Abstract
Understanding the signaling pathways that drive aggressive breast cancers is critical to the development of effective therapeutics. The oncogene MET is associated with decreased survival in breast cancer, yet the role that MET plays in the various breast cancer subtypes is unclear. We describe a knockin mouse with mutationally activated Met (Metmut) that develops a high incidence of diverse mammary tumors with basal characteristics, including metaplasia, absence of progesterone receptor and ERBB2 expression, and expression of cytokeratin 5. With gene expression and tissue microarray analysis, we show that high MET expression in human breast cancers significantly correlated with estrogen receptor negative/ERBB2 negative tumors and with basal breast cancers. Few treatment options exist for breast cancers of the basal or trastuzumab-resistant ERBB2 subtypes. We conclude from these studies that MET may play a critical role in the development of the most aggressive breast cancers and may be a rational therapeutic target.
Keywords: ErbB2, mouse model
Breast cancer affects >200,000 women in the United States each year, yet the mortality rate is rapidly declining with earlier detection and the availability of targeted therapies. However, tumors that do not express estrogen receptor (ER), progesterone receptor (PR), or both are less differentiated, more clinically aggressive, and less likely to respond to hormone-targeted therapy (1, 2). Moreover, 20–30% of all breast cancers overexpress human epidermal growth factor receptor-2 (HER2/ERBB2) and are associated with reduced survival (3). Those tumors that no longer depend on ER/PR and ERBB2 signaling have the poorest prognosis and no targeted therapeutic options (4). Recently, gene expression studies have identified several distinct breast cancer subtypes that correlate with clinical outcome (5, 6). These molecular subtypes include three main groups of ER negative tumors, including basal, ERBB2, and the normal-like/unclassified subtype, and at least two types of ER positive tumors (luminal A and luminal B). Patients with tumors classified as basal or ERBB2 have significantly shorter survival rates compared with those with the ER+ luminal A subtype. Metaplastic breast carcinomas have various combinations of adenocarcinoma, mesenchymal, and epithelial features and are associated with the basal subtype (7, 8). These classifications have improved our understanding of the unique molecular signatures present in breast cancer and how they may relate to patient prognosis.
The receptor tyrosine kinase Met is a well-known oncogene that is involved in the progression and metastasis of most solid human cancers (www.vai.org/metandcancer) (9). Under normal physiological conditions, Met is expressed by epithelial cells and activated through paracrine ligand binding of hepatocyte growth factor/scatter factor (HGF/SF). In neoplastic conditions, aberrant Met activity occurs through numerous mechanisms, including overexpression of Met, HGF/SF, or both, autocrine signaling, or mutational activation. In breast cancer, MET is overexpressed in 20–30% of cases, is a strong, independent predictor of decreased survival (10–12), and correlates with poor patient outcome, independent of HER2/ERBB2 expression (13). Met signaling may therefore have a unique function in breast cancer progression distinct from ERBB2 signaling.
We investigated the role of Met in mammary tumorigenesis, through a mouse model of mutationally activated Met. This model contains activating missense mutations within the tyrosine kinase domain (M1248T/L1193V) that have been knocked into the endogenous Met locus and are referred to as Metmut (14). Here, we report that activated Met (on a FVB/N background) induced a high incidence of mammary tumors with diverse histopathological phenotypes. High levels of Met amplification were present as extrachromosomal double minutes in the tumors. Most notable was that several mice developed multiple mammary tumors with distinct histopathologies and ER/PR/ErbB2 signatures. Metmut tumors were ER+/PR−, half did not express ErbB2, and several expressed the basal marker cytokeratin 5 (CK5). These results led us to examine MET expression in human breast cancer subtypes. Our analyses of human breast cancer expression data and tissue microarrays revealed that high MET expression correlates with ER−/ERBB2− tumors and the basal subtype.
Here, we show that a single oncogene, such as Met, is able to induce a spectrum of mammary cancers (solid adenocarcinomas, adenosquamous carcinomas, and myoepitheliomas) with histological, cytogenetic, and ER/ErbB2 characteristics that are present in aggressive human breast cancers. This is a valuable model for aggressive human breast cancers that express high levels of MET. Moreover, our mouse model, in conjunction with the human breast cancer analysis, indicates that MET may be a significant therapeutic target for one of the most aggressive breast cancer subtypes.
