C-terminal portion of the TPL is significant in transporting the preprotein from the ER into the chlorarachniophyte plastids, and that several positively charged amino acids within the TPL are also necessary, if the preprotein is to pass through the 2 innermost plastid membranes. We also carried out comparative analyses of bipartite targeting peptides among chlorarachniophytes and a few other organisms with secondary plastids. Here, we discuss the functional diversity among the TPLs for targeting preproteins into various secondary plastids.
Results and Discussion
Confirmation of the Plastid-Targeting Ability of Putative N-Terminal Bipartite Targeting Peptides in Chlorarachniophytes.
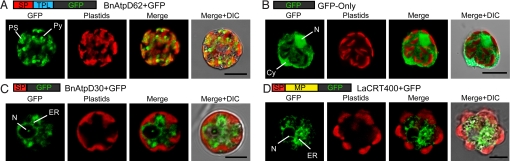
We obtained the cDNA sequences of 3 plastid-targeted protein genes (BnatpD, Bnfdx1, and BnrpL28) from a chlorarachniophyte, B. natans (29). N-terminal bipartite targeting peptides in the deduced preprotein sequences of those genes were predicted by the signal peptide prediction server SignalP (www.cbs.dtu.dk/services/SignalP) (30), and the chloroplast TP prediction server ChloroP (www.cbs.dtu.dk/services/ChloroP) (31). All 3 plastid-targeted preproteins were predicted to have N-terminal bipartite targeting peptides, an ER-targeting signal peptide (SP) followed by a TPL sequence (Fig. S1). The transient transformation system for the chlorarachniophyte Lotharella amoebiformis (32) was used to analyze the plastid-targeting abilities of these putative N-terminal targeting sequences in vivo. We prepared 3 plasmid constructs that expressed each of the putative bipartite targeting peptide regions of BnAtpD, BnFdx1, and BnRpL28 preproteins, fused with GFP; the cells were then transformed with each of these plasmids. In all 3 constructs (i.e., BnAtpD62+GFP, BnFdx94+GFP, and BnRpL88+GFP), the transformants exhibited GFP fluorescence in the plastid stroma and pyrenoid, which is a protein-rich structure in the plastid stroma (Fig. 1A and Fig. S2 A and B), and these GFP localizations were clearly different from the control, the GFP-Only (Fig. 1B). Therefore, these putative bipartite targeting peptides successfully delivered the GFP into the plastids across 4 plastid membranes. This result confirms that the bipartite targeting peptide of BnAtpD, BnFdx1, and BnRpL28 are sufficient for targeting these preproteins into the plastids. The chlorarachniophytes use a bipartite targeting signal system that is similarly used in other secondary plastid-bearing eukaryotic groups, including heterokonts, cryptophytes, and apicomplexans (17, 20, 33). It is interesting to note that bipartite targeting peptides are commonly used for targeting proteins into the secondary plastids of diverse groups, even though the origins of their plastids are different.
Fig. 1.
Confocal images of transformed L. amoebiformis cells, with GFP fluorescence. The images labeled GFP show GFP localization (green), and those labeled plastids show chlorophyll-autofluorescence (red). (A) A cell transformed with pBnAtpD62+GFP showing the GFP localization in the stromas and pyrenoids of plastids. (B) A cell transformed with pGFP-Only (a control), showing the GFP localization in the nucleus and cytoplasm. (C) A cell transformed with pBnAtpD30+GFP, showing the GFP localization in the putative ER. (D) A cell transformed with pLaCRT400+GFP, showing the GFP in the ER. (Scale bar, 5 μm.) DIC, differential interference contrast; Cy, cytoplasm; N, nucleus; PS, plastid stroma; Py, pyrenoid; MP, mature protein.
ER-to-Plastid Transport Signal in the TPL.
For detailed functional analyses of the N-terminal bipartite targeting peptide, we used a plastid-targeted preprotein, BnAtpD, as a model. First, to determine whether the putative SP is sufficient for targeting the preprotein into the ER, we created a plasmid construct that expresses the putative SP fused with GFP (BnAtpD30+GFP), and L. amoebiformis cells were transformed with this plasmid. In the transformed cells, GFP fluorescence was observed around the nucleus and in the cytoplasmic region (Fig. 1C). This localization was similar to that of GFP fused with an ER luminal protein, calreticulin (CRT), which had been isolated from L. amoebiformis (Fig. 1D and Fig. S2C). It appears that the GFP fused with the putative SP of BnAtpD was transported into the ER, but did not go into the plastids. This result demonstrates that, as in other cases, the SP is sufficient for delivering preproteins into the ER in the chlorarachniophytes. Combined with the fact that the whole bipartite targeting peptide of the BnAtpD preprotein can correctly deliver GFP into chlorarachniophyte plastids (Fig. 1A), these findings indicate that, in chlorarachniophytes, TPL is necessary for targeting preproteins from the ER into the plastids.
