Abstract
The entry of carbon from sucrose into cellular metabolism in plants can potentially be catalyzed by either sucrose synthase (SUS) or invertase (INV). These 2 routes have different implications for cellular metabolism in general and for the production of key metabolites, including the cell-wall precursor UDPglucose. To examine the importance of these 2 routes of sucrose catabolism in Arabidopsis thaliana (L.), we generated mutant plants that lack 4 of the 6 isoforms of SUS. These mutants (sus1/sus2/sus3/sus4 mutants) lack SUS activity in all cell types except the phloem. Surprisingly, the mutant plants are normal with respect to starch and sugar content, seed weight and lipid content, cellulose content, and cell-wall structure. Plants lacking the remaining 2 isoforms of SUS (sus5/sus6 mutants), which are expressed specifically in the phloem, have reduced amounts of callose in the sieve plates of the sieve elements. To discover whether sucrose catabolism in Arabidopsis requires INVs rather than SUSs, we further generated plants deficient in 2 closely related isoforms of neutral INV predicted to be the main cytosolic forms in the root (cinv1/cinv2 mutants). The mutant plants have severely reduced growth rates. We discuss the implications of these findings for our understanding of carbon supply to the nonphotosynthetic cells of plants.
Most plant cells receive essentially all of their carbon as sucrose. Sucrose catabolism in plants is one of the largest metabolic fluxes on the planet, second only to fluxes in primary carbon assimilation. Only 2 enzymes can catalyze sucrose catabolism under physiological conditions: sucrose synthase (SUS) and invertase (INV); thus, most plant biomass is derived via 1 of these 2 routes. However, despite their central role in carbon partitioning and biomass accumulation, the precise roles and relative importance of these enzymes remain largely unknown.
SUS and INV both occur as multiple, distinct isoforms. INV catalyzes the effectively irreversible hydrolysis of sucrose to glucose and fructose. Isoforms in the cell wall and vacuole (acid INV) differ in structure from those predicted to be in the cytosol, mitochondria and plastids (neutral/alkaline INV). SUS catalyzes the reversible conversion of sucrose to fructose and UDPglucose; SUS isoforms are believed to be cytosolic.
Several lines of evidence indicate a predominant role for SUS in the entry of carbon into metabolism in nonphotosynthetic cells. Individual isoforms are needed for normal development in some plant organs, including potato tuber, pea and maize seed, tomato fruit, and cotton fibers (1–5). SUS is held to be important in determining sink strength, and in phloem loading (1, 6, 7). It is also proposed to have specific roles in cellulose synthesis, and in starch synthesis in leaves. In the widely cited model for cellulose synthesis, the substrate UDPglucose is channeled to the cellulose synthase complex in the plasma membrane via a SUS associated with the inner face of the complex (8, 9). Consistent with this idea, some SUS activity is associated with the plasma membrane (10–12). Leaf starch synthesis is generally believed to occur via a pathway in which the substrate ADPglucose is generated inside the chloroplast, without involvement of SUS. However, a recent alternative proposal is that ADPglucose is generated via SUS from sucrose in the cytosol, then imported into the chloroplast (13). Evidence for this pathway includes parallel alterations in starch levels in leaves of transgenic potato plants in which SUS activity has been altered (14). If correct, this proposal gives SUS a central role in photosynthetic carbon assimilation and partitioning.
Most of the roles proposed for INVs are specific to particular developmental stages. Vacuolar INV is involved in mobilization of vacuolar sucrose in sucrose-storing organs (15, 16). It is required for normal root elongation in Arabidopsis, probably through its impact on vacuolar osmotic potential and, thus, on water uptake (17). Cell-wall INV activity is high after wounding and pathogen attack (18, 19), and in early seed development (20), and is required for normal kernel development in maize (21) and pollen tube extension (22). The functions of neutral INV are not known, but loss of 1 of the 6 isoforms in rice (OsCYT-INV1), or 1 of the 7 in Lotus japonicus (LjINV1), strongly affects plant growth and development (23, 41). Loss of 1 of the 9 isoforms in Arabidopsis (CINV1 or CYT-INV1) has much less pronounced effects. It reduces primary root extension by ≈30% and can reduce leaf and silique expansion (24, 25).
The route of sucrose catabolism has important implications for energy conservation and carbon allocation in nonphotosynthetic cells. Conversion of sucrose to hexose phosphates via SUS uses only half the ATP needed for conversion via INV. The reversibility of the reactions of the SUS route means flux via this route is sensitive to hexose phosphate levels and, thus, to demand for glycolytic intermediates (26, 27). The requirement for PPi (as a substrate for UDPglucose pyrophosphorylase) links sucrose catabolism via SUS to other PPi-requiring processes, including flux over the reversible glycolytic enzyme PPi-dependent fructose 6-phosphate phosphotransferase. In contrast, the INV-catalyzed reaction is effectively irreversible, and INV isoforms have no reported properties that would allow coordination of sucrose catabolism with carbon demand in nonphotosynthetic cells.
Despite the accepted importance of SUS, we recently showed that none of the 6 isoforms in Arabidopsis is individually required for normal growth and reproduction, neither are any of the 3 pairs of most closely related isoforms (SUS1/SUS4, SUS2/SUS3, and SUS5/SUS6) (28). Thus, there must either be a high level of redundancy within the SUS family in Arabidopsis, or INV isoforms must be able to compensate for loss of SUS in this species. To explore the implications of this finding, we have generated a quadruple mutant (the sus1/sus2/sus3/sus4 mutant) that has no detectable soluble or membrane-bound SUS activity. Remarkably, the mutant is normal with respect to growth and development, metabolite levels, seed composition, and the composition of cell walls. In marked contrast, loss of 2 of the 9 isoforms of neutral INV (the cinv1/cinv2 mutant) results in severe inhibition of growth. We discuss the implications of these results for understanding of sucrose catabolism in the nonphotosynthetic cells of plants.