Results
Mutationally Activated Met Induces Diverse Mammary Adenocarcinomas.
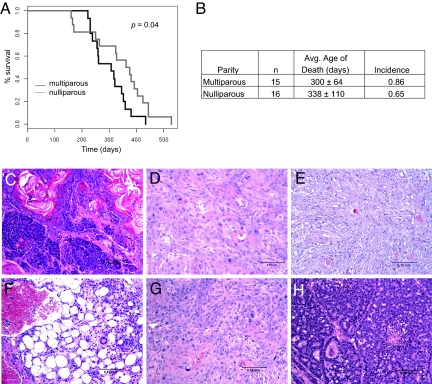
We wished to investigate the role of the Met oncogene in mammary tumorigenesis. Earlier our lab created a transgenic model of mutationally activated Met that developed metastatic mammary tumors in the founder mice, yet they failed to transmit (15). We previously described a knockin mouse model of mutationally activated Met generated on a C57BL/6;129/SV background that developed sarcomas, lymphomas, and carcinomas, but failed to generate mammary tumors (14). Because parous FVB/N females commonly develop hyperplasia (16), we bred the M1248T/L1193V mutant line congenically onto the FVB/N background. This model is referred to as Metmut and contains activating missense mutations within the tyrosine kinase domain (M1248T/L1193V). These mutations are knocked into the genomic Met locus and, therefore, expressed under the endogenous Met promoter. When maintained as heterozygotes, these mice develop aggressive mammary tumors. Both nulliparous (n = 16) and multiparous females (n = 15) developed mammary tumors with high penetrance, and Kaplan–Meier analysis revealed only a slight difference in the average age of tumor onset (Fig. 1 A and B).
Fig. 1.
Mutationally activated Met induces diverse mammary tumors. (A) Kaplan–Meier analysis demonstrated a slight difference in age of death between multiparous and nulliparous females (P = 0.04). (B) A high incidence of mammary tumor development was observed in both multiparous and nulliparous females. (C) Mammary adenosquamous carcinoma with solid and tubular patterns (animal 28). (D) Mammary squamous cell carcinoma (animal 6) that had significant karyomegaly. (E) Mammary myoepithelioma (animal 2). (F) Hemangiosarcoma (animal 22). (G) Mammary adenocarcinoma with squamous metaplasia and (H) an adenocarcinoma with solid patterns both observed in different mammary glands of animal 13 (solid patterns not shown in image).
Analysis of mammary pads from 6- and 8-month-old females showed no evidence of preneoplastic growth, indicating that tumor growth is extremely rapid after initiation. A surprising variety of histopathological phenotypes were identified (Fig. 1 C–H and Table S1). Solid features were present in 30% of the tumors, but in several cases the solid phenotype was intermixed with squamous metaplastic cells (Fig. 1C). Significant squamous metaplasia was observed in 65% of mammary tumors (Fig. 1D), and three myoepitheliomas were identified (Fig. 1E). In addition to mammary neoplasia, five mice developed undifferentiated sarcomas (Fig. 1F), as was observed on the B6 background (14). Interestingly, four mice developed two tumors with quite different histopathological phenotypes. For example, one animal developed both an adenocarcinoma with squamous metaplasia and an adenocarcinoma with solid patterns (Fig. 1 G and H). This may be explained if Met mediates transformation in early stages of mammary development and variation in the cell lineages allows for the development of distinct tumor types.
Met Amplification Is a Primary Chromosomal Alteration in Metmut Mammary Adenocarcinomas.