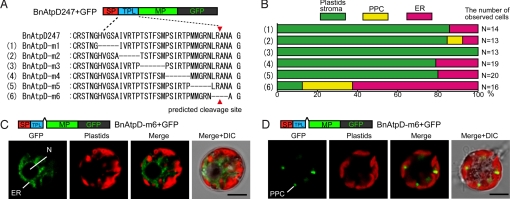
Although the requirement of TPL for the transport of preproteins from the ER to the inside of plastids has been confirmed in several eukaryotic groups with secondary plastids (11), it is still unclear whether the functional domain of chlorarachniophyte TPLs is similar or unique compared with TPLs of other secondary plastids. To detect the functional domain within the TPL of BnAtpD preprotein, we performed deletion analyses of GFP fusion proteins in vivo. The TPL region was divided into 6 portions (m1, m2, m3, m4, m5, and m6) consisting of 4–6 amino acids; each of these portions was deleted from the BnAtpD247+GFP (BnAtpD full-length preprotein + GFP), as shown in Fig. 2A. When the cells were transformed with each of pBnAtpD-m1, pBnAtpD-m2, pBnAtpD-m3, pBnAtpD-m4, and pBnAtpD-m5, the GFP fusion proteins were transported into the plastid stromas, and the obstruction to plastid-targeting was not observed in most transformed cells (Fig. 2B). However, when the BnAtpD-m6 fusion proteins (i.e., the products of the pBnAtpD-m6 deletion construct) were expressed in the cells, GFP fluorescence was observed in the putative ER in ≈60% of the transformed cells, and the ratio of the cells in which the GFP was transported into the plastid stroma was only 10% (Fig. 2 B and C). In the rest of the transformants, GFP was localized near the central region of each plastid, but the GFP fluorescence did not merge with chlorophyll autofluorescence (Fig. 2D). This localization pattern was similar to the localization of GFP fusion proteins targeted into PPC, as discussed in the following section (see Fig. 4). These results indicate that the m6 portion, which is in the vicinity of the putative cleavage site of the TPL, is important for targeting proteins from the ER into the plastids.
Fig. 2.
In vivo targeting of GFP fused with BnAtpD precursors possessing various deletions in the TPL. (A) Amino acid sequences of 6 constructs used for the deletion analyses. (A) Dashed line indicates a deleted portion in the TPL of BnAtpD. Red arrowheads indicate the predicted TPL cleavage site. (B) In vivo targeting-efficiencies of GFP fusion proteins, when cells were transformed with each plasmid construct. (C) Confocal images of a cell transformed with pBnAtpD-m6+GFP, showing the GFP localization in the putative ER. (D) Another transformed cell, showing the GFP localization in the possible PPC.
Fig. 4.
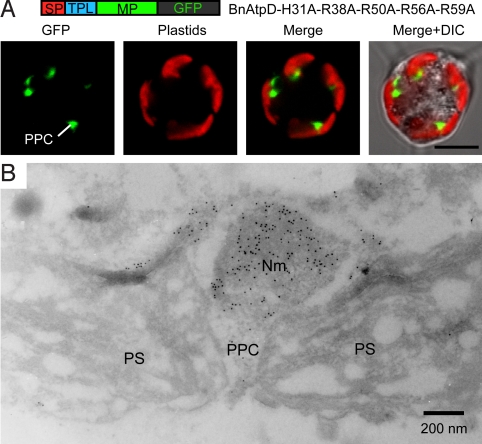
Immnocytochemical localization of a GFP fusion protein in the PPC. (A) Confocal images of a L. amoebiformis cell transformed with pBnAtpD-H31-R38A-R50A-R56A-R59A+GFP, showing the GFP localization in the PPC. (Scale bar, 5 μm.) (B) An immunoelectron micrograph of a plastid in a cell transformed with the pBnAtpD-H31A-R38A-R50A-R56A-R59A construct, showing the accumulation of conjugated gold particles (10 nm) in the PPC including a nucleomorph. Nm, nucleomorph.
