Abstract
Interspersed repeat composition and distribution in mammals have been best characterized in the human and mouse genomes. The bovine genome contains typical eutherian mammal repeats, but also has a significant number of long interspersed nuclear element RTE (BovB) elements proposed to have been horizontally transferred from squamata. Our analysis of the BovB repeats has indicated that only a few of them are currently likely to retrotranspose in cattle. However, bovine L1 repeats (L1 BT) have many likely active copies. Comparison of substitution rates for BovB and L1 BT indicates that L1 BT is a younger repeat family than BovB. In contrast to mouse and human, L1 occurrence is not negatively correlated with G+C content. However, BovB, Bov A2, ART2A, and Bov-tA are negatively correlated with G+C, although Bov-tAs correlation is weaker. Also, by performing genome wide correlation analysis of interspersed and simple sequence repeats, we have identified genome territories by repeat content that appear to define ancestral vs. ruminant-specific genomic regions. These ancestral regions, enriched with L2 and MIR repeats, are largely conserved between bovine and human.
Keywords: cow, BovB, microsatellite, repetitive DNA
Interspersed repeats are the largest class of sequences in mammalian genomes, accounting for 40 to 50% of the total length of these genomes (1–4). The most common interspersed repeats are retrotransposons, also known as retroposons, elements that replicate and jump throughout the genome in a manner similar to retroviruses (1). Although many retroposons are common to all mammals and are, thus, presumably of ancestral origin (5), every species/clade seems to have 1 or more unique kind of short interspersed nuclear element (SINE), which contribute heavily to species specific genome sequences (3). Although many retrotransposons are no longer active, species- and lineage-specific repeats serve to remodel genomes by interrupting and often outnumbering ancestral repeats during their phase of rapid transposition/expansion (6–8). Actively transposing repeats are believed to be responsible for 10% of mutations in rodents (9), whereas less active repeats in humans appear to account for a small fraction of new mutations (10). The accumulation of interspersed repeats within or near genes has been studied (11), and there is evidence that insertions within or near promoters can alter gene expression, whereas insertions into exons are often incorporated into existing protein-coding genes (12). Therefore, it is clear that interspersed repeats are major drivers of genome evolution.
In mammals, long (L)INE L1 repeats are the dominant retrotransposon type both in the common ancestor and in extant species (2, 4, 13). Few mammals have active non-LTR LINEs other than L1 that contribute significantly to repeat composition, with the exception of the LINE RTE repeats in ruminants and marsupials (14).
SINEs require LINEs for their transposition. In primates, LINE L1 repeats encode the machinery to transpose SINE Alu repeats (15). Ancestral L2 LINEs are believed to have encoded the machinery to transpose SINE mammalian-wide interspersed repeat (MIR) (3). In ruminants and marsupials, LINE RTEs encode the machinery to transpose SINE BovA (BOV-A2, Bov-tA1,2,3)/SINE ART2A or SINE RTE, respectively (16). RTE LINEs contain BovB repeats that are believed to have been horizontally transmitted from reptiles to ruminants (17, 18) and to marsupials (14).
In this report, we describe an analysis of the overall repeat content of the bovine genome. We also revisit the evidence for horizontal transfer of LINE RTE (BovB) repeats to ruminants, based on their evolution in cattle. Last, we show that there is evidence for spatial accumulation/segregation of repeats based on pairwise correlations of repeat abundance. Thanks to the unique presence of ruminant-specific LINE RTE (BovB) and associated SINE in cattle (19, 20), we will show evidence that these spatial correlations can differentiate ancestral vs. novel genomic territories on the basis of repeat content.
Results
Repeat Content.
We did not identify major new classes of repeats, but did construct improved consensus sequences for repeat masking. Based on these sequences, the total interspersed repeat content of the bovine genome is 46.5% (Table 1), with >24% made up of lineage-specific repeats. The vast majority of cattle-specific repeats are non-LTR LINE RTE (BovB) and BovB-derived SINEs.
Table 1.