Results
Location of SUS Isoforms SUS5 and SUS6.
We were previously unable to detect SUS5 or SUS6 proteins in soluble and membrane fractions of Arabidopsis plants (28). Surprisingly, both were present in insoluble (mainly cell-wall) material from roots and hypocotyls, and SUS6 was present in this material from stems (Fig. 1A). Neither protein was detected in insoluble material from plants of a double mutant lacking both SUS5 and SUS6 (the sus5/sus6 mutant) (28). SUS activity in insoluble material from roots and stems was reduced by ≈65% in sus5/sus6 mutant plants (Table 1).
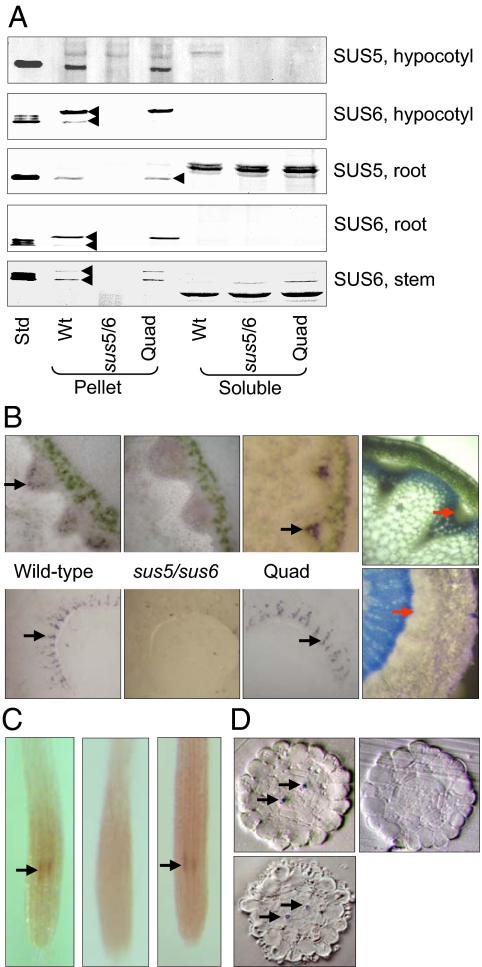
Fig. 1.
Location of SUS6 transcript and protein in WT and quadruple mutant plants. Pictures are representative of multiple plants for each genotype and treatment. (A) Blots of hypocotyl extracts (from 11-week-old plants grown in short days), stems (as in Table 1), whole root systems (from 5-week-old plants grown in an inert medium), and purified SUS proteins (28) (Std, SUS5 protein for SUS5 antiserum; SUS6 protein for SUS6 antiserum) were probed with SUS5 or SUS6 antiserum (28). In each panel, lanes are from the same gel and blot. Equal fractions of soluble and pellet material were loaded. Note that bands recognized by the SUS5 antiserum in root soluble fractions are not SUS5 protein. They migrate more slowly than authentic SUS5 (arrowed), and are present in the sus5/sus6 mutant. The SUS6 antiserum recognizes 2 bands in extracts (arrowed in WT). Both are missing in the sus5/sus6 mutant, so both are probably SUS6 protein. (B) Tissue prints of stem (Upper) and hypocotyl (Lower) sections probed with SUS6 antiserum. (Left to Right) WT, sus5/sus6 mutant, sus1/sus2/sus3/sus4 (Quad) mutant, and a stem section stained with toluidine blue. Black arrows show typical antiserum reactions; red arrows show the equivalent region on the section. (C) Location of SUS6 transcript in roots by whole-mount in situ RNA hybridization. Fixed and cleared roots of 4-day-old seedlings were treated with RNA probes for SUS6. (Left) WT with SUS6 antisense probe; (Center) WT with SUS6 sense probe; (Right) sus1/sus2/sus3/sus4 mutant with SUS6 antisense probe. The appearance of sus6 mutant roots with SUS6 antisense probe was the same as WT roots with SUS6 sense probe. Arrows show RNA hybridization. Root diameter is ≈130 μm. (D) Location of SUS transcript in root sections. Roots as in C were embedded and sectioned at the level at which RNA hybridization was observed. Arrows show hybridization. (Upper Left and Right) Sections of WT roots with SUS6 antisense probe and SUS6 sense probe respectively; (Lower Left) a sus1/sus2/sus3/sus4 mutant root with SUS6 antisense probe.
Table 1.