The amino acid sequence of the m6 portion in the BnAtpD preprotein was L-R-A-N-A. To know whether other plastid-targeted preproteins of chlorarachniophytes generally have this sequence in their putative TPLs, we searched for the L-R-A-N-A sequence in the TPL sequences of 36 plastid-targeted preproteins using the Teiresias algorithm (http://cbcsrv.watson.ibm.com/Tspd.html) (34). This exact sequence was not found in other TPLs, but 12 of the 36 TPLs possessed sequences that were similar to the L-R-A-N-A sequence in chemical nature [i.e., (I/L/M/V)-X-(A/G)-X-(A/G)]. Intriguingly, 8 of the 12 TPLs had the motifs near the putative cleavage sites of the putative TPLs (Fig. S3). It is possible, at least for several proteins, that the C-terminal portion of TPL is involved in targeting a plastid preprotein from the ER into the chlorarachniophyte plastid, although further study is needed to determine what information (e.g., primary sequence, secondary structure, or chemical nature) in the functional portion is important for plastid targeting.
Because the plastids of chlorarachniophytes are not connected with any endomembrane in the cell, plastid preproteins are assumed to be transported via vesicular transports from the ER to the plastids (35). It is possible that the C-terminal functional domain of the chlorarachniophyte TPLs is involved in this transport step. This type of functional domain in TPL has not been known in any other plastid-bearing eukaryotic groups. The uniqueness of this TPL functional domain may reflect the differences of plastid membrane structure and protein transport pathway between the chlorarachniophytes and other groups. Unlike the chlorarachniophyte plastids, the outermost plastid membrane is continuous with the ER in the plastids of heterokonts, cryptophytes, and haptophytes, and plastid preproteins are believed to be transported from the ER into the plastids via the membrane connection (9, 35). It has been demonstrated in the heterokonts that the TPL functional domain localizes in the N-terminal portion (33). In euglenophytes and dinoflagellates, plastids are surrounded only by 3 membranes, and plastid preproteins are known to be transported from the ER to the plastids via Golgi bodies (8). The TPLs of these preproteins typically possess a remarkable hydrophobic region, which is known to act as a stop-transfer membrane anchor during the vesicular transportation via the Golgi bodies (14, 15).
The plastids of apicomplexans, apicoplasts, are surrounded by 4 smooth membranes, which is structurally similar to the chlorarachniophyte plastids, and plastid preproteins are transported from the ER to the apicoplasts directly via vesicular transports (36). However, in apicomplexans, a possible Hsp70 binding site and a net positive charge in the TPLs are both involved in targeting preproteins from the ER into the apicoplasts (20, 21). The difference in the feature of TPL functional domain between the apiconplexans and the chlorarachniophytes suggests that functional domains do not always correlate with the plastid membrane structures and protein transport pathways.
PPC-to-Stroma Transport Signal in the TPL.
Based on the amino acid composition of TPLs in chlorarachniophyte plastid-targeted preproteins, TPL sequences typically contain several basic amino acids and very few acidic amino acids (27, 28). To demonstrate the role of basic amino acids in plastid-targeting, we performed substitution analyses on those amino acids within the BnAtpD preprotein. The BnAtpD TPL contains a weak basic amino acid (histidine at the 31st position) and 4 strong basic amino acids (arginine at the 38th, 50th, 56th, and 59th positions) from the N-terminal end of this preprotein.