Repeat content of the bovine genome
| Group | No. | Total bp | Percentage coverage of genome |
||
|---|---|---|---|---|---|
| Bos taurus | Human | Mouse | |||
| Non-LTR retrotransposons (LINE) | |||||
| L1 | 616,259 | 328,664,804 | 11.26352 | 17.07 | 19.14 |
| RTE (BovB) | 376,067 | 313,409,818 | 10.74072 | NA | 0.02 |
| L2 | 132,485 | 34,553,185 | 1.18416 | 3.07 | 0.37 |
| CR1 | 14,524 | 3,083,954 | 0.10569 | 0.27 | 0.06 |
| Total | 1,139,335 | 679,711,761 | 23.29409 | 20.40 | 19.59 |
| SINEs | |||||
| BOV-A2 | 377,697 | 68,880,046 | 2.360556 | NA | NA |
| Bov-tA | 1,461,800 | 225,579,571 | 7.730733 | NA | NA |
| ART2A | 348,768 | 121,997,595 | 4.18092 | NA | NA |
| tRNA | 388,920 | 57,981,206 | 1.98705 | NA | 0.00 |
| MIR | 301,335 | 40,569,445 | 1.39034 | 2.43 | 0.55 |
| Other | 4,322 | 432,334 | 0.01482 | 10.68 | 6.78 |
| Total | 2,882,842 | 515,440,197 | 17.66441 | 13.11 | 7.34 |
| ERVs | 277,632 | 93,363,384 | 3.19961 | 8.56 | 9.84 |
| DNA transposons | 244,174 | 57,157,641 | 1.95882 | 3.00 | 0.89 |
| LTR other | 34,352 | 12,395,410 | 0.42480 | 0.00 | 0.01 |
| Interspersed repeat total | 4,578,335 | 1,358,068,393 | 46.54174 | 45.08 | 37.65 |
| SSR total | 5,653,575 | 66,275,552 | 2.27130 | 0.78 | 4.16 |
NA, not applicable. Bold denotes repeat group totals.
Lineage Specific LINE.
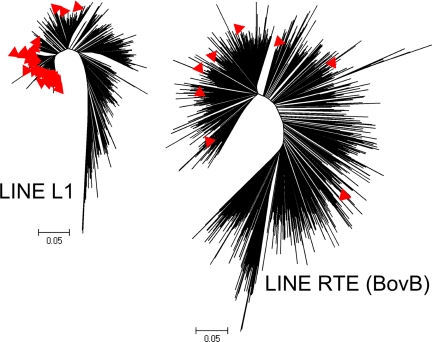
In view of the proposed horizontal transfer of BovB from squamates to an ancestral ruminant ≈50 Mya (18), we examined the divergence of BovB repeats within the bovine genome. Previous comparisons of BovB consensus sequences from a number of taxa (14) indicate that the simplest explanation for the BovBs found in marsupials and cattle is horizontal gene transfer. We identified 1,248 intact BovB by aligning our improved BovB consensus sequence to the bovine genome assembly v4 and extracting all full-length (≥90%) matching sequences that were ≥70% identical. These intact BovB were used to construct a maximum likelihood tree (Fig. 1). BovB repeats should have a single, large (≈1,000 aa) ORF encoding a protein with a reverse transcriptase domain and an endonuclease/exonuclease/phosphatase domain (21). We identified all of the ORFs in the 1,248 highly conserved BovB sequences to find potentially active BovB retroposons. Only 9 of these BovB sequences contained a large ORF meeting the domain criteria. The lack of potentially active BovB suggests that new mutations caused by lineage-specific repeat insertions are infrequent.
Fig. 1.
Intact LINE trees. Maximum Likelihood trees derived from global alignments of all intact/full-length LINE sequences. Red triangles indicate potentially active LINEs based on their intact ORF content.
L1 LINE is the second most prevalent type of interspersed repeat, based on insertion events (after the lineage-specific SINE Bov-tA repeats; see ref. 22) in the bovine genome. The counts for each repeat group are given in Table 1. Bovine L1 LINE sequences are highly conserved in a core region that spans the 2 ORFs found in these repeats, but are variable in the length and composition of their 5′ and 3′ regions. By using our consensus bovine L1 sequence (L1 BT), we identified 811 intact L1s in the bovine genome, which we used to construct a maximum likelihood tree (Fig. 1). Seventy-three of these L1 are potentially active based on their ORF content. This result indicated that L1 elements are probably more active than BovB.
We identified the potentially active BovB and L1 elements on our trees (Fig. 1), and found that active L1 elements clearly congregated in regions of short terminal branches, whereas putative active BovB did not. BovB repeats had a substitution rate of 0.065 ± 0.002 substitutions/site, whereas L1 BT repeats had a substitution rate of 0.031 ± 0.001.
Correlation Analysis.