SUS activity, metabolite contents, and seed weight of WT and mutant plants
| Genotype | Sucrose synthase activity, nmol min−1 g−1 fresh weight | |||
|---|---|---|---|---|
| Roots |
Stems |
|||
| Soluble | Pellet | Soluble | Pellet | |
| WT | 49.8 ± 3.2 | 5.3 ± 0.1 | 27.5 ± 6.6 | 1.8 ± 0.2 |
| sus5/sus6 | 49.8 ± 4.9 | 1.9 ± 0.5 | 41.6 ± 6.2 | 0.6 ± 0.5 |
| sus1/sus2/sus3/sus4 | 0.1 ± 0.1 | 3.4 ± 0.4 | 0.4 ± 0.3 | 1.6 ± 0.1 |
| Carbohydrate content, μmol g−1 fresh weight | ||||||||
|---|---|---|---|---|---|---|---|---|
| Leaves | Roots | |||||||
| End of day |
End of night |
End of day |
End of night |
|||||
| WT | Quad | WT | Quad | WT | Quad | WT | Quad | |
| Glucose | 0.59 ± 0.09 (7) | 0.64 ± 0.10 (7) | 0.22 ± 0.02 (7) | 0.14 ± 0.02 (7) | 1.5 ± 0.2 (7) | 1.9 ± 0.2 (7) | 0.48 ± 0.05 (6) | 0.58 ± 0.06 (5) |
| Fructose | 0.15 ± 0.03 (6) | 0.17 ± 0.02 (6) | 0.041 ± 0.012 (6) | 0.025 ± 0.004 (5) | 0.094 ± 0.006 (6) | 1.05 ± 0.07 (6) | 0.25 ± 0.04 (6) | 0.31 ± 0.03 (6) |
| Sucrose | 1.01 ± 0.19 (7) | 1.07 ± 0.19 (7) | 0.35 ± 0.04 (7) | 0.45 ± 0.03 (7) | 1.95 ± 0.22 (7) | 2.18 ± 0.34 (7) | 0.76 ± 0.10 (6) | 0.57 ± 0.08 (6) |
| Starch | 31.5 ± 1.3 (7) | 31.4 ± 1.9 (5) | 2.4 ± 0.3 (7) | 2.6 ± 0.3 (7) | 0.20 ± 0.02 (7) | 0.21 ± 0.02 (7) | 0.18 ± 0.02 (6) | 0.15 ± 0.01 (6) |
| Genotype | Seed weight, mg | Percentage oil | ||||||
| WT | 0.0165 ± 0.0007 | 33.62 ± 1.99 | ||||||
| sus1/sus2/sus3/sus4 | 0.0171 ± 0.0013 | 33.45 ± 1.38 | ||||||
SUS activity, metabolite contents, seed weight, and lipid content of WT and mutant plants. Roots were from 4- or 5-week-old plants grown in an inert medium. Stems were 4-cm sections from the base of the primary influorescence of 6-week-old plants grown in compost. Samples from the end of the day and the end of the night were a whole rosette (leaf) or a whole root system (root). Quad, sus1/sus2/sus3/sus4. Sus activities are means ± SD from 3 plants; metabolite values are means ± SD from the number of samples shown in parentheses; seed weights are means ± SD from 6 samples from 2 independent pools, each from 5 plants; lipid values are means ± SD from 6 independent pools, each from 5 plants.
Isoforms SUS1, SUS2, SUS3, and SUS4 were very largely soluble. The insoluble material contained a small fraction of the SUS1 protein (this fact may account for the SUS activity in insoluble material from the sus5/sus6 mutant), but SUS2, SUS3, and SUS4 were not detectable (Fig. S1). Soluble activity in sus5/sus6 mutant plants was the same as or greater than that in WT plants (Table 1), indicating that SUS5 and SUS6 do not contribute to soluble activity (the soluble fraction contains both free and membrane-associated SUS activity).
Tissue prints showed that SUS5 and SUS6 proteins were present specifically in the phloem region of the hypocotyl. SUS6 was also detected in this region in stems. No immunoreactive material was present in the sus5/sus6 mutant (Fig. 1B). Consistent with a phloem location for SUS6, we showed previously that expression of a GUS reporter gene driven by the SUS6 promoter occurred only in specific cell files within the stele of the root (28). In in situ hybridization experiments on roots, SUS6 transcript was detected only in the protophloem cells (Fig. 1 C and D), from which the phloem sieve elements arise. To investigate possible roles of SUS5 and SUS6, we examined sections of stems that were chemically fixed immediately after excision. An electron-translucent layer lining the pores of sieve plates was consistently thinner in the sus5/sus6 mutant than in WT plants (Fig. S2 A and B). ImmunoGold labeling with an anticallose antibody confirmed that this layer contains callose (Fig. S2C). In contrast, callose synthesis after wounding and callose associated with plasmodesmata in leaves appeared to be the same in WT and mutant plants (Fig. S3). Together, these results suggest that SUS5 and SUS6 are confined to sieve elements where they have a specific function in callose synthesis. Thus, a sus1/sus2/sus3/sus4 mutant would be expected to lack SUS activity in all cell types except sieve elements.
Phenotype of the sus1/sus2/sus3/sus4 Mutant.
We selected a sus1/sus2/sus3/sus4 mutant (the quadruple mutant) and confirmed that it lacked all 4 SUS proteins (Fig. S4). As expected, SUS5 and SUS6 proteins were in the same locations in quadruple mutant and WT plants (Fig. 1). SUS activity in the soluble fraction of roots and stems was below the level of detection (<2% WT levels; Table 1). Thus, the remaining SUS activity in the quadruple mutant is highly likely to be located exclusively in cell walls of phloem sieve elements.
Quadruple mutant plants were not visibly different from WT plants when grown in well-drained compost under natural daylight or short days (Fig. S5). WT and mutant plants were the same with respect to carbohydrate levels in roots, stems, siliques and leaves, seed weight, seed oil content (Table 1, Table S1, and Table S2), callose in sieve plates, and callose in plasmodesmata and after wounding (Fig. S2and S3). The only exception was a 50% higher starch content in siliques of mutant plants (Table S1 and Table S2).
We used several methods to examine cellulose location and content in the quadruple mutant. There was no difference between WT and quadruple mutant plants in the appearance of walls of mesophyll and stem xylem cells (Fig. 2 A and B), indicating that primary and secondary cell walls are not seriously disrupted. Mutants deficient in secondary cell-wall synthesis have a collapsed xylem phenotype (29). FTIR microspectroscopy on root cell-wall preparations revealed no major differences in the carbohydrate region of the spectrum between quadruple mutant and WT seedling roots (Fig. 2C; Fig. S6). Scanning FTIR array microscopy of stem sections revealed no differences at a wavelength at which the cell-wall IR spectrum is dominated by cellulose (Fig. 2D; Fig. S6). There was no difference in cellulose content between quadruple mutant and WT stems (42.4 ± 3.3 and 44.6 ± 4.5% of dry weight, respectively; means ± SD from 5 samples; dry weights 6 to 9 mg). We also found no differences between sus5/sus6 and WT plants with respect to any of the above analyses (cellulose content 42.1 ± 4.3%; Fig. S6).
