Abstract
Astrocyte elevated gene-1 (AEG-1) is overexpressed in >90% of human hepatocellular carcinoma (HCC) patients and plays a significant role in mediating aggressive progression of HCC. AEG-1 is known to augment invasion, metastasis, and angiogenesis, and we now demonstrate that AEG-1 directly contributes to another important hallmark of aggressive cancers, that is, resistance to chemotherapeutic drugs, such as 5-fluorouracil (5-FU). AEG-1 augments expression of the transcription factor LSF that regulates the expression of thymidylate synthase (TS), a target of 5-FU. In addition, AEG-1 enhances the expression of dihydropyrimidine dehydrogenase (DPYD) that catalyzes the initial and rate-limiting step in the catabolism of 5-FU. siRNA-mediated inhibition of AEG-1, LSF, or DPYD significantly increased the sensitivity of HCC cells to 5-FU in vitro and a lentivirus delivering AEG-1 siRNA in combination with 5-FU markedly inhibited growth of HCC cells xenotransplanted in athymic nude mice when compared to either agent alone. The present studies highlight 2 previously unidentified genes, AEG-1 and LSF, contributing to chemoresistance. Inhibition of AEG-1 might be exploited as a therapeutic strategy along with 5-FU-based combinatorial chemotherapy for HCC, a highly fatal cancer with currently very limited therapeutic options.
Keywords: 5-FU, AEG-1, chemoresistance, DPYD, LSF
Hepatocellular carcinoma (HCC) is a highly aggressive and vascular tumor (1, 2). The treatment options for HCC depend on the stages and grades of the disease (3). Surgical resection, radiofrequency ablation, and liver transplantations are the treatments of choice with localized disease (4, 5). However, most HCC patients present with advanced symptomatic tumors with underlying cirrhotic changes that are not amenable to surgical resection or transplantation. Transarterial chemoembolization (TACE) and systemic therapy with doxorubicin alone or a combination of cisplatin, IFN, doxorubicin, and 5-fluorouracil (5-FU) (PIAF) are being used for advanced disease with only moderate improvement (5–11). Small molecule inhibitors and monoclonal antibodies targeting specific signaling pathways are also being evaluated as potential therapeutics for HCC albeit also with limited success (4, 12–14). As such identification of master molecules regulating the aggressive progression of the disease will help develop targeted strategies to effectively combat this fatal malady.
We and others have demonstrated that the expression of astrocyte elevated gene-1 (AEG-1), discovered in our laboratory (15), is augmented in diverse tumors, such as breast and prostate cancer, melanoma, and malignant glioma (16–21). Our recent findings document that AEG-1 is frequently overexpressed in HCC compared to normal liver and that the AEG-1 gene is amplified in a significant proportion of HCC patients (22). More importantly AEG-1 expression increases with the stages and grades of HCC indicating that AEG-1 might control the aggressive progression of this cancer (22). AEG-1 expression is low in HCC cell lines that are less aggressive and do not form tumors in nude mice (such as HepG3), while it is conspicuously high in aggressive tumorigenic HCC cell lines (such as QGY-7703 and others) (22). Inhibition of AEG-1 by siRNA significantly inhibited growth of QGY-7703 tumors in nude mice while stable overexpression of AEG-1 in HepG3 cells (Hep-AEG-1–14 and Hep-AEG-1–8) increased invasion, anchorage-independent growth, and led to formation of highly aggressive, vascular tumors in nude mice (22). In HCC cells, AEG-1 activates multiple signal transduction pathways, such as MEK/ERK, NF-κB, and Wnt signaling, all known to play vital roles in HCC pathogenesis (22). AEG-1 also augments angiogenesis further contributing to the aggressiveness of this cancer. These findings indicate that AEG-1 might be a key molecule regulating HCC progression.
Global and pathway-specific gene expression analysis identified several AEG-1-downstream genes potentially involved in mediating its function. One gene that is highly induced by AEG-1 is LSF (late SV40 Factor) (22). LSF was first identified in HeLa cell extracts as a transcriptional activator of the late Simian Virus 40 promoter (23). There are 3 identified LSF subfamily genes in the human genome: LSF (chromosome 12q13), LBP-1a/b (chromosome 3), and LBP9 (chromosome 2) (24). While LSF and LBP-1a/b are ubiquitously expressed in the developing and adult mouse, LBP9 expression is restricted to specific tissues, such as placenta. Independent identification of LSF as a DNA-binding protein and transcriptional regulator of other viral and cellular promoters resulted in the additional names of LBP-1 or UBP-1 (on the HIV long-terminal repeat), CP2 (on the murine α-globin promoter), and SEF1 (on the murine serum amyloid A3 promoter) (25–29). LSF acts both as a transcriptional activator and repressor. It activates transcription of serum amyloid A3 (SAA3), IL-4, α-globin, α-A crystallin, thymidylate synthase (TS), and PAX6 in different vertebrate species (25, 29–33). In cell-free extracts, it activates RNA polymerase II transcription by binding to basal promoter factor TFIIB (34). LSF also inhibits transcription of HIV LTR by binding to YY1 and histone deacetylase 1 (HDAC1) (35). A major cellular target of LSF is the thymidylate synthase (TS) gene, which encodes the rate limiting enzyme in production of dTTP, required for DNA synthesis (30). LSF binds to the TS promoter and up-regulates TS mRNA at the G1/S transition. Inhibition of LSF by a dominant-negative construct (LSFdn) inhibits TS induction and induces apoptosis, while addition of thymidine in the medium protects the cells from inhibition of DNA synthesis and induction of apoptosis (30). As a consequence, LSF plays an important role in DNA synthesis and cell survival.