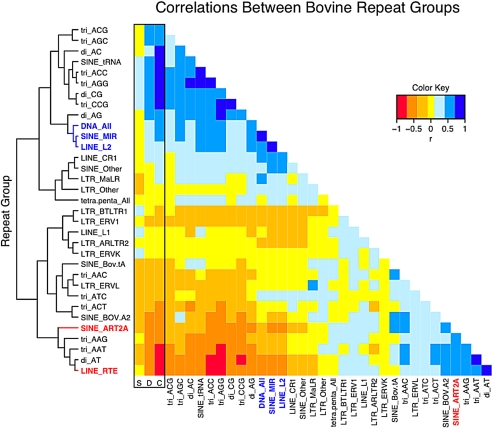
We captured 99% of the total chromosome scaffold sequence for our correlation analysis (1,750 bins). Spearman's rank correlation was calculated for each pairwise combination of the repeat types and between repeat types and G+C content, gene density, and segmental duplications. The correlations are depicted in Fig. 2, where repeat types are hierarchically clustered based on all their correlations. A number of striking relationships were observed, most notably, repeats belonging to LINE/SINE pairs, such as L2/MIR and LINE RTE (BovB)/ART2A, were highly positively correlated. Also, whereas ancestral repeats such as L2 and MIR were positively correlated with gene density and G+C content, ruminant-specific repeats BovB, Bov-tA, BOV-A2, and ART2A were negatively correlated with gene density and G+C content. Other recent repeats, such as tRNA derived repeats, were found to be highly positively correlated with both gene density and G+C content. L1 repeats were not strongly correlated with any features. Also, DNA All transposons clustered with L2 and MIR, even though they are not retroposons. Last, some simple sequence repeats (SSRs) were found to exhibit very strong correlations with G+C content and certain types of interspersed repeat.
Fig. 2.
Correlation analysis of repeat groups. Pairwise correlations among the repeat groups and between the repeat groups and segmental duplication (column S), gene density (column D), and G+C content (column C). Repeat groups are clustered based on all their correlations. Yellow cells have nonsignificant correlations (95% 2-tailed test after Bonferroni correction). Blue cells indicate significant positive correlations, and the orange/red cells indicate significant negative correlations.
Ancestral Vs. Ruminant-Specific Repeat Locations.
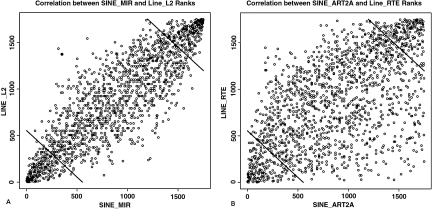
The strong positive correlations for both ancestral and ruminant-specific LINE/SINE pairs were further investigated to clarify the spatial relationships within and between the 2 correlations. The strength of the correlations is evident from both the high correlation coefficients (LINE RTE/SINE ART2A, r = 0.64; LINE L2/SINE MIR, r = 0.88) and from the plot of correlations (Fig. 3). These plots show rank correlations, although directly tied to specific repeat densities, and therefore, do not inform us directly about total repeat content. However, the rank correlations as plotted did allow us to identify the extreme density bins for L2/MIR and RTE/ART2A based on the 5% tails from the random distribution of the sum of the ranks. The extreme L2/MIR density bins (shown in Fig. 3A) defined genomic regions containing either a high or low density of ancestral repeats. Of the 1,750 bins, 261, 1,232, and 257 were classified as having low, medium, and high L2/MIR density, respectively. The medium density bins contained ≈2.5 times the number of MIR/L2 repeats than the low density bins, whereas the high density bins contained ≈2 times the MIR/L2 counts per bin as the medium density bins. The number of MIR and L2 repeats in each density group is given in Table 2. To determine any relationship between more recent ruminant-specific repeats (BovB/ART2A/BOV-A2) and the ancestral repeats, we compared the numbers of BovB, ART2A, and BOV-A2 repeats in high, medium, and low density ancestral bins. For each of the ruminant-specific repeats, there was a significant difference (Pr < 0.001) between the counts in the low and high L2/MIR density bins. In each case, the ruminant-specific repeats were present at a lower level in the high L2/MIR density bins than in the low-density bins. The medium L2/MIR density bins had medium levels of BovB and ART2A (probability of a quadratic trend 0.58 and 0.83, respectively), but high levels of BOV-A2 (Pr < 0.001) with a median of 202, which was higher than in the low-density L2/MIR bins (median 195). This analysis clearly showed that ruminant-specific repeats were present at a significantly lower density in high density ancestral repeat rich regions.
Fig. 3.
Rank correlations for ancestral and recent LINE/SINE pairs. (Left) Ranks of the ancestral LINE L2 and SINE MIR counts for each 1.5-Mbp bin. (Right) Ranks of recent LINE RTE (BovB) and SINE ART2A counts for each 1.5-Mbp bin. Lines in the upper right and lower left corners indicate the cut off for the high and low density bins, respectively, and are based on the expected 5% tails from the random distribution of the sum of the ranks.
Table 2.