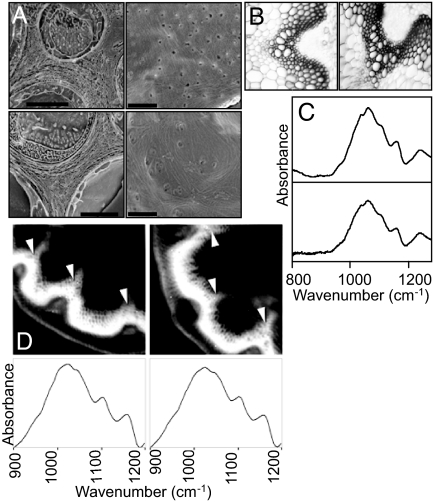
Fig. 2.
Anatomy and composition of WT and quadruple mutant plants. (A) Scanning electron micrographs of freeze-fractured leaves, showing cell walls in cross-section (Scale bars, 2 μm.) (Left); and face-on (Scale bars, 1 μm.) (Right). (Upper) WT leaves. (Lower) The sus1/sus2/sus3/sus4 mutant leaves. (B) Light micrographs of cross-sections of the basal 4 cm of flowering stems. (Left) WT; (Right) sus1/sus2/sus3/sus4 mutant. (C) FTIR spectra derived from the insoluble (cell wall) fraction of roots of 4-day-old seedlings. Spectra are averaged from analyses on 20 samples from a preparation of 40 primary roots. (Upper) WT roots; (Lower) sus1/sus2/sus3/sus4 mutant roots. Similar results were obtained with a different batch of seedlings. (D) Scanning FTIR array microscopy of stems (as in Table 1). (Upper) “Heatmap” images of 25-μm stem sections at a wavenumber dominated by cellulose (1,056 cm−1). White is the highest and black the lowest cellulose content. (Lower) Averaged spectra derived from xylem regions (arrowed). (Left) WT plant; (Right) sus1/sus2/sus3/sus4 mutant plant.
To examine further the capacity for sucrose catabolism in the quadruple mutant, we analyzed the expression of INV genes and the levels of hexose phosphates, UDPglucose, and a range of other primary metabolites. We found no differences between WT and quadruple mutant plants in these respects (Fig. S7, Table S1, and Table S2).
Phenotype of the Neutral INV Mutant cinv1/cinv2.
The data above showed that Arabidopsis plants lacking SUS in all cell types except sieve elements can grow and reproduce normally, without increased expression of INV genes or major perturbations of pool sizes of hexose phosphates and UDPglucose. Thus, it appeared that INV could substitute completely for SUS in sucrose catabolism in Arabidopsis, and indeed, that INV rather than SUS might be the dominant route for sucrose catabolism in WT plants. To test this idea, we selected a mutant lacking 2 isoforms of cytosolic INV [CINV1 (24), also called CYT-INV1 (25), encoded at At1g35580; CINV2 encoded at At4g09510]. These isoforms are the 2 most highly expressed neutral/alkaline INV predicted to be located in the cytosol of root cells (https://www.genevestigator.ethz.ch/gv/index.jsp) (30): the cytosol is the likely location for enzymes important in the entry of carbon from sucrose into cellular metabolism. Mutations in CINV1 are already known to have a relatively mild effect on Arabidopsis growth (24, 25). CINV1 and CINV2 are both closely related to a single isoform shown by mutational analysis to be essential for normal growth and development of L. japonicus plants (41). We established that the best available T-DNA insertion line for CINV2 (Sail_518_D02, designated cinv2) has only 10% of WT CINV2 transcript levels in its roots (Fig. S8).
The single mutants cinv1 and cinv2 appeared identical to WT plants on soil under our growth conditions. The cinv1/cinv2 plants flowered and set seed when grown on soil, but were much smaller in all respects than WT plants at maturity (Fig. 3A; Fig. S8). Seedlings of cinv1/cinv2 had relatively normal shoot growth, but drastically reduced root growth on solid medium without sugar. Whereas primary root extension over 7 days of growth was 60% of the WT value in cinv1 (24, 25), and 120% of the WT value in cinv2, it was only 17% of the WT value in cinv1/cinv2 [lengths: WT, 5.18 ± 0.16 cm (31); cinv1, 3.14 ± 0.11 cm (24); cinv2, 5.86 ± 0.14 cm (45); cinv1/cinv2, 0.88 ± 0.01 cm (55); mean ± SE, n in parentheses; Fig. 3].
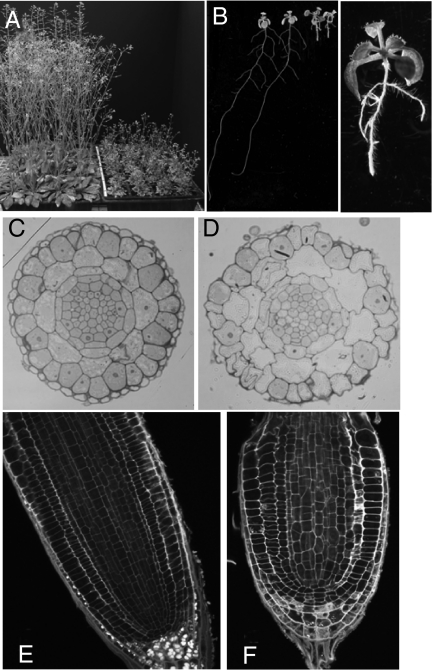
Fig. 3.