5-FU is a common chemotherapeutic for HCC (36). 5-FU is converted intracellularly into 5′-fluoro-2′-deoxyuridine by thymidine phosphorylase with subsequent phosphorylation by thymidine kinase into the active metabolite 5-fluoro-2′-deoxyuridine 5′-monophosphate (FdUMP) (37). FdUMP inhibits thymidylate synthase (TS), which reduces the thymidine pool and increases the uracil pool leading to the inhibition of DNA synthesis. 5-FU is converted into its inactive metabolite fluoro-5,6-dihydrouracil (FUH2) by dihydropyrimidine dehydrogenase (DPYD) (37). TS and DPYD gene expression and/or activity are major determinants of the efficacy of 5-FU (38, 39).
The present studies demonstrate that overexpression of AEG-1 increases resistance of HCC cells to 5-FU. This resistance is mediated by induction of LSF, with resultant increase in TS and DPYD by AEG-1. We thus establish another attribute of AEG-1, chemoresistance, conferring AEG-1-mediated aggressive progression of HCC and identify 2 previously unrecognized molecules, LSF and AEG-1, contributing to resistance to 5-FU.
Results
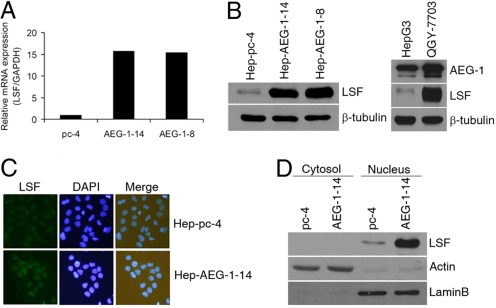
We have previously documented that HepG3 cells express low levels of AEG-1, and stable overexpression of AEG-1 in HepG3 cells (Hep-AEG-1–14 and Hep-AEG-1–8) significantly increases invasion and anchorage-independent growth (22). Additionally, while HepG3 cells do not form tumors in nude mice, Hep-AEG-1–14 and Hep-AEG-1–8 clones generate highly aggressive, vascular tumors in nude mice (22). To identify AEG-1-downstream genes, we performed Affymetrix cDNA microarray analysis between Hep-AEG-1–14 and Hep-pc-4 cells; the latter consists of HepG3 cells stably transformed with empty pcDNA3.1-hygro vector (22). A list of the modulated genes have been discussed in our previous publication (22) and the present manuscript focuses on 2 of them, LSF and dihydropyrimidine dehydrogenase (DPYD). LSF mRNA expression was identified to be 23-fold greater in the Hep-AEG-1–14 clone as compared to the Hep-PC-4. Taqman real-time PCR showed LSF expression to be 16- and 15-fold higher in Hep-AEG-1–14 and Hep-AEG-1–8 clones, respectively, compared to the Hep-pc-4 clone (Fig. 1A). In corollary, both Hep-AEG-1–14 and Hep-AEG-1–8 clones expressed markedly higher levels of LSF protein compared to Hep-pc-4 cells as determined by Western blot analysis (Fig. 1B Left). HepG3 cells that express low level of AEG-1 also express low level of LSF while QGY-7703 cells that express high level of AEG-1 also show high level expression of LSF indicating a direct correlation between AEG-1 and LSF expressions (Fig. 1B Right). As a transcription factor LSF is localized in the nucleus and immunofluorescence analysis confirmed overexpression of LSF in nucleus in the Hep-AEG-1–14 clone compared to the Hep-pc-4 clone (Fig. 1C). The nuclear localization of LSF was also confirmed by fractionating the cytosolic and nuclear compartments and analyzing LSF expression by Western blot. In Hep-AEG-1–14 clone LSF is highly expressed in the nucleus (Fig. 1D).
Fig. 1.