Ancestral and clade-specific repeat densities in MIR/L2 density group bins
|
MIR/L2 density bins |
||||
|---|---|---|---|---|
| Low | Medium | High | All | |
| Number of bins | 261 | 1,232 | 257 | 1,750 |
| MIR counts | 14,864 | 181,670 | 83,615 | 280,149 |
| MIR/Bin | 56.95 | 147.46 | 325.35 | 160.09 |
| L2 counts | 7,110 | 82,439 | 32,922 | 122,471 |
| L2/Bin | 27.24 | 66.91 | 128.10 | 69.98 |
| Median RTE | 258 | 183 | 139 | 181 |
| Median BOVA2 | 195 | 202 | 175 | 198 |
| Median ART2A | 207 | 176.5 | 149 | 176 |
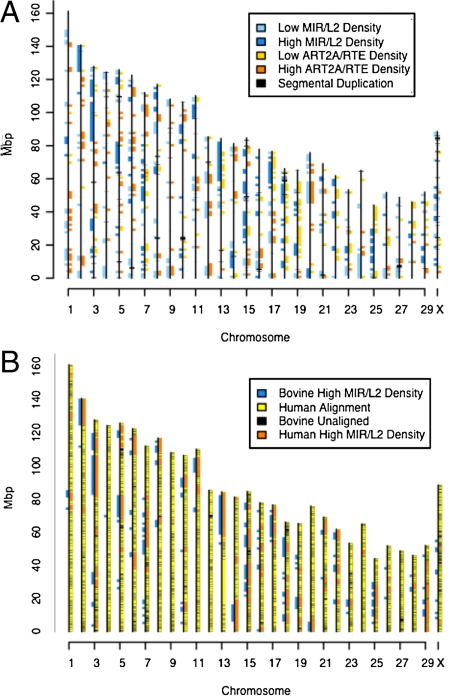
Because our analysis of extreme repeat density bins indicated that recent repeat insertions were underrepresented in ancestral repeat-rich bins, we sought to determine the location of these extreme density bins across the genome. When we plotted the coordinates (Table S1) of extreme density bins for L2/MIR correlations on the genome (Fig. 4A), it became apparent that the high-density bins were highly clustered and appeared to define particular regions. Also, although the high-density bins for ruminant-specific repeats did not cluster, none of them overlapped with the high-density ancestral regions, and thus, they defined mutually exclusive territories.
Fig. 4.
Ancestral and new repeat groups define different genomic territories. (A) Locations of 1.5-Mbp bins with extreme (high and low) ancestral (L2/MIR) and recent RTE/ART2A repeat densities are shown on the Btau4 assembly. Locations of the segmental duplications are plotted in black. Ancestral repeats tend to occur in blocks, whereas recent repeats generally do not. There is no overlap between high-density ancestral repeat blocks and bins with a high density of recent repeat blocks. Segmental duplications do not appear to colocalize with either high- or low-density regions of either repeat class. (B) Ancestral (L2/MIR) high-density bins for bovine and human are shown on the bovine assembly, along with the overall alignment from Cow-Net (23–25). Note that the “top” of the chromosomes corresponds to the end near the x axis on our plot. The y axis corresponds to nucleotide coordinates in mega base pairs (Mbp) from the bovine assembly Btau4.
To determine whether these ancestral repeat rich territories were evolutionarily conserved, we repeated our correlation analysis on the human genome assembly (hg18), and found that L2/MIR were very highly correlated in human (r = 0.86) as well. We identified the high-density L2/MIR bins for the human genome, and then, converted their coordinates to bovine genome locations using the University of California Santa Cruz (UCSC) Cow Net alignments (23–25). When we examined the overlaps of the bovine and human ancestral territories based on the bovine sequence, we found that 77% of the human bins overlapped with bovine bins, and 79% of bovine bins overlapped with human bins, indicating a very high degree of conservation. Fig. 4B shows the bovine and human ancestral domain overlaps, and reveal that they are particularly striking for the larger contiguous sets of bins that define large territories.
Discussion
To our knowledge, our hybrid method of detecting and classifying repeats is novel, and provides useful insight into the true state of repeats in mammalian genomes. Most of the repeats we identified de novo can be regarded as chimeric based on their apparent inclusion of multiple pieces of known repeats. Although many of these chimeric repeats probably result from multiple insertion events leading to nested, fragmented repeats, some may represent new or emerging repeats formed by the recombination of existing repeats. It will require considerable additional analytical effort to distinguish possible new hybrid repeats from fragmented repeats resulting from multiple insertion events.