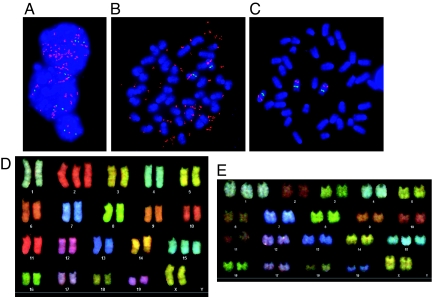
In both human papillary renal tumors and mice with germline-activating mutations, nonrandom duplication of the mutant Met allele was observed (14, 17). To determine if similar chromosomal events were present in the Metmut mammary tumors, we performed interphase dual-color FISH on six mammary tumors with various histologies. Using a BAC clone specific for the Met locus on chromosome (Chr) 6 and a control probe on Chr 6 allowed us to distinguish between a gain of Chr 6 and localized amplification of the Met locus. In six tumors examined, we observed up to 50 copies of Met (Fig. 2A and Table S2). The high levels of Met amplification were observed in cells with extremely large nuclei that composed 2–5% of the tumor. Trisomy and larger gains of Chr 6 also were detected. We performed metaphase FISH analysis on three primary tumors (Table S3) to assess if the amplification was extrachromosomal. In all cases, the majority of Met amplifications were present as extrachromosomal double minutes (Fig. 2B). Only one tumor showed intrachromosomal and extrachromosomal Met amplification by metaphase FISH (Table S3). However, we did identify several metaphases containing trisomy 6 and no double minutes (Fig. 2C). Curiously, we did observe a higher copy number of Chr 6 in interphase FISH compared with that observed in metaphase FISH, but some tetraploid metaphases did not spread sufficiently and were unanalyzable for metaphase FISH. Therefore, the amplification of Met and gain of Chr 6 may be higher than that detected by metaphase FISH.
Fig. 2.
Met amplification is a primary chromosomal alteration present in Metmut mammary tumors. (A) Relative copy number of Met was measured using FISH. Photomicrographs with Met probe (red), control probe (green), and DAPI (blue) are shown for animal 17. One nuclei (bottom) is diploid, whereas the other two nuclei have ≈18 (middle) and >50 (top) copies of Met. (B) Metaphase FISH of a mammary adenocarcinoma (animal 29) with two copies of Chr 6 and ≈49 copies of Met in double minute chromosomes. (C) Metaphase FISH of a mammary adenocarcinoma (animal 30) with trisomy of Chr 6. (D) Spectral karyotyping of a myoepithelioma from animal 29 with trisomy of Chr 2 and 15. (E) Spectral karyotyping of a mammary adenocarcinoma from animal 25 with trisomy of Chr 1 and 15.
We examined the chromosomal region around the Met locus and observed that the Wnt2 proto-oncogene is 0.5 Mb from Met. Interphase FISH analysis revealed that the double minutes also contained Wnt2. However, further analysis determined that Wnt2 expression was not increased in these tumors, yet Met expression was increased.
To determine what other chromosomal changes occur in Metmut tumors, we performed spectral karyotyping on three adenocarcinomas. We observed trisomy of Chr 1, 2, and 15 frequently (Fig. 2 D and E and Table S3) and a low occurrence of trisomy of Chr 3, 6, and 13. The lack of numerous chromosomal aberrations suggests that Met amplification is sufficient to avoid the need for extensive instability ordinarily associated with mammary tumorigenesis. These results and a study showing high levels of Met amplification in Brca1/Trp-53 mammary tumors (18) indicate that Met amplification may be a common event in murine mammary tumorigenesis. However, interphase FISH was performed on a human breast cancer tissue microarray, and Met amplification was not observed.
Met Is Highly Expressed in Tumor Cells that Are PR and ErbB2 Negative.