AEG-1 induces the expression of LSF. (A) Analysis of expression of LSF mRNA in Hep-pc-4 (pc-4), Hep-AEG-1–14 (AEG-1–14), and Hep-AEG-1–8 (AEG-1–8) cells by Taqman real-time PCR. (B) (Left) Analysis of LSF protein expression in the indicated cells by Western blot. (Right) Analysis of AEG-1 and LSF protein expression in HepG3 and QGY-7703 cells by Western blot. Expression of β-tubulin was used as a loading control in both panels. (C) Analysis of LSF expression by immunofluorescence in the indicated cells. (D) Subcellular localization of LSF. Hep-pc-4 (pc-4) and Hep-AEG-1–14 (AEG-1–14) cells were fractionated into cytosolic and nuclear fractions that were subjected to Western blot analysis for LSF expression. Expression of actin (for cytosol) and laminB (for nucleus) was analyzed to authenticate the purity of the individual fractions.
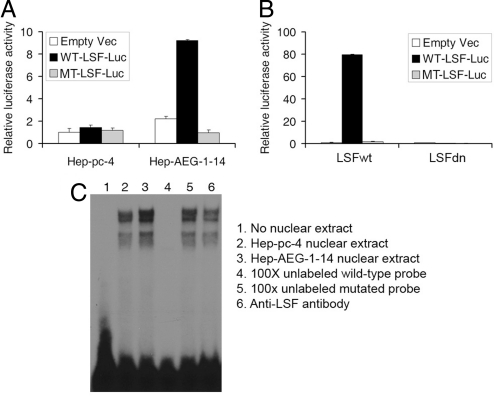
We next checked the transcriptional activity of the induced LSF in the Hep-AEG-1–14 clone. We transfected LSF-reporter luciferase construct, that contains the LSF-binding site upstream of the luciferase gene, in Hep-pc-4 and Hep-AEG-1–14 clones. The luciferase activity was significantly higher in the Hep-AEG-1–14 clone compared to the Hep-pc-4 clone. As a control, the activity of a construct with a mutated binding site upstream of luciferase gene did not show increased activity in Hep-AEG-1–14 clone (Fig. 2A). The specificity of LSF transcriptional activity in Hep-AEG-1–14 clone was confirmed by a dominant-negative LSF (LSFdn, a double amino acid substitution mutant of LSF initially named 234QL/236KE that is unable to bind DNA) that oligomerizes with wild-type LSF to also inhibit its DNA-binding activity (40). The activity of the LSF-luciferase reporter was markedly induced in the presence of wild-type LSF (LSFwt) while it was completely extinguished by LSFdn in the Hep-AEG-1–14 clone (Fig. 2B). Electrophoretic mobility shift assay (EMSA) analyzing LSF DNA binding to a radiolabeled consensus LSF-binding element further confirmed the transcriptional activity of LSF. LSF DNA binding was significantly higher in Hep-AEG-1–14 nuclear extract compared to Hep-pc-4 nuclear extract (Fig. 2C, lanes 3 and 2, respectively). The shifted bands could be effectively competed by 100× cold wild-type probe but not by mutant probe (Fig. 2C, lanes 4 and 5, respectively). Incubation with anti-LSF antibody did not supershift the band but reduced the intensity which might be due to recognition of the DNA-binding domain of LSF by this antibody (Fig. 2C, lane 6).
Fig. 2.
AEG-1 induces transcriptionally active LSF. (A) Hep-pc-4 and Hep-AEG-1–14 clones were transfected with empty pGL3-basic vector, WT-LSF-luc (pGL3B-WT4-E1b), and MT-LSF-luc (pGL3B-MT4-E1b) and luciferase activity was measured 48 h later. (B) Hep-pc-4 cells were transfected with empty pGL3-basic vector, WT-LSF-luc (pGL3B-WT4-E1b), and MT-LSF-luc (pGL3B-MT4-E1b) along with an expression plasmid expressing wild-type LSF (LSFwt) or a dominant-negative LSF (LSFdn) and luciferase activity was measured 48 h later. In both A and B, firefly luciferase activity was normalized by renilla luciferase activity and the activity of the empty pGL3-basic vector was considered as 1. The data represents mean ± SEM. (C) Electrophoretic mobility shift assay (EMSA) using 32P-labeled consensus LSF probe and nuclear extracts from Hep-pc-4 and Hep-AEG-1–14 cells. The lane numbers are described in the right panel. In lanes 4–6, Hep-AEG-1–14 nuclear extract was used.