Appearance of the cinv1/cinv2 mutant. (A) Mature WT (Left) and cinv1/cinv2 mutant (Right) plants of the same age, grown in the same conditions. (B) Seven-day-old seedlings grown vertically on solid medium. (Left) WT seedlings (at the left) and cinv1/cinv2 seedlings (at the right); (Right) a cinv1/cinv2 seedling. (C and D) Cross-sections of roots of WT (C) and cinv1/cinv2 (D) seedlings. Sections are close to the base of the root cap (outer layer of cells) for WT roots and at an equivalent position for mutant roots. Plants were from the same plate. Magnifications are the same. (E and F) Longitudinal sections of roots of WT (E) and cinv1/cinv2 (F) seedlings. Plants were from the same plate. Magnifications are the same. The partial collapse of the root was seen in all mutant roots, but no WT roots. (C–F) Results are typical of those for many seedlings.
Cells in the root expansion zone of cinv1/cinv2 mutants were enlarged and had a greater tendency to collapse during manipulation than those of WT and single-mutant plants. Abnormal cell divisions occurred in the stele, endodermis, and cortex (Fig. 3). Growth on glucose restored root extension in double mutants to approximately half of WT (Fig. S8). Neutral INV activity in roots was ≈40% lower in double mutant than in WT seedlings (WT, 50.8 ± 2.3; double mutant, 30.5 ± 0.7 nmol min−1 mg−1 protein; mean ± SD, n = 3).
Discussion
Our findings have important implications for understanding of plant primary metabolism. It has generally been assumed that SUS is more important than INV in catalyzing the entry of carbon into metabolism in many plant cells. As described above, this route requires less ATP, and allows for feedback regulation of sucrose catabolism, and reductions in SUS in organs of crop plants often have obvious, deleterious effects. However, our results show that soluble SUS is not required for normal growth in Arabidopsis under our experimental conditions, whereas cytosolic INV is indispensible.
The requirement for SUS in some tubers, seeds, and fruits of crop plants may reflect the dense and/or bulky nature of these organs, which results in low internal oxygen levels (32) and, thus, reduced ATP supply. Low oxygen levels promote patterns of metabolism that conserve energy, including reliance on SUS rather than INV as the main route of sucrose catabolism (27). In potato tubers, overexpression of INV reduces oxygen levels and inhibits starch synthesis (33). Similarly, loss of SUS activity in roots has little effect on growth in well-aerated conditions, but reduces viability in hypoxic conditions (28). The lack of an obvious phenotype in the Arabidopsis quadruple sus mutant when grown in well-aerated conditions may, thus, reflect the absence of organs with oxygen-limited metabolism in this species.
Our data show that SUS is not required for cellulose synthesis in Arabidopsis. Quadruple mutants are not deficient in cellulose in a range of organs and cell types, and show no lesions characteristic of mutants with cell-wall defects. Specific associations may exist between SUS and cellulose synthase in WT plants, but in quadruple mutants, cellulose synthase must either compete with other metabolic processes for the cytosolic pool of UDPglucose or, for example, associate with an enzyme that synthesizes UDPglucose from a hexose phosphate rather than from sucrose. Our results highlight the need for further research to discover the pathway by which carbon from sucrose is supplied to cellulose synthesis.
Contrary to recent proposals (13, 14), we can conclude unambiguously that SUS is not required for transitory starch synthesis in leaves. It remains possible that some ADPglucose is produced via SUS in WT leaves, but the normal levels of starch, ADPglucose, and other primary metabolites in leaves of the quadruple mutant indicate that any contribution by SUS is minor. In any case, the activity of SUS in Arabidopsis leaves (0.023 μmol min−1 g−1 fresh weight) (28) is too low to account for the rate of starch synthesis (0.1 μmol min−1 g−1 fresh weight) (34).
We provide evidence that SUS6, and probably SUS5, are confined to phloem sieve elements, and are involved in sieve-plate callose synthesis. The function of the callose lining of the sieve plate pores in intact plants is not known; the sus5/sus6 mutant provides a tool for studying this question.
In marked contrast to SUS, cytosolic INV activity is indispensable for normal growth in Arabidopsis. First, in the quadruple sus mutant, catabolism of sucrose via INV can apparently supply all of the carbon requirements of nonphotosynthetic cells without major changes in INV gene expression or in levels of sucrose catabolites. Second, loss of 2 closely related, cytosolic isoforms of neutral/alkaline INV dramatically retards growth. These results indicate that cytosolic INV may be the primary route by which carbon from sucrose is supplied to nonphotosynthetic cells in Arabidopsis.
The seedling phenotype of the cinv1/cinv2 mutant is consistent with general carbon starvation brought about by reduced capacity for sucrose catabolism in root cells. Evidence includes the extreme reduction in root growth, the loss of starch from the root cap (Fig. 3), and the enlargement and tendency to collapse of several cell types. The roots resemble those of mutants defective in cell-wall biosynthesis (35, 36), except that the phenotypes of cell-wall mutants are stronger at high sugar concentrations (37), whereas the phenotype of cinv1/cinv2 shows the opposite effect. We suggest that cell expansion in roots of cinv1/cinv2 is abnormal because of a lack of substrate for cell-wall synthesis, rather than a defect in a specific pathway of cell-wall synthesis. The strong promotion of root extension by exogenous glucose also indicates that the phenotype is in part due to carbon starvation.