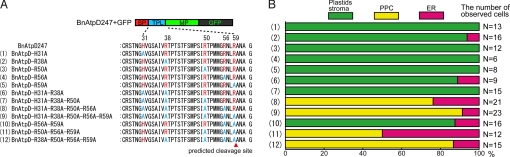
First, we created 5 plasmid constructs, each of which had an alanine substitution at each of the basic amino acids in BnAtpD247+GFP (Fig. 3A); L. amoebiformis cells were then transformed with each of these constructs. With all 5 constructs, GFP fusion proteins were correctly transported into the plastid stromas, and no plastid-targeting obstructions were observed (Fig. 3B, rows 1–5). Next, the numbers of alanine substitutions in the TPL were increased. When 2 to 3 basic amino acids from the N terminus were substituted, the GFP fusion proteins (BnAtpD-H31A-R38A and BnAtpD-H31A-R38A-R50A) were localized in the plastid stromas in almost all transformants (Fig. 3B, rows 6 and 7). In contrast, when 4 to 5 basic amino acids were substituted, the fusion proteins (BnAtpD-H31A-R38A-R50A-R56A and BnAtpD-H31A-R38A-R50A-R56A-R59A) were not transported into the plastid stromas at all, and >75% of the transformants showed GFP fluorescence in the possible PPC near the central part of each plastid (Fig. 3B, rows 8 and 9). Another GFP fusion protein, BnAtpD-R56A-R59A, in which 2 basic amino acids from the C terminus were substituted, was correctly transported into the plastid stroma (Fig. 3B, row 10). However, when 3 or 4 basic amino acids from the C terminus were substituted (i.e., BnAtpD-R50A-R56A-R59A and BnAtpD-R38A-R50A-R56A-R59A), GFP fluorescence was accumulated in the possible PPC regions in the majority of transformants (Fig. 3B, rows 11 and 12). To confirm the possible PPC localization of GFP, we performed an immunogold localization of GFP in the transformants with the pBnAtpD-H31A-R38A-R50A-R56A-R59A construct, using an anti-GFP antibody. The conjugated gold particles accumulated in the PPC including a nucleomorph in the transformed cells (Fig. 4); the average number of gold particles was 326 ± 66 μm−2 in the PPC, which was significantly higher than the count of cytoplasm and plastid stroma (5.5 ± 2.8 μm−2 and 6.6 ± 4.1 μm−2, respectively). In nontransformed cells (WT), no accumulation of gold particles was observed in the PPC (4.2 ± 1.2 μm−2).
Fig. 3.
In vivo targeting of GFP fused with BnAtpD preproteins possessing various substitutions in the TPL. (A) Amino acid sequences of 12 constructs used for the substitution analyses. Characters in blue indicate the alanine residues substituted at the positions of basic amino acids (red characters). Red arrowheads indicate the predicted TPL cleavage site. (B) In vivo targeting-efficiencies of GFP fusion proteins when cells were transformed with each plasmid construct.
The substitution experiments demonstrated that the GFP fusion proteins with at least 2 strong basic amino acids (arginine residues) in the TPL were correctly targeted into the plastid stroma; however, the use of >4 substitutions was found to impede plastid-targeting, and the GFP fusion proteins consequently accumulated in the PPC. This result suggests that the number of strong basic amino acids, positively charged amino acids, in the TPL is significant, rather than their position. The net positive charge of TPLs might be important for preproteins in passing through the 2 innermost plastid membranes of chlorarachniophytes.
In green plants (i.e., land plants and green algae) with primary plastids surrounded by 2 envelope membranes, the TPs of plastid-targeted preproteins are comparatively depleted of negatively charged amino acids, and show an overall positive charge (11, 12). In the process of transporting these preproteins, the net positive charges of TPs are thought to assist the preproteins to interact electrostatically with the negatively charged plastid outer membrane and import receptors (37). The fact that positively charged amino acids are involved in protein transport across the 2 envelope membranes in both green plants and chlorarachniophytes leads us to believed that these 2 groups use a similar mechanism for protein translocation. This idea is further supported by the discovery of a few genes for the translocons at the outer and inner envelope membranes of chloroplast (TOC and TIC) in a chlorarachniophyte nucleomorph genome (26). It is likely that the portion of the protein transport mechanism in the 2 innermost plastid membranes in the chlorarachniophyte directly evolved from the green algal endosymbiont, the ancestor of the chlorarachniophyte plastids, in the process of secondary endosymbiosis.
In cryptophytes, heterokonts, and perhaps apicomplexans, an N-terminal aromatic amino acid of TPL is involved in transporting preproteins across the 2 innermost plastid membranes (11, 16–19). These groups have red algal-derived secondary plastids, and it has been known that the TPs of red algae also possess a conserved aromatic amino acid at the N termini (11). In both primary and secondary plastids of red algal-lineages, the N-terminal aromatic amino acid of TPs and TPLs is believed to be essential for transporting preproteins into the plastids across the 2 innermost plastid membranes. These facts and our study imply that TPL functional domains for passing through the 2 innermost membranes of secondary plastids have generally derived from the TPs of the ancestor of their plastids.
Functional Compatibility of Bipartite Plastid-Targeting Peptide Among Eukaryotic Groups with Secondary Plastids.