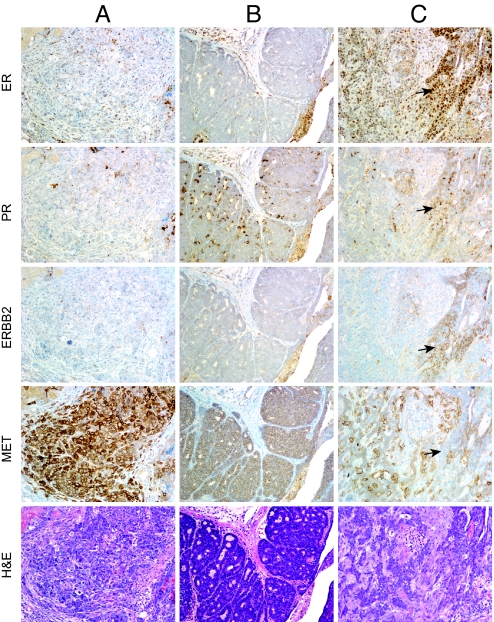
To determine the ER/PR/ErbB2 profile of Met-induced tumors, we performed immunohistochemical staining on 12 tumors with various histologies and normal tissue sections. All of the tumors expressed high levels of Met (Fig. 3), which was confirmed through Western blot analysis (Fig. S1). The majority of tumors (11 of 12) were ER+ (Fig. 3 and Table S4). This result was unexpected given that genetically engineered mouse models commonly develop ER− mammary tumors (19). In addition, we observed that all Metmut tumors lacked PR expression. The absence of PR may reflect a lack of ER activity or the up-regulation of growth factor signaling pathways (20). When we examined ErbB2 expression, we found that half of the tumors were ErbB2 negative. We also observed that a single animal could develop tumors with unique ER/PR/ErbB2 profiles. For instance, animal 26 developed two carcinomas, one which was ER+/PR−/ErbB2− (Fig. 3A) and one which was ER+/PR−/ErB2+ (Fig. 3B). Remarkably, a single oncogene, such as Met, is able to induce diverse tumors with unique histologic features and signaling requirements within a single animal.
Fig. 3.
Immunohistochemical analysis of ER, PR, ErbB2, and Met in Metmut tumors. In animal 26, two tumors developed with diverse profiles. Tumor 26a (A) was ER+/PR−/ErbB2−, and Tumor 26b (B) was ER−/PR−/ErbB2+. In Tumor 26a, we observed intense Met staining in individual tumor cells compared with the uniform staining observed in Tumor 26b. In animal 13a (C), Met staining was undetectable in a hyperplastic duct (see arrows) where ER, PR, and ErbB2 were strongly expressed. Positive background staining of PR sections was observed in some sections (B).
Within the Metmut mammary tumors, we often observed ductal structures that appeared more differentiated. When we carefully evaluated these structures compared with adjacent anaplastic regions, we observed an inverse relationship between Met and ER/PR/ErbB2 expression within individual cells. As seen in Fig. 3C, high ER, PR, and ErbB2 expression was observed in a hyperplastic ductal structure, yet Met expression was negligible. Conversely, high levels of Met expression were observed in the surrounding tumor regions, whereas PR and ErbB2 expression was diminished and ER expression decreased. This inverse relationship with Met and ER/PR/ErbB2 expression occurred only in hyperplastic regions within the tumors. Expression of Met, in addition to ER, PR, and ErbB2, was observed in hyperplastic ducts outside the tumor borders (Fig. S2). We hypothesize that increased Met signaling compensates for the decrease or absence of ER, PR, or ErbB2 signaling within tumorigenic cells.
MET Expression Is Enhanced in Basal and ErbB2 Breast Cancer Subtypes.
Basal-like breast carcinomas are heterogeneous and have distinct histological features, such as myoepithelial and metaplastic differentiation. In addition, basal-like tumors typically express genes characteristic of basal epithelial cells (i.e., cytokeratins 5, 14, 15, and 17) (21). Because Metmut tumors consist of myoepithelial and metaplastic features and are often PR−/ErbB2−, we examined Metmut tumors for expression of the basal marker CK5. We detected CK5 expression in the majority of the Metmut tumors but not in solid regions within the tumors (Fig. S3). The above results suggested that the Metmut tumors may have basal-like characteristics and led us to examine MET gene expression in human basal and nonbasal cancers.