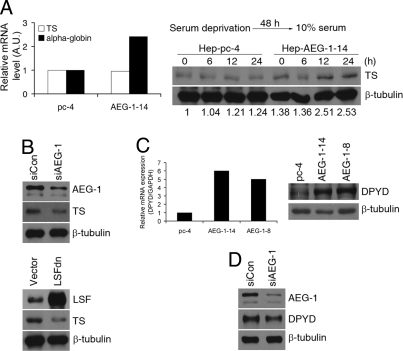
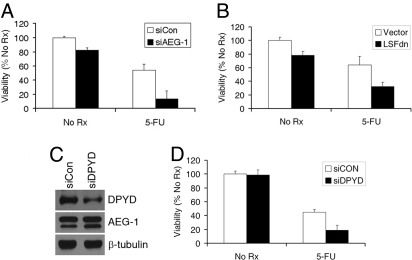
To assay the transcriptional activity of LSF on endogenous genes, we checked the expression level of 2 known LSF-downstream genes, α-globin and thymidylate synthase (TS), by Taqman real-time PCR. While we detected an approximate 3-fold increase in steady-state expression of α-globin mRNA in Hep-AEG-1–14 clone compared to the Hep-pc-4 clone, we did not detect any difference in TS gene expression between the 2 clones (Fig. 3A Left). Under steady-state condition, TS protein expression also showed very little increase in the Hep-AEG-1–14 clone compared to the Hep-pc-4 clone (Fig. S1). TS expression is cell-cycle dependent and we reasoned that analysis of steady-state expression of TS mRNA or protein might mask its cell-cycle-dependent changes. Accordingly, we serum-starved the cells for 48 h to synchronize them in G0/G1 phase and then allowed them to continue progression through the cell cycle by the addition of serum. TS protein expression was significantly increased in the Hep-AEG-1–4 clone compared to the Hep-pc-4 clone 12 h after release from serum starvation and it persisted until 24 h (Fig. 3A Right). The role of AEG-1 and LSF in regulating TS expression was confirmed by AEG-1 siRNA and LSFdn. Both AEG-1siRNA and LSFdn significantly decreased TS protein expression (Fig. 3B).
Fig. 3.
AEG-1 induces thymidylate synthase (TS) and dihydropyrimidine dehydrogenase (DPYD) expression. (A) (Left) Analysis of TS and α-globin mRNA in Hep-pc-4 (pc-4) and Hep-AEG-1–14 (AEG-1–14) clones by Taqman real-time PCR. (Right) Analysis of TS protein expression in the indicated cells by Western blot. Cells were cultured in the absence of serum for 48 h and then incubated in 10%-serum containing media for 6, 12, and 24 h. Expression of β-tubulin was used as a loading control. The numbers at the bottom represent TS/β-tubulin expression ratio for each lane when Hep-pc-4 at 0 h was considered as 1. (B) (Upper) Hep-AEG-1–14 cells were transfected with control siRNA (siCon) or AEG-1 siRNA (siAEG-1) and expression of AEG-1, TS, and β-tubulin was analyzed by Western blot. (Lower) Hep-AEG-1–14 cells were transfected with empty vector or LSFdn and expression of LSF, TS, and β-tubulin was analyzed by Western blot. (C) (Left) Analysis of expression of DPYD mRNA in Hep-pc-4 (pc-4), Hep-AEG-1–14 (AEG-1–14), and Hep-AEG-1–8 (AEG-1–8) cells by Taqman real-time PCR. (Right) Analysis of DPYD and β-tubulin protein expression in Hep-pc-4 (pc-4), Hep-AEG-1–14 (AEG-1–14), and Hep-AEG-1–8 (AEG-1–8) cells by Western blot. (D) Hep-AEG-1–14 cells were transfected with control siRNA (siCon) or AEG-1 siRNA (siAEG-1) and expression of AEG-1, DPYD, and β-tubulin was analyzed by Western blot.
In addition to LSF, the microarray analysis identified DPYD to be 25-fold up-regulated in the Hep-AEG-1–14 clone compared to the Hep-pc-4 clone. Taqman real-time PCR confirmed that in Hep-AEG-1–14 and Hep-AEG-1–8 clones DPYD expression was 6- and 5-fold higher, respectively, compared to the Hep-pc-4 clone (Fig. 3C Left). As a corollary, the expression of DPYD protein was significantly higher in Hep-AEG-1 clones compared to Hep-pc-4 cells (Fig. 3C Right). AEG-1 siRNA down-regulated DPYD protein level in the Hep-AEG-1–14 clone further confirming that DPYD is downstream of AEG-1 (3D).