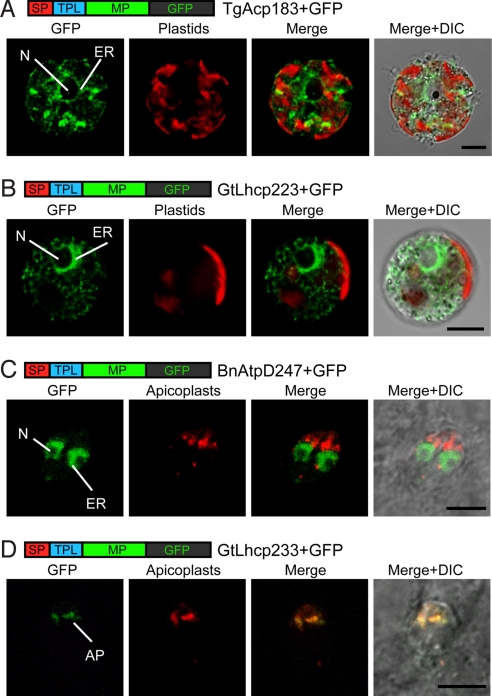
The functional domains of TPLs are different among groups of organisms with secondary plastids. To understand the functional compatibility of TPLs among some of these groups, we tested the in vivo transports of heterologous preproteins, using the transformation systems of the apicomplexan Toxoplasma gondii and the chlorarachniophyte L. amoebiformis. First, we created 2 plasmid constructs that expressed the GFP fused with a plastid-targeted preprotein, the acyl carrier protein of T. gondii (TgAcp) or the light harvesting complex protein of the cryptophyte Guillardia theta (GtLhcp); the L. amoebiformis cells were then transformed with each of these plasmids. In the transformed cells, neither of the GFP fusion proteins (TgAcp183+GFP and GtLhcp233+GFP) was transported into the chlorarachniophyte plastids, and in both cases, GFP fluorescence was observed in the putative ER (Fig. 5 A and B). Next we created 2 more plasmid constructs that expressed the GFP fused with the BnAtpD or GtLhcp preprotein, and the T. gondii cells were transformed with each plasmid. In the transformed cells with the pBnAtpD247+GFP, the GFP fusion proteins were localized in the putative ER, and no GFP fluorescence was observed in the apicomplexan plastid, the apicoplast (Fig. 5C). However, another GFP fusion protein, GtLhcp233+GFP, was targeted into the apicoplast (Fig. 5D).
Fig. 5.
Confirmation of functional compatibility of bipartite plastid-targeting peptides among the chlorarachniophytes, cryptophytes, and apicomplexans. Confocal images of transformed cells expressing heterologous plastid preproteins fused with GFP. (A) Localization of GFP fused with acyl carrier protein of T. gondii (TgAcp183+GFP) in a L. amoebiformis cell. (B) Localization of GFP fused with light harvesting complex protein of G. theta (GtLhcp233+GFP) in a L. amoebiformis cell. (C) Localization of GFP fused with an ATP synthase delta subunit of B. natans (BnAtpD247+GFP) in T. gondii cells. (D) Localization of the GtLhcp233+GFP protein in T. gondii cells. To detect apicoplasts in T. gondii cells, DsRed (red fluorescent protein) fused with an apicoplast-targeted preprotein of T. gondii (TgAcp183+DsRed) was used. AP, apicoplast. (Scale bar, 5 μm.)
These results indicate that the SPs of plastid-targeted preproteins have functional compatibility among cryptophytes, chlorarachniophytes, and apicomplexans, but the TPLs have no compatibility between chlorarachnophytes and apicomplexans, or between chlorarachniophytes and cryptophytes. In contrast, the plastid-targeting peptide of the cryptophytes is able to deliver the GFP to the plastid of the apicomplexans. This result is very interesting, because the plastids of apicomplexans and cryptophytes are different, in terms of the structure of the envelope membranes surrounding them. Also, these findings imply that the plastid-targeting peptide is functionally compatible among the photosynthetic lineages of the large phylogenetic group “chromalveolates,” which further implies that all photosynthetic lineages of the chromalveolates (namely, the apicomplexans, cryptophytes, heterokonts, haptophytes, and dinoflagellates) possibly share a common plastid origin (38). This functional compatibility is also supported by 2 previous findings. The existence of an ER-associated degradation (ERAD)-like system for transporting preproteins across the second-from-the-outside of the 4 plastid-surrounding membranes in cryptophytes, heterokonts, and apicomplexans (39); and the capability of cryptophyte plastid-targeting peptide to deliver GFP into the plastids of the heterokont Phaeodactylum tricornutum (17). It appears that photosynthetic chromalveolates with secondary plastids use, at least in part, the protein-targeting signals for plastids of the red algal endosymbiont, which eventually became their own plastids.