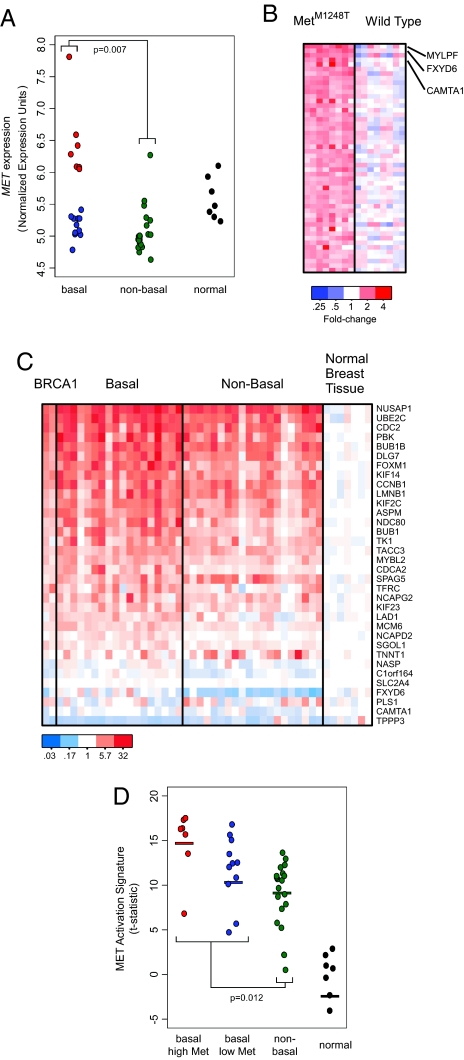
Examination of MET expression in an existing human breast cancer expression dataset (22) indicated that MET was significantly expressed in basal-like cancers compared with nonbasal cancers (P = 0.007, Fig. 4A). In addition, there were two subsets in the basal-like category: one with increased expression of MET (Fig. 4A, red) and one with MET expression levels similar to those of the nonbasal cancers (Fig. 4A, blue). To determine if increased expression of MET had functional consequences in these cancers, we generated a gene signature of MET activation from mouse embryonic fibroblasts (MEFs) and used a gene set enrichment approach to evaluate MET activation in the breast cancer gene expression data (Fig. 4 B and C and SI Methods) (23). Although a signature of MET activation was identified in all of the breast tumors, activation of MET was most strongly associated with basal-like cancers (P = 0.012, Fig. 4D).
Fig. 4.
MET expression significantly correlates with basal breast cancers. (A) Gene expression data show that MET expression correlates with basal breast cancers (P = 0.007) (21). (B) Generation of gene expression signatures associated with MET activation. Gene expression profiling data as obtained from WT and MetM1248T MEFs. The 50 most significantly overexpressed genes in the MetM1248T samples were identified, and the expression values were plotted as a heat map. Red indicates that the gene has increased expression compared with the median of the WT samples; blue indicates decreased expression. (C) Gene expression signature of MET activation in human breast cancer. Genes from the MET activation signature shown in (B) were converted from mouse gene identifiers to human gene identifiers using the HomoloGene database, and the expression values were plotted as a heat map. Of the 50 genes shown in (B), 34 genes (68%) unambiguously mapped between mice and humans. (D) A MET expression signature significantly correlates with basal breast cancers (P = 0.012). Mean values are represented by bars: basal high MET = 14.79; basal low MET = 11.76; nonbasal = 10.00; normal = −4.06.
We examined the correlation between clinical outcome and MET expression and the MET signature in an additional dataset that contained survival data. Analysis revealed that increased MET expression (P = 0.036) and the MET gene expression signature (P = 0.00078) associated with poor clinical outcome. Moreover, both MET expression and the MET signature associated with poor outcome independent of ER status or the ERBB2 subtype.
We further examined MET expression in human breast cancer subtypes using a breast cancer tissue microarray (Table S5) (21). MET staining was performed using an antibody specifically designed for staining formalin-fixed tissues (24), which revealed distinct patterns of cytoplasmic and membrane expression (Fig. 5 A and B). Tumor cores were evaluated similar to Lindemann et al. (25): 0, no immunoreactivity; 1, weak immunoreactivity; 2, moderately strong immunoreactivity; 3, strong immunoreactivity. Immunohistochemistry for ER, PR, ERBB2, EGF receptor (EGFR), and CK5/6 also was performed, and subtypes were determined as described (21, 26).
Fig. 5.