Caution must be exercised in further interpretation of the strong phenotype of the double cinv mutant. The stunted growth may reflect a direct requirement for cytosolic INV in many organs, but it might also be a consequence of a strong inhibition of root growth alone, or of other pleiotropic effects on gene expression and on sugar signaling pathways (as suggested for the cinv1 mutant) (24). The fact that neutral INV activity is reduced by only 40% in the double mutant is also difficult to interpret. The residual activity may be due to other cytosolic isoforms and/or the isoforms of unknown function located in organelles. We found that that loss of CINV1 expression increases CINV2 transcript levels (Fig. S8A); thus, there may be pleiotropic effects on INV activity in the cinv1/cinv2 mutant.
Together with recent reports for rice (23) and Lotus (41), our results show that cytosolic INV is essential for normal plant growth and development. Little is known about this class of enzymes. It is not clear whether they have regulatory properties that would allow flux of carbon out of sucrose via this route to be coordinated with the metabolic demands of the cell, or why SUS cannot compensate for loss of cytosolic INV isoforms. It is interesting to note that AtCINV1 interacts in yeast 2-hybrid experiments with the phosphatidylinositol monophosphate 5-kinase AtPIP5K9, and is coimmunoprecipitated with PIP5K9 from extracts of transgenic plants overexpressing both proteins (24). PIP5K9 is a component of phosphatidylinositol signaling pathways that are necessary for normal root growth. These observations suggest that cytosolic INV may be a target for signaling pathways that coordinate carbohydrate availability with growth and development in nonphotosynthetic organs.
Methods
Plant Material.
The sus mutant lines were as in ref. 28. The quadruple mutant was created by crossing sus1/sus4 with sus2/sus3. For the cinv1 mutant (24), absence of CINV1 transcript was confirmed by RT-PCR.
Unless otherwise stated, plants were grown in compost at 20 °C with 12-h light, 12-h dark, 150–200 μmol quanta PAR m−2 s−1, and 75% relative humidity. Roots of mature plants were harvested from a 1:1 mixture of sand and Terragreen (Oil-Dri) with a slow-release fertilizer. Hypocotyls were harvested after 11 weeks growth in compost at 22 °C with 9-h light, 15-h dark, 95 μmol quanta m−2 s−1, and 65% relative humidity. For seedling roots, seeds were surface sterilized (70% vol/vol aq. ethanol 5 min, ≈1% vol/vol Na hypochlorite 10 min), rinsed with water, then sown on plates of solid medium (Phytagel, Sigma; 5 g l−1 plus inorganic nutrients; see ref. 38) with or without 1% (wt/vol) glucose. After 3 days at 4 °C, plates were placed vertically at 22 °C with 6-h dark, 18-h light at 85 μmol quanta PAR m−2 s−1.
Starch, Metabolite, and Oil Measurements.
For starch and metabolite measurements, samples were transferred to liquid nitrogen immediately after cutting. Roots were rapidly rinsed and blotted before freezing. For starch, sugars, hexose phosphates, and UDPglucose, frozen samples were extracted in 0.7 M perchloric acid, and metabolites assayed enzymatically (34). The UDPglucose assay contained UDPglucose pyrophosphorylase, phosphoglucomutase and NAD-glucose 6-phosphate dehydrogenase. For other metabolites, extraction was in chloroform-methanol and analysis by LC-MS/MS (39). Oil content of mature seeds was measured by NMR spectroscopy (28). Cellulose was assayed chemically on tissue samples powdered in liquid nitrogen before extraction (29).
Immunoblotting, Enzyme Assays, and Tissue Printing.
Frozen tissue was powdered then extracted at 4 °C in either 50 mM Na-Hepes (pH 7.5), 5 mM MgCl2, 1 mM EDTA, 5 mM DTT, and 10 mL L−1 protease inhibitor mixture (Sigma) (for SUS assays), or 50 mM Bis-Tris (pH 7.0), 5 mM ascorbic acid, 5 mM DTT, 2 mM EDTA, 1 mM EGTA, 50 mM NaCl, 10 mL L−1 protease inhibitor mixture, and 10 mL L−1 phosphatase inhibitor mixture 1 (Sigma) (for immunoblotting). The homogenate was subjected to centrifugation for 10 min at 10,000 × g and 4 °C. The supernatant is referred to as the soluble fraction. The pellet was washed twice by resuspension in extraction medium using a glass homogenizer, followed by centrifugation, then resuspended either in extraction medium (for assays) or in SDS sample buffer (for SDS/PAGE). The soluble fraction was precipitated with 10% (wt/vol) trichloroacetic acid, then solubilized in SDS sample buffer. SDS/PAGE, immunoblotting, and assays were as described (28). Tissue prints were made by gently pressing cut stems or hypocotyls onto nitrocellulose membrane, then developing as for immunoblotting.
In Situ Hybridization.
A DNA template for the last exon of SUS6 was generated from full-length cDNA by PCR by using the following primers:
5′- AAGAAAGTGACAATCCCGGAAGATAAACCTC-3′
5′- CCTCGATCCAAGGTCAAACTTTTTAATACTC-3′
T3 (sense) and T7 (antisense) promoter sequences were incorporated by PCR
(5′ – AATTAACCCTCACTAAAGGGAAGAAAGTGA C AATC-3′;
5′ – TAATACGACTCACTATAGGGCTCGATCCAA G GTC-3′).
PCR-product purification was carried out by using a QIAquick PCR purification kit (QIAGEN). DIG-labeling of the PCR product was done by using the Riboprobe in vitro Transcription System (Promega). Whole mount in situ hybridization was done on 4-day-old seedlings (40).
Microscopy.
For light microscopy.