The present study clearly demonstrates that the functional domains of TPLs are different among plastid-bearing organisms, which contributes to providing a better interpretation of the evolution of diverse plastid-targeting peptides. However, further detailed study is required to understand the true diversity and evolution of protein-targeting mechanisms in secondary plastids, given that the detailed function of plastid-targeting peptides is still unclear in several secondary algal groups such as euglenophytes where even different classes of TPLs exist (15), haptophytes, and dinoflagellates. The development of good genetic transformation systems for these algal groups is likely essential to the advancement of the studies in this field.
Methods
Plasmid Constructions.
To transform L. amoebiformis cells, cDNA fragments of BnatpD (AY267652), Bnfdx1 (AY267628), BnrpL28 (AY267644), LaCRT (FJ209028), Tgacp (AF038925), and Gtlhcp (AM491789) with artificial restriction sites at both ends were amplified by PCR and inserted between the HindIII and NcoI sites of the pLaRGfp+mc vector generated from the pLaRGfp (32). To introduce mutations (i.e., substitutions or deletions) into the BnatpD fragments, we used a PCR-based site-directed mutagenesis technique (40).
To transform T. gondii cells, the fragments of BnatpD or Gtlhcp fused with the egfp gene were amplified by PCR, and these fragments were inserted between the EcoRI and PacI sites of the pSAG1/1 CAT vector (41), replacing the CAT gene to generate pSAG-BnAtpD247+GFP and pSAG-GtLhcp233+GFP, respectively. A chimera fragment of Tgacp fused with the DsRed-Express gene (Clontech) was generated using the splicing by overlapped extension by PCR (42); it was then inserted into the pSAG1/1 CAT, replacing CAT gene. The resulting fragment, which contained the SAG1 promoter, Tgacp fused with DsRed, and the SAG1 terminator, was ligated into the SacII site of pTub5CATSag1 vector. All constructs were subsequently sequenced to ensure correct construction.
Transient Transformation.
L. amoebiformis (CCMP2058) (43) was grown at 20 °C, under white illumination (80–100 μmol photons·m−2·s−1) with a 12-h light/12-h dark cycle, in 500 mL Erlenmeyer flasks containing 300 mL of ESM medium (44). L. amoebiformis cells were transformed with plasmid vectors using the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad), as previously described (32). T. gondii (RH strain) was maintained by growth in a monolayer of Vero cells in RPMI1640 medium with 3% FCS at 37 °C. The transformation of T. gondii was carried out via an electroporation method, using a Gene Pulser Xcell (Bio-Rad) (45). After transformation, those cells were incubated under the conditions described above.
Observation of GFP Localizations.
Twenty-four to 48 h after transformation, the transient transformed cells expressing reporter genes were observed under an inverted Zeiss LSM 510 laser scanning microscope (Carl Zeiss); confocal imaging was performed using the single-track mode. GFP fluorescence was detected with a 505–530 nm band pass filter, and plastid autofluorescence and DsRed fluorescence were detected with 585- and 560-nm-long pass filters, respectively, in the excitation line of a 488 nm argon laser and a 543 nm He/Ne laser. The localization of GFP in the transformed cells was also observed using an immunogold localization method under a transmission electron microscope (SI Methods).
Supplementary Material
Acknowledgments.
We thank Dr. L. David Sibley (Washington University, St. Louis, MO) for the pTub5CATSag1 vector and Dr. Shuhei Ota (Station Biologique de Roscoff, France) for technical help using the electron microscope. This work was supported by the Grant-in-Aid for Scientific Research (KAKENHI) on Priority Area “Comparative Genomics” and by Ministry of Education, Culture, Sport, Science, and Technology of Japan Special Coordination Funds for Promoting Science and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.M.A. is a guest editor invited by the Editorial Board.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. FJ209028).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902578106/DCSupplemental.