Interspersed, repetitive DNA elements of all classes account for 46.5% of the bovine genome. This percentage is similar to human repetitive content (≈45%), but significantly greater than mouse (≈38%) and dog (35%) (2, 4, 13). Also, the bovine genome is unusual, because its repeat composition is more similar to that of the opossum than to other nonruminant eutheria. This composition is attributable to LINE RTE type BovB and its associated SINE elements (BovA2, BOV-tA, and ART2A), which together account for 25% of the bovine genome. However, even in the opossum, LINE RTE and SINE RTE together account for only ≈5% of the genome (14). Therefore, the bovine genome is very different in composition to all other sequenced mammalian genomes, implying that its repeat-related evolution has been largely driven by the expansion of ruminant-specific repeats.
The origin of BovB in ruminants is probably a result of horizontal transfer. It is unlikely to have originated from an ancestral repeat that has completely died out in most mammalian lineages. To explain the current phylogenetic distribution of BovB in mammals requires 3 separate horizontal transfer events; once to monotremes ≈146 Mya, once to marsupials over 100 Mya, and once to ruminants ≈50 Mya. This type of horizontal transfer has been observed before for space invaders (SPIN) transposons (26). Although to our knowledge BovB does not appear to be present in nonruminant artiodactyls (27), BovB and derived repeats have contributed significantly to bovine evolution based on the proportion of the genome they represent. In European cattle, BovB is possibly no longer active despite many of apparently intact copies. Less than 10 of the intact repeats are plausibly active. Therefore, we conclude that BovB is potentially no longer a source of significant repeat site insertion polymorphism or mutation. Compared with cattle, the opossum has half the number of LINE RTEs, and they cover half the number of base pairs, but only 2% are intact, and none are potentially active, confirming that BovB repeats are older in the opossum than in the cow.
LINE L1 repeats are not only the most abundant type of LINE in the bovine genome, but are also the most abundant LINE in other sequenced mammalian genomes. In fact, LINE L1 evolution is characterized by a rapid rise and fall and replacement of subfamilies leading to dominant, lineage specific LINE L1 (28). From our data, most L1 in cattle belong to the L1 BT subfamily. Although cattle have only 60% of the L1 found in humans, they appear to be more active based on the 1.6-fold greater number of intact, potentially functional copies (there are ≈45 active L1 in humans; see ref. 29) present in cattle. Therefore, in contrast to BovB, L1 BT elements are probably still quite active in the bovine genome. Also, the substitution rate for BovB was twice that for L1 BT. Based on these results, we conclude that BovB repeats are older compared with the L1 BT subfamily of repeats, which have retained almost an order of magnitude more active copies.
Our comprehensive, pairwise correlation analysis is a previously undescribed method of globally profiling the association of repeats and other genomic features. Our identification of spatially correlated repeat type pairs indicated that a number of simple and interspersed repeats may share some type of previously uncharacterized target sequence bias or accumulation bias. By using repeat correlations, we were able to identify regions of the genome with distinct repeat compositions that were not readily identified by looking solely at counts. Our observation that some recent repeats BovB, Bov A2, ART2A, and Bov-tA are negatively correlated with G+C, whereas tRNA derived repeats are strongly positively correlated with G+C and L1 show no correlation with G+C, is in contrast to positive correlations with G+C for recent repeats such as Alu in human and mouse and negative correlations with G+C for L1 (2, 4). Therefore, although LINE/SINE pairs show similar correlation with G+C in cattle, they are oppositely correlated in human and mouse. Many of the strong correlations we observed involved SSRs, in particular correlated with gene density and G+C content. It is not clear why SSRs should be spatially correlated with any genomic features. However, some may serve to influence replication via effects on DNA secondary structure (30).
The bovine genome is particularly informative for this type of repeat correlation analysis, because it is unique among fully sequenced eutheria in possessing 2 distinct LINE/SINE pairs, one of which is ancestral and inactive, and one of recent origin and possibly still active. Spatial correlations within these pairs allowed us to identify regions of the genome with distinct repeat compositions that corresponded to ancient vs. recent genomic domains. This domain structure is a previously undescribed observation, whose significance is unclear at this time. The ancestral repeat-enriched regions were predominantly clustered as opposed to the new, ruminant-specific repeat regions, which were scattered. One possible explanation for the observed clustering of ancestral repeats into domains could be that certain regions of the genome tended to resist the invasion and accumulation of new, ruminant-specific repeats. This explanation seems likely in view of the overwhelming conservation of these ancestral repeat enriched regions in human, which diverged from cattle ≈92 Mya (31). Conservation of ancient repeats across taxa could imply potential functional consequences in terms of recombination hotspots, gene expression domains, or segmental duplication. Because the first 2 types of data are not available for cattle, we were only able to compare the locations of segmental duplications with these ancestral domains. Segmental duplications were not associated with ancestral or ruminant-specific repeat-rich regions, but did show weakly positive, significant correlations with L1, some LTR containing repeats and SINE tRNA (Fig. 2). Similar analyses in human (hg17) revealed no strong correlations between repeats and segmental duplications, recombination hot spots, or evolutionary breakpoints. So although our overall correlations supported the idea that some interspersed repeats serve as drivers of segmental duplication via nonallelic homologous recombination (32, 33), our observation of ancestral repeat-rich domains did not have any obvious significance in this context.