Stem sections (30 μm) were cut on a vibratome and stained with 0.02% (wt/vol) toluidine blue O. For root transverse sections (Fig. 3 C and D), roots were cut into 2.5% (vol/vol) glutaraldehyde in 0.05 M Na cacodylate, pH 7.3, vacuum infiltrated and left overnight in fresh fixative. Samples were washed in 0.05 M Na cacodylate, postfixed in 1% (wt/vol) OsO4 in 0.05 M Na cacodylate for 1 h, washed in distilled water, and dehydrated through an alcohol series. Dehydrated samples were infiltrated with LR White resin by successive changes of resin/ethanol mixes over 5 days at room temperature, then transferred to fresh resin. The resin was polymerized at 60 °C for 16 h. Sections (0.5 μm) were dried onto glass slides and stained with 0.5% (wt/vol) toluidine blue O in 0.5% (wt/vol) borax. For root longitudinal sections (Fig. 3 E and F), roots were fixed overnight in 4% paraformaldehyde at 4 °C, then subjected to a modified pseudoSchiff propidium iodide staining (31). Samples were viewed with a confocal microscope at excitation wavelength 488 nm.
Scanning Electron Microscopy.
Samples were mounted on an aluminum stub by using O.C.T. compound (BDH), plunged into liquid nitrogen slush, then transferred onto the cryostage of an ALTO 2500 cryo-transfer system (Gatan) attached to a Zeiss Supra 55 VP FEG scanning electron microscope. After fracturing at −100 °C, samples were sputter-coated with platinum (90 s at 10 mA, <−110 °C), then imaged at 3 kV on the cryo-stage in the main chamber of the microscope at ≈−130 °C.
FTIR Analyses.
Cell walls from primary roots were prepared and analyzed as previously (35). Details of FTIR techniques are provided in Fig. S6.
Supplementary Material
Acknowledgments.
We thank Trevor Wang and Tracey Welham (John Innes Centre) for suggesting selection of the cinv1/cinv2 mutant and other valuable discussions; Keith Roberts and Irmi Horst (John Innes Centre), Nick Kruger (University of Oxford, Oxford), and Bob Furbank (Commonwealth Scientific and Industrial Research Organisation, Canberra, Australia) for advice and discussions; Grant Calder and Sue Bunnewell (John Innes Centre) for expert assistance with microscopy; and Alexander Graf (John Innes Centre) for help with qRT-PCR experiments. This work was supported by a core strategic grant from the Biotechnology and Biological Sciences Research Council to the John Innes Centre.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900689106/DCSupplemental.
References
- 1.Zrenner R, Salanoubat M, Willmitzer L, Sonnewald U. Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.) Plant J. 1995;7:97–107. doi: 10.1046/j.1365-313x.1995.07010097.x. [DOI] [PubMed] [Google Scholar]
- 2.D'Aoust MA, Yelle S, Nguyen-Quoc B. Antisense inhibition of tomato fruit sucrose synthase decreases fruit setting and the sucrose unloading capacity of young fruit. Plant Cell. 1999;11:2407–2418. doi: 10.1105/tpc.11.12.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruan YL, Llewellyn DJ, Furbank RT. Suppression of sucrose synthase gene expression represses cotton fibre cell initiation, elongation, and seed development. Plant Cell. 2003;15:952–964. doi: 10.1105/tpc.010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chourey PS, Taliercio EW, Carlson SJ, Ruan YL. Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol Gen Genet. 1998;259:88–96. doi: 10.1007/s004380050792. [DOI] [PubMed] [Google Scholar]
- 5.Craig J, et al. Mutations at the rug4 locus alter the carbon and nitrogen metabolism of pea plants through an effect on sucrose synthase. Plant J. 1999;17:353–362. [Google Scholar]
- 6.Sun J, Loboda T, Sung SJS, Black CC. Sucrose synthase in wild tomato, Lycopersicon chmielewskii, and tomato fruit sink strength. Plant Physiol. 1992;98:1163–1169. doi: 10.1104/pp.98.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lerchl J, Geigenberger P, Stitt M, Sonnewald U. Impaired photoassimilate partitioning caused by phloem-specific removal of pyrophosphate can be complemented by a phloem-specific yeast-derived invertase in transgenic plants. Plant Cell. 1995;7:259–270. doi: 10.1105/tpc.7.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpita N, McCann M. In: Biochemistry and Molecular Biology of Plants. Buchanan BB, Gruissem W, Jones RL, editors. Rockville, MD: Amer Soc Plant Physiol; 2000. pp. 52–108. [Google Scholar]
- 9.Koch K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol. 2004;7:235–246. doi: 10.1016/j.pbi.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Winter H, Huber JL, Huber SC. Membrane association of sucrose synthase: Changes during the graviresponse and possible control by protein phosphorylation. FEBS Lett. 1997;420:151–155. doi: 10.1016/s0014-5793(97)01506-8. [DOI] [PubMed] [Google Scholar]
- 11.Haigler CH, et al. Carbon partitioning to cellulose. Plant Mol Biol. 2001;47:29–51. [PubMed] [Google Scholar]
- 12.Salnikov VV, Grimson MJ, Seagull RW, Haigler CH. Localization of sucrose synthase and callose in freeze-substituted secondary-cell-wall stage cotton fibres. Protoplasma. 2003;221:175–184. doi: 10.1007/s00709-002-0079-7. [DOI] [PubMed] [Google Scholar]
- 13.Baroja-Fernández E, Muñoz FJ, Akazawa T, Pozueta-Romero J. Reappraisal of the currently prevailing model of starch biosynthesis in photosynthetic tissues. Plant Cell Physiol. 2001;42:1311–1320. doi: 10.1093/pcp/pce175. [DOI] [PubMed] [Google Scholar]
- 14.Muñoz FJ, et al. Sucrose synthase controls both intracellular ADP glucose levels and transitory starch biosynthesis in source leaves. Plant Cell Physiol. 2005;46:1366–1376. doi: 10.1093/pcp/pci148. [DOI] [PubMed] [Google Scholar]