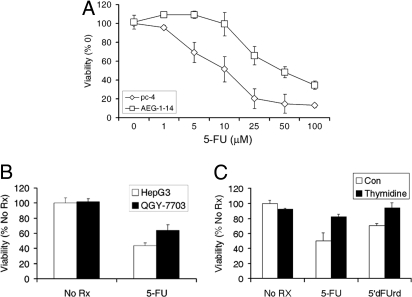
The observation that TS and DPYD, 2 important proteins determining sensitivity to 5-FU, are increased in Hep-AEG-1 clones prompted us to analyze the sensitivity of Hep-AEG-1 clones to 5-FU. Hep-AEG-1–14 clone is more resistant to 5-FU compared to the Hep-pc-4 clone (Fig. 4A). The IC50 for 5-FU shifted from approximately 10 μM in Hep-pc-4 cells to ≈50 μM in Hep-AEG-14 cells. QGY-7703 cells that express more AEG-1 also showed more resistance to 5-FU compared to HepG3 cells (Fig. 4B). 5′-deoxy-5-fluorouridine (5′dFUrd) is converted to 5-FU by the enzyme thymidine phosphorylase (TP). The 5-FU- and 5′dFUrd-mediated cell death could be rescued by addition of exogenous thymidine indicating that 5-FU-mediated killing involves TS inhibition (Fig. 4C). Inhibition of AEG-1 by siRNA or LSF by LSFdn significantly increased 5-FU-mediated killing in the Hep-AEG-1–14 clone (Fig. 5 A and B, respectively). It should be noted that AEG-1 siRNA had a more pronounced effect on 5-FU-mediated killing compared to LSFdn indicating that AEG-1 controls multiple effectors mediating 5-FU killing. A similar finding was also observed in QGY-7703 cells. We next checked the effect of DPYD siRNA on 5-FU sensitivity. DPYD siRNA significantly reduced DPYD protein level but not AEG-1 protein level indicating that DPYD is downstream of AEG-1 (Fig. 5C). DPYD siRNA also potentiated 5-FU killing both in the Hep-AEG-1–14 clone (Fig. 5D) and in QGY-7703 cells.
Fig. 4.
AEG-1 confers resistance to 5-FU. (A) Hep-pc-4 (pc-4) and Hep-AEG-1–14 (AEG-1–14) cells were treated with the indicated increasing concentrations of 5-FU (in μM). (B) HepG3 and QGY-7703 cells were treated with 5-FU (50 μM). (C) Hep-AEG-1–14 cells were treated with 5-FU (50 μM) or 5′dFUrd (10 μM) in the absence or presence of thymidine (20 μM). In A–C, cell viability was analyzed by standard MTT assay on day 7. The data represents mean ± SEM.
Fig. 5.
AEG-1-induced resistance to 5-FU is mediated by LSF and DPYD. (A) Hep-AEG-1–14 cells were transfected with either control siRNA (siCon) or AEG-1 siRNA (siAEG-1) and then treated with 5-FU (50 μM). (B) Hep-AEG-1–14 cells were transfected with either empty vector or LSFdn construct and then treated with 5-FU (50 μM). In A and B, cell viability was analyzed by standard MTT assay on day 7. The data represents mean ± SEM. (C) Hep-AEG-1–14 cells were transfected with control siRNA (siCon) or DPYD siRNA (siDPYD) and expression of AEG-1, DPYD, and β-tubulin was analyzed by Western blot. (D) Hep-AEG-1–14 cells were transfected with control siRNA (siCon) or DPYD siRNA (siDPYD) and then treated with 5-FU (50 μM). Cell viability was analyzed by standard MTT assay on day 7. The data represents mean ± SEM.
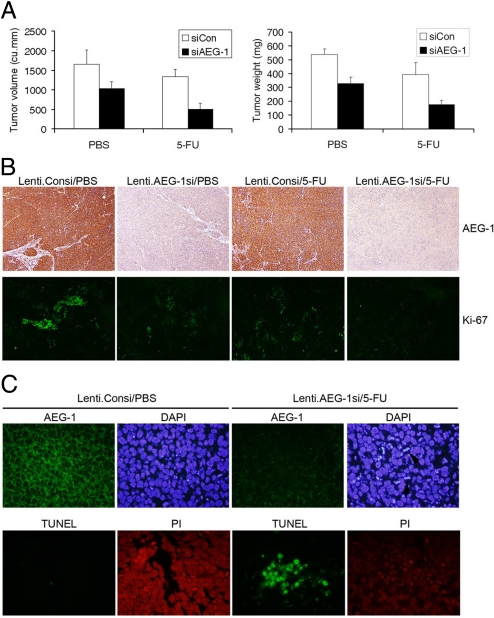
The in vitro findings were corroborated using nude mice xenograft studies. QGY-7703 cells were ex vivo transduced with a lentivirus expressing control siRNA (Lenti.Consi) or AEG-1 siRNA (Lenti.AEG-1si), and 2 days later the cells were s.c. implanted on the flanks of athymic nude mice. After establishment of the tumor (≈100 mm3 requiring about a week) the animals received i.p. injection of either PBS or 5-FU (30 mg/kg) 3 days/week for 2 weeks. The animals were maintained for another 2 weeks. At this time point the control animals had to be killed because of large tumor burden (≈2,000 mm3) according to IACUC protocol. Inhibition of AEG-1 alone resulted in significant inhibition of tumor growth (Fig. 6A). While 5-FU treatment alone also resulted in inhibition of tumor growth, the combination of AEG-1 inhibition and 5-FU treatment provided an additive effect on tumor growth inhibition versus either agent alone. The combination treatment reduced the tumor volume and tumor weight approximately 70% compared to the control animals. Analysis of tumor sections revealed that Lenti.AEG-1si treatment resulted in profound down-regulation of AEG-1 protein compared to Lenti.Consi treatment in combination with PBS or 5-FU (Fig. 6B Upper). Staining for Ki-67, a proliferation marker, showed patches of highly proliferating cells in Lenti.Consi/PBS-treated tumors (Fig. 6B Lower). Treatment with Lenti.AEG-1si/PBS significantly down-regulated Ki-67 staining. Although Lenti.Consi/5-FU treatment also down-regulated Ki-67 staining with Lenti.AEG-1si/5-FU treatment Ki-67 staining virtually disappeared indicating profound inhibition of proliferation with this combination treatment.