By identifying regions of the genome enriched with ancient repeats and others with recent repeats, we have highlighted genomic regions that differ in their degree of retroposon-mediated remodelling. Our method of identifying ancient genomic territories determined by repeat composition has highlighted ancestral conserved regions of the genome in bovine and human, and may provide a new way of viewing genome evolution in mammals, as an alternative to efforts to reconstruct the mammalian ancestral karyotype by breakpoint mapping (34, 35). Because the ancient repeat-enriched regions cluster into conserved domains, we conclude that these domains may be correlated with other types of genomic/chromosomal domains already identified for gene expression (36), nuclear localization (37), or DNA methylation (38). The latter is intriguing, and prompts us to speculate that genomic, or epigenomic methylation aimed at repressing retroposon activity, might affect not only retroposon transcription, but also retroposon insertion. Alternatively, these clusters may be indicative of other, as yet unknown structural or functional genomic domains.
Methods
De Novo Repeat Identification.
Bovine genome assembly v4 was used for repeat identification. Repeats were identified independently using 2 methods and then subsequently pooled for annotation. First, repeats were identified using a pipeline comprised of PALS/PILER/MUSCLE (39–42). PALS output files were concatenated chromosomewise and used as input to PILER and MUSCLE, which generated consensus sequences. The jobs were run in parallel on an SGI ALTIX supercluster. Second, RepeatScout (43) was used to identify repeats from individual chromosome scaffolds, using default settings and build_lmer_table –l 14. Consensus sequences from PILER and RepeatScout output were generated by identifying globally alignable sets of sequences with blastclust (available at http://www.ncbi.nlm.nih.gov/BLAST/docs/blastclust.htm) at S = 90% and L = 0.95. Each cluster was then globally aligned using MUSCLE and a consensus generated using PILER.
Repeat Annotation.
Identifiable repeats were annotated by masking with RepeatMasker. Also, WU-BLAST (44) was used with a comprehensive retroviral and retroposon protein database assembled from National Center for Biotechnology Information resources (42) to further annotate repeats, and with swissprot (45) to identify known protein-coding genes from large gene families inappropriately included in the repeat set. Consensus sequences identified as similar to protein-coding sequences, but not similar to retroposon or endogenous retrovirus protein-coding sequences, were removed from the consensus set.
Identification of, and Tree Construction for, Intact LINE Elements.
Intact L1 and BovB gff coordinates were retrieved from the bovine genome assembly using PALS, with a minimum length of 90% of the query sequence, and a minimum of 70% identity. Because we were unable to retrieve any intact BovB from the opossum genome mondom4 using this method and the MD RTE consensus sequence, they were retrieved based on RepeatMasker coordinates for repeats >90% as long as the MD RTE RepeatMasker consensus. Sequences were globally aligned using MUSCLE (40), and the alignments used to create maximum likelihood trees using RAxML (46) with the GTRCAT substitution model, and an initial 200 bootstraps followed by a thorough ML search. To avoid the confounding effect of G+C content (47), and the biasing that would be introduced because of the negative correlation of BovB with G+C content, as seen in Fig. 2, a bin-balanced subset of 162 intact, full-length BovBs, and L1 BTs were used to calculate their Jukes–Cantor substitution rates (48). All positions containing gaps and missing data were omitted, and standard errors were estimated by a bootstrap procedure with 500 repeats. Calculation of the Jukes–Cantor substitution rates and editing the trees for appearance were performed by using MEGA v4 (49). Potentially active elements were identified by scanning for ORF of the appropriate length using CLC Sequence Viewer 5 (CLC Bio).
Correlation Analysis.