- 15.Husain SE, James C, Shields R, Foyer CH. Manipulation of fruit sugar composition but not content in Lycopersicon esculentum fruit by introgression of an acid invertase gene from Lycopersicon pimpinellifolium. New Phytol. 2001;150:65–72. [Google Scholar]
- 16.Yau YY, Simon PW. A 2.5-kb insert eliminates acid soluble invertase isozyme II transcript in carrot (Daucus carota L.) roots, causing high sucrose accumulation. Plant Mol Biol. 2003;53:151–162. doi: 10.1023/B:PLAN.0000009272.44958.13. [DOI] [PubMed] [Google Scholar]
- 17.Sergeeva LI, et al. Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proc Natl Acad Sci USA. 2006;103:2994–2999. doi: 10.1073/pnas.0511015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sturm A, Chrispeels MJ. cDNA cloning of carrot extracellular β-fructosidase and its expression in response to wounding and bacterial infection. Plant Cell. 1990;2:1107–1119. doi: 10.1105/tpc.2.11.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swarbrick PA, Schulze-Lefert P, Scholes JD. Metabolic consequences of susceptibility and resistance (race-specific and broad-spectrum) in barley leaves challenged with powdery mildew. Plant Cell Env. 2006;29:1061–1076. doi: 10.1111/j.1365-3040.2005.01472.x. [DOI] [PubMed] [Google Scholar]
- 20.Weber H, Borisjuk L, Heim U, Buchner P, Wobus U. Seed coat-associated invertases of fava bean control both unloading and storage functions. Plant Cell. 1995;7:1835–18466. doi: 10.1105/tpc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng WH, Taliercio EW, Chourey PS. The minature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell. 1996;8:971–983. doi: 10.1105/tpc.8.6.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goetz M, et al. Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc Natl Acad Sci USA. 2001;98:6522–6527. doi: 10.1073/pnas.091097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia L, et al. OsCYT-INV1 for alkaline/neutral invertase is involved in root cell development and reproductivity in rice (Oryza sativa L.) Planta. 2008;288:51–59. doi: 10.1007/s00425-008-0718-0. [DOI] [PubMed] [Google Scholar]
- 24.Lou Y, Gou JY, Xue HW. PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell. 2007;19:163–181. doi: 10.1105/tpc.106.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi X, et al. AtCYT-INV1, a neutral invertase, is involved in osmotic stress-induced inhibition on lateral root growth in Arabidopsis. Plant Mol Biol. 2007;64:575–587. doi: 10.1007/s11103-007-9177-4. [DOI] [PubMed] [Google Scholar]
- 26.Geigenberger P, Stitt M. Sucrose synthase catalyses a readily reversible reaction in vivo in developing potato tubers and other plant tissues. Planta. 1993;189:329–339. doi: 10.1007/BF00194429. [DOI] [PubMed] [Google Scholar]
- 27.Stitt M. Pyrophosphate as an energy donor in the cytosol of plant cells; an enigmatic alternative to ATP. Bot Acta. 1998;111:167–175. [Google Scholar]
- 28.Bieniawska Z, et al. Analysis of the sucrose synthase gene family in Arabidopsis. Plant J. 2007;49:810–828. doi: 10.1111/j.1365-313X.2006.03011.x. [DOI] [PubMed] [Google Scholar]
- 29.Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell. 1997;9:689–70142. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murayama S, Handa H. Genes for alkaline/neutral invertase in rice: Alkaline/neutral invertases are located in plant mitochondria and also in plastids. Planta. 2007;225:1193–1203. doi: 10.1007/s00425-006-0430-x. [DOI] [PubMed] [Google Scholar]
- 31.Truernit E, et al. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell. 2008;20:1494–1503. doi: 10.1105/tpc.107.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geigenberger P. Response of plant metabolism to too little oxygen. Curr Opin Plant Biol. 2003;6:247–256. doi: 10.1016/s1369-5266(03)00038-4. [DOI] [PubMed] [Google Scholar]
- 33.Bologa KL, Fernie AR, Leisse A, Loureiro ME, Geigenberger P. A bypass of sucrose synthase leads to low internal oxygen and impaired metabolic performance in growing potato tubers. Plant Physiol. 2003;132:2058–2072. doi: 10.1104/pp.103.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chia T, et al. A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J. 2004;37:853–863. doi: 10.1111/j.1365-313x.2003.02012.x. [DOI] [PubMed] [Google Scholar]
- 35.Schindelman G, et al. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 2001;15:1115–1127. doi: 10.1101/gad.879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng L, Hocart CH, Redmond JW, Williamson RE. Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta. 2000;211:406–411. doi: 10.1007/s004250000301. [DOI] [PubMed] [Google Scholar]
- 37.Hauser MT, Morikama A, Benfey PN. Conditional root expansion mutants of Arabidopsis. Development (Cambridge, U.K.) 1995;121:1237–1252. doi: 10.1242/dev.121.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma Z, Bielenberg DG, Brown KM, Lynch JP. Regulation of root hair density by phosphorous availability in Arabidopsis thaliana. Plant Cell Env. 2001;24:459–467. [Google Scholar]
- 39.Lunn JE, et al. Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J. 2006;397:139–148. doi: 10.1042/BJ20060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carol RJ, et al. A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature. 2005;438:1013–1016. doi: 10.1038/nature04198. [DOI] [PubMed] [Google Scholar]
- 41.Welham T, et al. A cytosolic invertase is required for normal growth and cell development in the model legume, Lotus japonicus. J Exp Bot. 2009 doi: 10.1093/jxb/erp169. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.