References
- 1.McFadden GI. Primary and secondary endosymbiosis and the origin of plastids. J Phycol. 2001;37:951–959. [Google Scholar]
- 2.Keeling PJ, Archibald JM, Fast NM, Palmer JD. Comment on “The evolution of modern eukaryotic phytoplankton”. Science. 2004;306:2191b. doi: 10.1126/science.1103879. [DOI] [PubMed] [Google Scholar]
- 3.Moreira D, Guyader HL, Philippe H. The origin of red algae and the evolution of chloroplasts. Nature. 2000;405:69–72. doi: 10.1038/35011054. [DOI] [PubMed] [Google Scholar]
- 4.Rodríguez-Ezpeleta N, et al. Monophyly of primary photosynthetic eukaryotes: Green plants, red algae and glaucophytes. Curr Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 5.Cavalier-Smith T. Membrane heredity and early chloroplast evolution. Trends Plants Sci. 2000;5:174–182. doi: 10.1016/s1360-1385(00)01598-3. [DOI] [PubMed] [Google Scholar]
- 6.Martin W, et al. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- 7.Bock R, Timmis JN. Reconstructing evolution: Gene transfer from plastids to the nucleus. BioEssays. 2008;30:556–566. doi: 10.1002/bies.20761. [DOI] [PubMed] [Google Scholar]
- 8.van Dooren GG, Schwartzbach SD, Osafune T, McFadden GI. Translocation of proteins across the multiple membranes of complex plastids. Biochim Biophys Acta. 2001;1541:34–53. doi: 10.1016/s0167-4889(01)00154-9. [DOI] [PubMed] [Google Scholar]
- 9.Ishida K. Protein targeting into plastids: A key to understanding the symbiogenetic acquisitions of plastids. J Plant Res. 2005;118:237–245. doi: 10.1007/s10265-005-0218-2. [DOI] [PubMed] [Google Scholar]
- 10.Nassoury N, Morse D. Protein targeting to the chloroplasts of photosynthetic eukaryotes: Getting there is half the fun. Biochim Biophys Acta. 2005;1743:5–19. doi: 10.1016/j.bbamcr.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Patron NJ, Waller RF. Transit peptide diversity and divergence: A global analysis of plastid targeting signals. BioEssays. 2007;29:1048–1058. doi: 10.1002/bies.20638. [DOI] [PubMed] [Google Scholar]
- 12.Bruce BD. The paradox of plastid transit peptide: Conservation of function despite divergence in primary structure. Biochim Biophys Acta. 2001;1541:2–21. doi: 10.1016/s0167-4889(01)00149-5. [DOI] [PubMed] [Google Scholar]
- 13.Steiner JM, Yusa F, Pompe JA, Löffelhaedt W. Homologous protein import machineries in chloroplasts and cyanelles. Plant J. 2005;44:646–652. doi: 10.1111/j.1365-313X.2005.02559.x. [DOI] [PubMed] [Google Scholar]
- 14.Patron NJ, Waller RF, Archibald JM, Keeling PJ. Complex protein targeting to dinoflagellate plastids. J Mol Biol. 2005;348:1015–1024. doi: 10.1016/j.jmb.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 15.Durnford DG, Gray MW. Analysis of Euglena gracilis plastid-targeted proteins reveals different classes of transit sequences. Eukaryot Cell. 2006;5:2079–2091. doi: 10.1128/EC.00222-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilian O, Kroth PG. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 2005;41:175–183. doi: 10.1111/j.1365-313X.2004.02294.x. [DOI] [PubMed] [Google Scholar]
- 17.Gould SB, et al. Protein targeting into the complex plastid of cryptophytes. J Mol Evol. 2006;62:674–681. doi: 10.1007/s00239-005-0099-y. [DOI] [PubMed] [Google Scholar]
- 18.Gould SB, et al. Nucleus-to-nucleus gene transfer and protein retargeting into a remnant cytoplasm of cryptophytes and diatoms. Mol Biol Evol. 2006;23:2413–2422. doi: 10.1093/molbev/msl113. [DOI] [PubMed] [Google Scholar]
- 19.Gruber A, et al. Protein targeting into complex diatom plastid: Functional characterisation of a specific targeting motif. Plant Mol Biol. 2007;64:519–530. doi: 10.1007/s11103-007-9171-x. [DOI] [PubMed] [Google Scholar]
- 20.Foth BJ, et al. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science. 2003;299:705–708. doi: 10.1126/science.1078599. [DOI] [PubMed] [Google Scholar]
- 21.Tonkin CJ, Roos DS, McFadden GI. N-terminal positively charged amino acids, but not their exact position, are important for apicoplast transit peptide fidelity in Toxoplasma gondii. Mol Biochem Parasitol. 2006;150:192–200. doi: 10.1016/j.molbiopara.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Tonkin CJ, et al. Evolution of malaria parasite plastid targeting sequences. Proc Natl Acad Sci USA. 2008;105:4781–4785. doi: 10.1073/pnas.0707827105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishida K, Green BR, Cavalier-Smith T. Diversification of a chimaeric algal group, the chlorarachniophytes: Phylogeny of nuclear and nucleomorph small-subunit rRNA genes. Mol Biol Evol. 1999;16:321–331. [Google Scholar]