Fig. 6.
Combination of AEG-1 inhibition and 5-FU inhibits growth of QGY-7703 cells in athymic nude mice. (A) QGY-7703 cells were ex vivo transduced with Lenti.Consi or Lenti.AEG-1si. Two days later, the cells were s.c. implanted onto the flanks of athymic nude mice. After the establishment of the tumors the animals were treated with 5-FU for 2 weeks. Tumor volume (Left) and tumor weight (Right) was measured at the end of the study (4 weeks after the initiation of 5-FU treatment). (B) Tumor sections from the indicate treatment groups were stained for AEG-1 (Upper) or Ki-67 (Lower) as described in the materials and methods. (C) Tumor sections from the indicated treatment groups were stained for AEG-1 and nuclei were stained with DAPI (Upper). TUNEL was performed in the sections of the same treatment group and the nuclei were stained with propidium iodide (PI) (Lower).
Immunofluorescence staining for AEG-1 in the tumor samples also showed profound down-regulation of AEG-1 protein in Lenti.AEG-1si/5-FU-treated tumors compared to Lenti.Consi/PBS-treated tumors (Fig. 6C Upper). Nuclear staining with DAPI showed marked increase in cells containing fragmented DNA, indicative of apoptosis, in Lenti.AEG-1si/5-FU-treated tumors (indicated by arrows in the rightmost upper panel) compared to Lenti.Consi/PBS-treated tumors. The induction of apoptosis was further confirmed by TUNEL staining. A significant increase in TUNEL-positive cells was observed in Lenti.AEG-1si/5-FU-treated tumors compared to Lenti.Consi/PBS-treated tumors (Fig. 6C Lower).
Discussion
In this manuscript, we demonstrate that AEG-1 confers resistance to 5-FU by inducing the expression of 2 key genes LSF and DPYD. AEG-1 has been shown to facilitate invasion and metastasis (17, 18, 20, 22, 41). Additionally, it protects normal cells from serum-starvation induced apoptosis and it cooperates with Ha-ras to transform normal melanocytes and astrocytes (16, 42, 43). Thus AEG-1 contributes to tumor progression by evoking multiple changes in cellular physiology. Our present studies demonstrate that AEG-1 can directly contribute to another important feature of aggressive cancers, that is, resistance to chemotherapeutic drugs, such as 5-FU. Nude mice xenograft studies using HCC cells transduced with Lenti.AEG-1si indicated that inhibition of AEG-1 resulted in almost complete cessation of cell proliferation, documented by Ki-67 staining, demonstrating that AEG-1 is essential in activating growth promoting signals in HCC cells (Fig. 6). Indeed, AEG-1 activates a plethora of pro-proliferation signaling, such as MEK/ERK, PI3K/Akt, Wnt and NF-κB, and profound inhibition of AEG-1 by Lenti.AEG-1si, as observed in immunohistochemistry in tumor samples, might lead to cessation of cell proliferation (17, 18, 22, 41–43). Collectively these findings point to a central role of AEG-1 in HCC development and progression.
Analysis of AEG-1 expression in clinical samples from multiple tumor indications has demonstrated that AEG-1 is frequently overexpressed in a large proportion of tumor patients (17, 21, 22). We have shown that AEG-1 is overexpressed in >90% of HCC cases compared to normal liver and the expression level of AEG-1 shows direct correlation to the stages and grades of HCC (22). The high expression of AEG-1 in HCC and its ability to confer chemoresistance might explain why HCC are notoriously resistant to chemotherapy. In this context, AEG-1 expression might provide a useful marker for stratifying patients receiving chemotherapy. Moreover, targeted down-regulation of AEG-1 might be an effective means of sensitizing HCC patients to 5-FU therapy. The ability to exploit this potentially effective combinatorial approach will be contingent on developing ways of specifically inhibiting AEG-1 expression in patients. In the future, this may be achieved using small molecule inhibitors of AEG-1. Another alternative approach might involve transarterial delivery (through the hepatic artery) of Lenti.AEG-1si or nanoparticles complexed with AEG-1 siRNA in combination with chemotherapy. Our observation that Lenti.AEG-1si could reverse the resistance of QGY-7703 cells to 5-FU in a nude mouse xenograft model further validates the utility of evaluation of this approach in not only HCC patients but also in other cancer indications in which 5-FU is extensively used such as colorectal carcinoma.