Interspersed repeat coordinates were obtained from RepeatMasker using a custom library comprising the consequence sequences and RepeatMasker's mammal library. SSR coordinates were obtained from Phobos output (50). The interspersed repeats were grouped according to the classification given by RepeatMasker, except for CHR/tRNA/tRNA-Glu, which we amalgamated as tRNA, BTLTR1, and ARLTR2, which were each given their own family due to their predominance in LTR Other, and SINE BovA, which was divided into BOV-A2 and Bov-tA (comprising Bov-tA1, 2, and 3) families, because these repeats are thought to have different evolutionary histories (51). The SSRs with repeating pattern length 4 or 5 were combined into one group (tetra/penta All), whereas those with repeating pattern lengths of 2 or 3 were grouped according to their repeating pattern. Each chromosome was divided into 1.5-Mbp segments (bins) beginning at the 5′ end. For each bin, we calculated the number of repeats from each repeat group that were entirely within the bin, the number of GLEAN 5 gene models that started in the bin (gene density), the G+C content, and the number of segmental duplications entirely within the bin. All bins with at least 1-Mbp non-N specified base pair were used to calculate Spearman rank correlations between each repeat group and the other repeat groups, as well as gene density, G+C content, and segmental duplication. To control for multiple testing among our many correlations, we used Bonferroni corrected P values as a measure of statistical significance. The repeat groups were clustered based on the correlations among the repeat groups, gene density, G+C content, and segmental duplication.
Identification of Extreme Density Bins and Repeat Content Analysis.
The bins were classified as having low, medium, or high MIR/L2 density. The cutoff between the groups was the 2-tail 10% significance level cutoff for the sum of the MIR and L2 ranks. For the LINE RTE, SINE ART2A, and SINE BOV-A2 repeat groups, the statistical package R was used to perform Wilcoxon rank sum tests with continuity correction between the high and low density groups, and between the medium group and the high and low groups combined, to test for linear and quadratic trends, respectively. The high-density MIR/L2 bins for human (hg18) were obtained as for the bovine, except the RepeatMasker library for human was used. These human bins were mapped to bovine bins where there was an overlap of at least 200,000 bp, using UCSC Cow-Net coordinates (23).
Supplementary Material
Acknowledgments.
We thank the Bovine Genome Sequencing Project for providing segmental duplication data (E. Eichler, Seattle, WA), GLEAN gene models (C. Elsik, Washington, DC), and their coordinates; and the anonymous reviewers who helped improve this report.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901282106/DCSupplemental.
References
- 1.Smit AF. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Jurka J, Kapitonov VV, Kohany O, Jurka MV. Repetitive sequences in complex genomes: Structure and evolution. Annu Rev Genom Hum G. 2007;8:241–259. doi: 10.1146/annurev.genom.8.080706.092416. [DOI] [PubMed] [Google Scholar]
- 4.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 5.Smit AFA, Riggs AD. MIRs are classic, transfer-RNA-derived SINEs that amplified before the mammalian radiation. Nucleic Acids Res. 1995;23:98–102. doi: 10.1093/nar/23.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Giordano J, et al. Evolutionary History of Mammalian Transposons Determined by Genome-Wide Defragmentation. PLoS Comput Biol. 2007;3:e137. doi: 10.1371/journal.pcbi.0030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kazazian HH., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 9.Kazazian HH., Jr Mobile elements and disease. Curr Opin Genet Dev. 1998;8:343–350. doi: 10.1016/s0959-437x(98)80092-0. [DOI] [PubMed] [Google Scholar]
- 10.Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 11.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krull M, Petrusma M, Makalowski W, Brosius J, Schmitz J. Functional persistence of exonized mammalian-wide interspersed repeat elements (MIRs) Genome Res. 2007;17:1139–1145. doi: 10.1101/gr.6320607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 14.Gentles AJ, et al. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 2007;17:992–1004. doi: 10.1101/gr.6070707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 16.Ohshima K, Okada N. SINEs and LINEs: Symbionts of eukaryotic genomes with a common tail. Cytogenet Genome Res. 2005;110:475–490. doi: 10.1159/000084981. [DOI] [PubMed] [Google Scholar]
- 17.Kordis D, Gubensek F. Horizontal transfer of non-LTR retrotransposons in vertebrates. Genetica. 1999;107:121–128. doi: 10.1023/a:1004082906518. [DOI] [PubMed] [Google Scholar]
- 18.Kordis D, Gubensek F. Unusual horizontal transfer of a long interspersed nuclear element between distant vertebrate classes. Proc Natl Acad Sci USA. 1998;95:10704–10709. doi: 10.1073/pnas.95.18.10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimamura M, Abe H, Nikaido M, Ohshima K, Okada N. Genealogy of families of SINEs in cetaceans and artiodactyls: The presence of a huge superfamily of tRNA(Glu)-derived families of SINEs. Mol Biol Evol. 1999;16:1046–1060. doi: 10.1093/oxfordjournals.molbev.a026194. [DOI] [PubMed] [Google Scholar]