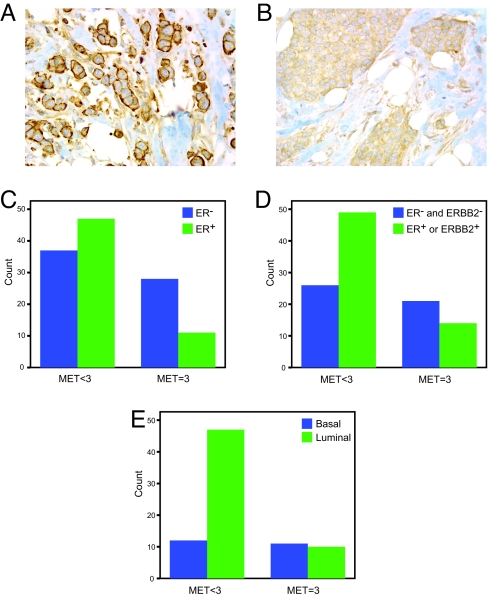
High MET staining (score = 3) correlates with ER−, ER−/ERBB2−, and basal cancers. (A) Strong membrane staining and (B) strong cytoplasmic staining of MET observed in breast cancer tissue microarray cores (Magnification: 400×). (C) High MET staining (score = 3) negatively correlates with ER staining (P = 0.016). (D) High MET staining (score = 3) negatively correlates with ER and ERBB2 staining (P = 0.024). (E) High MET staining (score = 3) significantly associates with the basal subtype by immunohistochemistry (P = 0.015).
Correlative analysis was performed to determine if MET levels were associated with ER, ERBB2, luminal, or basal breast cancer subtypes. Although MET expression was observed in all subtypes, MET levels ≥2 were not associated with ER expression, ERBB2 expression, or a specific subtype. However, the highest MET staining (score = 3) was enriched in ER− and ER−/ERBB2− cancers (Fig. 5 C and D and Table S6). Furthermore, high MET expression (MET = 3) correlated with the basal immunohistochemical subtype (Fig. 5E and Table S6). These findings show that MET is expressed in the majority of breast cancers and highest levels of MET are present more frequently in aggressive subtypes.
Discussion
Several studies have demonstrated that Met and HGF/SF are associated with poor prognosis in breast cancer (www.vai.org/metandcancer), but we still have a limited understanding for how and when Met influences mammary tumorigenesis. Here, we report that mutationally activated Met on the FVB/N background is able to induce a high incidence of diverse breast carcinomas in a knockin mouse model. Other mouse studies support the tumorigenic effect that Met activation has on the mammary epithelium (15, 27). In this issue, Ponzo et al. describe a transgenic model of mutationally activated Met that develops mammary carcinomas (28). The development of diverse histological phenotypes with basal characteristics in both models suggests that Met may initiate tumorigenesis in an early progenitor cell. This is contrary to the conventional thought of Met primarily being involved in the later stages of tumor progression but consistent with the Wang et al. transgenic hepatocellular carcinoma model (29). Metmut mice require both mutation and Met amplification for the development of aggressive mammary cancers, yet MET has not been shown to be mutated or amplified in human breast cancer. We hypothesize that Metmut bypasses the genetic instability required for sporadic human breast cancer and genetic amplification is key to the high penetrance observed in the mouse.
Understanding the signaling pathways that drive tumorigenic growth in the absence of hormone and ErbB2 signaling is critical for the development of effective therapies. Several reports indicate that up-regulation of growth factor signaling pathways may compensate for decreased estrogen and progesterone signaling (2). More recent work has shown that MET amplification occurs in lung cancers that develop resistance to gefitinib or erlotinib treatment (30). Metmut mammary tumors are ER+/PR−, suggesting that Met activation may overcome decreased hormone signaling. In addition, high MET expression was associated with ER−/ERBB2− human breast cancers. These results corroborate the finding in human breast cancers that MET is a prognostic factor independent of ERBB2 and suggest that MET signaling may be a critical component for driving tumor progression in the absence of ER, PR, or ERBB2 expression.
Molecular subtypes of breast cancer have given us a new understanding of factors that affect prognosis, yet our comprehension of these subtypes is still somewhat limited to their hormone receptor and ERBB2 status. Recent work has connected EGFR and CK5 expression to the basal subtype, but an independent predictor has not been defined (31). More likely, several genes will be needed to predict each subtype. This is the first report of a receptor tyrosine kinase mouse model that develops tumors with basal characteristics In addition, we demonstrate that high MET expression is associated with human basal cancers. These results are strongly supported by both the mouse model and the human breast cancer analysis reported by Ponzo et al. Importantly, these studies indicate that MET may serve as a therapeutic target for those patients with the most aggressive tumors and currently the fewest therapeutic options.