- 24.Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: Evidence for independent origin of chlorarachniophyte and euglenid secondary endosymbionts. Mol Biol Evol. 2007;24:54–62. doi: 10.1093/molbev/msl129. [DOI] [PubMed] [Google Scholar]
- 25.McFadden GI, Gilson PR, Hofmann CJB, Adcock GJ, Maier UG. Evidence that an amoeba acquired a chloroplast by retaining part of an engulfed eukaryotic alga. Proc Natl Acad Sci USA. 1994;91:3690–3694. doi: 10.1073/pnas.91.9.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilson PR, et al. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: Nature's smallest nucleus. Proc Natl Acad Sci USA. 2006;103:9566–9571. doi: 10.1073/pnas.0600707103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers MB, et al. Plastid-targeting peptide from the chlorarachniophyte Bigelowiella natans. J Eukaryot Microbiol. 2004;51:529–535. doi: 10.1111/j.1550-7408.2004.tb00288.x. [DOI] [PubMed] [Google Scholar]
- 28.Gile GH, Keeling PJ. Nucleus-encoded periplastid-targeted EFL in chlorarachniophytes. Mol Biol Evol. 2008;25:1967–1977. doi: 10.1093/molbev/msn147. [DOI] [PubMed] [Google Scholar]
- 29.Archibald JM, Rogers MB, Toop M, Ishida K, Keeling PJ. Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc Natl Acad Sci USA. 2003;100:7678–7683. doi: 10.1073/pnas.1230951100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 31.Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirakawa Y, Kofuji R, Ishida K. Transient transformation of a chlorarachniophyte alga, Lotharella amoebiformis (Chlorarachniophyceae), with uidA and egfp reporter genes. J Phycol. 2008;44:814–820. doi: 10.1111/j.1529-8817.2008.00513.x. [DOI] [PubMed] [Google Scholar]
- 33.Apt KE, et al. In vivo characterization of diatom multipartite plastid targeting signals. J Cell Sci. 2002;115:4061–4069. doi: 10.1242/jcs.00092. [DOI] [PubMed] [Google Scholar]
- 34.Rigoutsos I, Floratos A. Combinatorial pattern discovery in biological sequences: The TEIRESIAS algorithm. Bioinformatics. 1998;14:55–67. doi: 10.1093/bioinformatics/14.1.55. [DOI] [PubMed] [Google Scholar]
- 35.Bolte K, et al. Protein targeting into secondary plastids. J Eukaryot Microbiol. 2009;56:9–15. doi: 10.1111/j.1550-7408.2008.00370.x. [DOI] [PubMed] [Google Scholar]
- 36.Tonkin CJ, Struck NS, Mullin KA, Stimmler LM, McFadden GI. Evidence for Golgi-independent transport from the early secretory pathway to the plastid in malaria parasites. Mol Microbiol. 2006;61:614–630. doi: 10.1111/j.1365-2958.2006.05244.x. [DOI] [PubMed] [Google Scholar]
- 37.Jarvis P, Robinson C. Mechanisms of protein import and routing in chloroplasts. Curr Biol. 2004;14:R1064–R1077. doi: 10.1016/j.cub.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 38.Cavalier-Smith T. Principle of protein and lipid targeting in secondary symbiogenesis: Euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- 39.Sommer MS, et al. Der1-mediated preprotein import into the periplastid compartment of chromalveolates? Mol Biol Evol. 2007;24:918–928. doi: 10.1093/molbev/msm008. [DOI] [PubMed] [Google Scholar]
- 40.Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interaction. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soldati D, Boothroyd JC. Transient transfection and expression in the obligate intercellular parasite Toxoplasma gondii. Science. 1993;260:349–352. doi: 10.1126/science.8469986. [DOI] [PubMed] [Google Scholar]
- 42.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 43.Ishida K, Ishida N, Hara Y. Lotharella amoeboformis sp nov: A new species of chlorarachniophytes from Japan. Phycol Res. 2000;48:221–229. [Google Scholar]
- 44.Kasai F, Kawachi M, Erata M, Watanabe MM. NIES-Collection List of Strains: Microalgae and Protozoa. 7th Ed. Tsukuba, Japan: Microbial Culture Collection, National Institute for Environmental Studies; 2004. p. 54. [Google Scholar]
- 45.Karsten V, Qi H, Beckers CJ, Joiner K. Targeting the secretory pathway of Toxoplasma gondii. Methods. 1997;13:103–111. doi: 10.1006/meth.1997.0503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.