DPYD plays an essential role in inactivating 5-FU by catalyzing conversion of 5-FU to fluoro-5,6-dihydrouracil (FUH2) (44). Indeed DPYD deficiency, for example, by inactivating mutation or by polymorphism, directly contributes to 5-FU toxicity in patients (45, 46). On the other hand, increased DPYD activity in HCC patient samples has been attributed to the inherent resistance of HCC to 5-FU and the relatively low level of DPYD in colorectal cancer explains the reason why colorectal cancer is sensitive to 5-FU (47, 48). Additionally, the expression of TS is higher in HCC samples compared to matched normal liver (49). Currently, no study has analyzed the expression of LSF, controlling TS expression, in HCC patients. We identify both LSF and DPYD as downstream targets of AEG-1, and we have previously shown a correlation between AEG-1 expression with LSF and DPYD expression levels in patient-derived HCC samples (22). In our microarray analysis, we did not observe an increase in other 5-FU metabolizing enzymes, such as dihydropyrimidinase and β-ureidopropionase following AEG-1 overexpression (22). Thus, the effect of AEG-1 might be selective for LSF and DPYD. We previously demonstrated that AEG-1 interacts with CBP and thus might function as a transcriptional co-activator (18). As such AEG-1 might directly induce the transcription of LSF and DPYD and analysis of the promoter region of these 2 genes will provide relevant insights into the molecular mechanism by which AEG-1 regulates their transcription. The DPYD promoter is known to be regulated by AP-1 (50). Studies in prostate cancer cells demonstrate augmentation of AP-1 activity by AEG-1 (17) that might result in the induction of DPYD. The transcriptional regulation of LSF gene is not well-defined. Our initial studies demonstrate that AEG-1 augments LSF promoter activity, and we are currently analyzing the molecular mechanism of LSF induction by AEG-1.
In summary, we have identified several potential important molecules, namely AEG-1 and LSF, the targeted inhibition of which might be exploited as an effective adjuvant therapy for HCC. Liver is an ideal organ for targeted gene therapy because in vivo administered viral-based vectors will first be sequestered by the liver. In this context, Lenti.AEG-1si in combination with 5-FU might be evaluated in transgenic animal models of HCC for preclinical evaluation as a prelude to the translation of this approach into the clinics. Successful completion of these studies could provide a rational basis for developing an effective combinatorial therapeutic approach for liver cancer.
Materials and Methods
Cell Lines, Culture Conditions, and Viability Assays.
HepG3, Hep-pc-4, Hep-AEG-1–14, Hep-AEG-1–8 and QGY-7703 human HCC cell were cultured as described (22). Cell viability was determined by standard MTT assays as described (22).
Plasmids, siRNAs, and Lentiviruses.
LSFwt and LSFdn constructs have been described before (40). All siRNAs were obtained from Santa Cruz Biotechnology. The 19-bp AEG-1 sequence used to generate AEG-1 shRNA is 5′ CAGAAGAAGAAGAACCGGA 3′. Detailed description of lentivirus vector production is described previously (51).
Transient Transfection and Luciferase Assay.
Transient transfection and luciferase assays were performed using Dual Luciferase Reporter Assay kit (Promega) as described (18). The plasmids used were: empty vector (pGL3-basic), pGL3B-WT4-E1b (luciferase reporter plasmid containing 4 tandem LSF-binding sites), or pgL3B-MT4-E1b (luciferase reporter plasmid containing 4 tandem mutated LSF-binding sites) and renilla luciferase expression plasmid for transfection control. To knock down AEG-1 or DPYD, cells were cultured for 2 days after transfection of 20 pmol of siRNA for AEG-1 or DPYD, respectively.
Preparation of Whole Cell Lysates and Western Blot Analyses.
Preparation of whole cell lysates and Western blot analyses was performed as described (22). The primary antibodies used were anti-AEG-1 (1:500), anti-LSF (1:2,000), anti-actin (1:1,000), anti-laminB (1:1,000), anti-TS (1:1,000), anti-DPYD (1:1,000), and anti-β-tubulin (1:2,000).
Extraction of Total RNA and Real-Time PCR.
Total RNA was extracted using a Qiagen mRNAeasy mini kit (Qiagen). Real time PCR was performed using ABI 7900 fast real time PCR system and Taqman gene expression assays (Applied Biosystems).
Immunofluorescence and Immunohistochemical Analyses.