Methods
Survival Curves.
Survival was determined using the Kaplan and Meier survival function. Pairwise comparisons of survival curves were done using a log-rank test.
Tumor Analysis.
Mice were examined biweekly for tumor development and euthanized when tumors were between 1 and 2 cm3. Tumor samples were surgically isolated and fixed in 4% paraformaldehyde/PBS for 24 h. Fixed tissues were dehydrated, paraffin-embedded, and cut into 5-μm sections. Experiments using mice were approved by the Van Andel Research Institute Institutional Animal Care and Use Committee.
Met FISH.
Interphase FISH was performed on tumor touch preparations as described in ref. 14. Detailed procedures are provided in the SI Methods.
Immunohistochemical Analysis.
Antigen retrieval and detection were performed using a Discovery XT (Ventana Medical Systems). Detailed procedures are provided in the SI Methods.
Breast Tissue Microarray Construction and Immunohistochemical Subtype Scoring.
Human breast tissue microarrays consisting of duplicate 0.6-mm cores (Beecher Instruments) were constructed from archival tumor blocks from 137 patients with invasive breast cancer after surgical intervention at Washington University and Barnes-Jewish Hospital in St. Louis from 1997 to 2003. Immunohistochemistry for ER, PR, EGFR, Ck5/6, and HER2 and FISH for HER2 were performed as described in refs. 21 and 26. Detailed procedures are provided in the SI Methods.
Gene Expression Analysis.
Detailed procedures are provided in the SI Methods.
Supplementary Material
Acknowledgments.
We thank B. Berghuis and Van Andel Research Institute Histology for their expertise, the members of the B. Williams laboratory for advice, B. Eagleson and the Van Andel Research Institute Vivarium staff for technical assistance, and S. Moshkovitz for critical reading of the manuscript. We thank R. D. Cardiff for pathology consultation and T. O. Nielsen for assistance with the FISH studies and critical review of the manuscript. This work was supported by National Institutes of Health (NIH) Ruth L. Kirschstein National Research Service Award F32CA105748, NIH Grant U01 CA114722), and the Breast Cancer Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810403106/DCSupplemental.
References
- 1.Lapidus RG, Nass SJ, Davidson NE. The loss of estrogen and progesterone receptor gene expression in human breast cancer. J Mammary Gland Biol Neoplasia. 1998;3:85–94. doi: 10.1023/a:1018778403001. [DOI] [PubMed] [Google Scholar]
- 2.Cui X, Schiff R, Arpino G, Osborne CK, Lee AV. Biology of progesterone receptor loss in breast cancer and its implications for endocrine therapy. J Clin Oncol. 2005;23:7721–7735. doi: 10.1200/JCO.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Lohrisch C, Piccart M. HER2/neu as a predictive factor in breast cancer. Clin Breast Cancer. 2001;2:129–135. doi: 10.3816/CBC.2001.n.017. discussion 136–127. [DOI] [PubMed] [Google Scholar]
- 4.Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: Therapeutic options. Lancet Oncol. 2007;8:235–244. doi: 10.1016/S1470-2045(07)70074-8. [DOI] [PubMed] [Google Scholar]
- 5.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 6.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reis-Filho JS, et al. Metaplastic breast carcinomas are basal-like tumours. Histopathology. 2006;49:10–21. doi: 10.1111/j.1365-2559.2006.02467.x. [DOI] [PubMed] [Google Scholar]
- 8.Pezzi CM, et al. Characteristics and treatment of metaplastic breast cancer: Analysis of 892 cases from the National Cancer Data Base. Ann Surg Oncol. 2007;14:166–173. doi: 10.1245/s10434-006-9124-7. [DOI] [PubMed] [Google Scholar]