- 20.Jobse C, et al. Evolution and recombination of bovine DNA repeats. J Mol Evol. 1995;41:277–283. [PubMed] [Google Scholar]
- 21.Malik HS, Eickbush TH. The RTE class of non-LTR retrotransposons is widely distributed in animals and is the origin of many SINEs. Mol Biol Evol. 1998;15:1123–1134. doi: 10.1093/oxfordjournals.molbev.a026020. [DOI] [PubMed] [Google Scholar]
- 22.Lenstra JA, van Boxtel JA, Zwaagstra KA, Schwerin M. Short interspersed nuclear element (SINE) sequences of the Bovidae. Anim Genet. 1993;24:33–39. doi: 10.1111/j.1365-2052.1993.tb00916.x. [DOI] [PubMed] [Google Scholar]
- 23.Kent WJ, Baertsch R. The UCSC Genome Browser Database. 2009. Available at http://genome.ucsc.edu/goldenPath/credits.html#cow_credits. [DOI] [PMC free article] [PubMed]
- 24.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution's cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz S, et al. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pace JK, II, Gilbert C, Clark MS, Feschotte C. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc Natl Acad Sci USA. 2008;105:17023–17028. doi: 10.1073/pnas.0806548105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwashita S, et al. A Tandem Gene Duplication Followed by Recruitment of a Retrotransposon Created the Paralogous Bucentaur Gene (bcntp97) in the Ancestral Ruminant. Mol Biol Evol. 2006;23:798–806. doi: 10.1093/molbev/msj088. [DOI] [PubMed] [Google Scholar]
- 28.Waters PD, Dobigny G, Waddell PJ, Robinson TJ. Evolutionary History of LINE-1 in the Major Clades of Placental Mammals. PLoS ONE. 2007;2:e158. doi: 10.1371/journal.pone.0000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sassaman DM, et al. Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 30.Nakagama H, et al. Molecular mechanisms for maintenance of G-rich short tandem repeats capable of adopting G4 DNA structures. Mutat Res-Fund Mol M. 2006;598:120–131. doi: 10.1016/j.mrfmmm.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 31.Liu GE, Matukumalli LK, Sonstegard TS, Shade LL, Van Tassell CP. Genomic divergences among cattle, dog and human estimated from large-scale alignments of genomic sequences. BMC Genomics. 2006;7:140. doi: 10.1186/1471-2164-7-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue K, Lupski JR. Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet. 2002;3:199–242. doi: 10.1146/annurev.genom.3.032802.120023. [DOI] [PubMed] [Google Scholar]
- 33.Lupski JR, Stankiewicz P. Genomic Disorders: Molecular Mechanisms for Rearrangements and Conveyed Phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy WJ, Bourque G, Tesler G, Pevzner P, O'Brien SJ. Reconstructing the genomic architecture of mammalian ancestors using multispecies comparative maps. Hum Genomics. 2003;1:30–40. doi: 10.1186/1479-7364-1-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy WJ, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- 36.Cohen BA, Mitra RD, Hughes JD, Church GM. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet. 2000;26:183–186. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
- 37.Kupper K, et al. Radial chromatin positioning is shaped by local gene density, not by gene expression. Chromosoma. 2007;116:285–306. doi: 10.1007/s00412-007-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trasler JM. Gamete imprinting: Setting epigenetic patterns for the next generation. Reprod Fertil Dev. 2006;18:63–69. doi: 10.1071/rd05118. [DOI] [PubMed] [Google Scholar]
- 39.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC, Myers EW. PILER: Identification and classification of genomic repeats. Bioinformatics. 2005;21:I152–I158. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- 42.Smith CD, et al. Improved repeat identification and masking in Dipterans. Gene. 2007;389:1–9. doi: 10.1016/j.gene.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:I351–I358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 44.Gish W, States DJ. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 45.The UniProt C. The Universal Protein Resource (UniProt) Nucl. Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 47.Arndt PF, Hwa T, Petrov DA. Substantial regional variation in substitution rates in the human genome: Importance of GC content, gene density, and telomere-specific effects. J Mol Evol. 2005;60:748–763. doi: 10.1007/s00239-004-0222-5. [DOI] [PubMed] [Google Scholar]
- 48.Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism. New York: Academic; 1969. pp. 21–123. [Google Scholar]
- 49.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 50.Mayer C. PHOBOS. 2008 Available at http://www.ruhr-uni-bochum.de/spezzoo/cm/cm_phobos.htm. [Google Scholar]
- 51.Nijman IJ, Lenstra JA. Mutation and recombination in cattle satellite DNA: A feedback model for the evolution of satellite DNA repeats. J Mol Evol. 2001;52:361–371. doi: 10.1007/s002390010166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.