- 9.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 10.Ghoussoub RA, et al. Expression of c-met is a strong independent prognostic factor in breast carcinoma. Cancer. 1998;82:1513–1520. doi: 10.1002/(sici)1097-0142(19980415)82:8<1513::aid-cncr13>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 11.Camp RL, Rimm EB, Rimm DL. Met expression is associated with poor outcome in patients with axillary lymph node negative breast carcinoma. Cancer. 1999;86:2259–2265. doi: 10.1002/(sici)1097-0142(19991201)86:11<2259::aid-cncr13>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 12.Ocal IT, Dolled-Filhart M, D'Aquila T, Camp RL, Rimm DL. Tissue microarray-based studies of patients with lymph node negative breast carcinoma show that met expression is associated with worse outcome but is not correlated with epidermal growth factor family receptors. Cancer. 2003;97:1841–1847. doi: 10.1002/cncr.11335. [DOI] [PubMed] [Google Scholar]
- 13.Lengyel E, et al. C-Met overexpression in node-positive breast cancer identifies patients with poor clinical outcome independent of Her2/neu. Int J Cancer. 2005;113:678–682. doi: 10.1002/ijc.20598. [DOI] [PubMed] [Google Scholar]
- 14.Graveel C, et al. Activating Met mutations produce unique tumor profiles in mice with selective duplication of the mutant allele. Proc Natl Acad Sci USA. 2004;101:17198–17202. doi: 10.1073/pnas.0407651101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeffers M, et al. The mutationally activated Met receptor mediates motility and metastasis. Proc Natl Acad Sci USA. 1998;95:14417–14422. doi: 10.1073/pnas.95.24.14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nieto AI, et al. Persistent mammary hyperplasia in FVB/N mice. Comp Med. 2003;53:433–438. [PubMed] [Google Scholar]
- 17.Schmidt L, et al. Germline and somatic mutations in the tyrosine kinase domain of the Met proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 18.Smolen GA, et al. Frequent met oncogene amplification in a Brca1/Trp53 mouse model of mammary tumorigenesis. Cancer Res. 2006;66:3452–3455. doi: 10.1158/0008-5472.CAN-05-4181. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, et al. Estrogen receptor positivity in mammary tumors of Wnt-1 transgenic mice is influenced by collaborating oncogenic mutations. Oncogene. 2005;24:4220–4231. doi: 10.1038/sj.onc.1208597. [DOI] [PubMed] [Google Scholar]
- 20.Kim HJ, Cui X, Hilsenbeck SG, Lee AV. Progesterone receptor loss correlates with human epidermal growth factor receptor 2 overexpression in estrogen receptor-positive breast cancer. Clin Cancer Res. 2006;12:1013s–1018s. doi: 10.1158/1078-0432.CCR-05-2128. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen TO, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 22.Richardson AL, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 23.Sweet-Cordero A, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 24.Knudsen BS, et al. A novel multipurpose monoclonal antibody for evaluating human c-Met expression in preclinical and clinical settings. Appl Immunohistochem Mol Morphol. 2009;17:57–67. doi: 10.1097/PAI.0b013e3181816ae2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindemann K, et al. Differential expression of c-Met, its ligand HGF/SF and HER2/neu in DCIS and adjacent normal breast tissue. Histopathology. 2007;51:54–62. doi: 10.1111/j.1365-2559.2007.02732.x. [DOI] [PubMed] [Google Scholar]
- 26.Cheang MC, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 27.Gallego MI, Bierie B, Hennighausen L. Targeted expression of HGF/SF in mouse mammary epithelium leads to metastatic adenosquamous carcinomas through the activation of multiple signal transduction pathways. Oncogene. 2003;22:8498–8508. doi: 10.1038/sj.onc.1207063. [DOI] [PubMed] [Google Scholar]
- 28.Ponzo MG, et al. Met induces mammary tumors with multiple pathologies and is associated with both poor outcome and basal-type breast cancers. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0810402106. doi/ 10.1073/pnas.0810402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R, Ferrell LD, Faouzi S, Maher JJ, Bishop JM. Activation of the Met receptor by cell attachment induces and sustains hepatocellular carcinomas in transgenic mice. J Cell Biol. 2001;153:1023–1034. doi: 10.1083/jcb.153.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 31.Yehiely F, Moyano JV, Evans JR, Nielsen TO, Cryns VL. Deconstructing the molecular portrait of basal-like breast cancer. Trends Mol Med. 2006;12:537–544. doi: 10.1016/j.molmed.2006.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.