Immunofluorescence studies in cells and tumor sections and immunohistochemical studies in tumor sections were performed as described (22). For immunofluorescence the primary antibodies used were: anti-LSF (1:100), anti-AEG-1 (1:400), and anti-Ki67 (1:200). For immunohistochemistry, anti-AEG-1 antibody was used at 1:200 dilution.
Preparation of Cytosolic and Nuclear Extracts and Electrophoretic Mobility Shift Assay.
Fractionation of cytosolic and nuclear extracts was performed using the nuclear extract kit (ActiveMotif), according to the manufacturer's protocol. EMSA was performed using Gel Shift Assay System (Promega) as described (41). The sequences of the wild-type probe are, sense: 5′-ANA ACT GGG TNG AGC CNG C- 3′ and antisense: 5″- G CNG GCT CNA CCC AGT TNT-3′ and that of the mutated probe are, sense: 5′-TAT GGG TNG AGA CNG C-3′ and antisense: 5′- G CNG TCT CNA CCC ATA TNT-3′.
Nude Mice Xenograft Studies.
QGY-7703 cells were transduced with either a lentivirus expressing control (scrambled) shRNA or a lentivirus expressing AEG-1 shRNA at a concentration of 2 MOI per cell for 48 h. One million cells were s.c. implanted in the flanks of athymic nude mice. 5-FU was injected 3 times/week for 2 weeks at a dose of 30 mg/kg. Tumor diameter was measured with calipers at 2 weeks later after the last 5-FU injection, and the tumor volume in mm3 was calculated by the formula: (width)2 × length/2.
TUNEL Assay.
TUNEL assay was performed using ApoAlert DNA fragmentation assay kit (Clontech) according to the manufacturer's protocol.
Statistical Analysis.
Data were represented as the mean ± standard error of mean (SEM) and analyzed for statistical significance using 1-way analysis of variance (ANOVA) followed by Newman-Keuls test as a post-hoc test. A P value of <0.05 was considered significant.
Supporting Information.
More detailed materials and methods are available in SI Materials and Methods.
Supplementary Material
Acknowledgments.
The present study was supported in part by a Goldhirsh Foundation grant and a Dana Foundation grant (to D.S.) and National Institutes of Health Grants P01 NS31492 and R01 CA035675 (to P.B.F.). D.S. is the Harrison Endowed Scholar in Cancer Research. P.B.F. holds the Thelma Newmeyer Corman Chair in Cancer Research and is a Samuel Waxman Cancer Research Foundation Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.R.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901451106/DCSupplemental.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Pang RW, et al. Biology of hepatocellular carcinoma. Ann Surg Oncol. 2008;15:962–971. doi: 10.1245/s10434-007-9730-z. [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Bru C, Bruix J. Prognosis of hepatocellular carcinoma: The BCLC staging classification. Semin Liver Dis. 1999;19:329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 4.O'Neil BH, Venook AP. Hepatocellular carcinoma: The role of the North American GI Steering Committee Hepatobiliary Task Force and the advent of effective drug therapy. Oncologist. 2007;12:1425–1432. doi: 10.1634/theoncologist.12-12-1425. [DOI] [PubMed] [Google Scholar]
- 5.Georgiades CS, Hong K, Geschwind JF. Radiofrequency ablation and chemoembolization for hepatocellular carcinoma. Cancer J. 2008;14:117–122. doi: 10.1097/PPO.0b013e31816a0fac. [DOI] [PubMed] [Google Scholar]
- 6.Yeo W, et al. A randomized phase III study of doxorubicin versus cisplatin/interferon alpha-2b/doxorubicin/fluorouracil (PIAF) combination chemotherapy for unresectable hepatocellular carcinoma. J Natl Cancer Inst. 2005;97:1532–1538. doi: 10.1093/jnci/dji315. [DOI] [PubMed] [Google Scholar]
- 7.Llovet JM, Bruix J. Systematic review of randomized trials for unresectable hepatocellular carcinoma: Chemoembolization improves survival. Hepatology. 2003;37:429–442. doi: 10.1053/jhep.2003.50047. [DOI] [PubMed] [Google Scholar]
- 8.Llovet JM, et al. Arterial embolisation or chemoembolisation versus symptomatic treatment in patients with unresectable hepatocellular carcinoma: A randomised controlled trial. Lancet. 2002;359:1734–1739. doi: 10.1016/S0140-6736(02)08649-X. [DOI] [PubMed] [Google Scholar]
- 9.Nerenstone S, Friedman M. Medical treatment of hepatocellular carcinoma. Gastroenterol Clin North Am. 1987;16:603–612. [PubMed] [Google Scholar]
- 10.Leung TW, et al. Complete pathological remission is possible with systemic combination chemotherapy for inoperable hepatocellular carcinoma. Clin Cancer Res. 1999;5:1676–1681. [PubMed] [Google Scholar]
- 11.Patt YZ, et al. Durable clinical and pathologic response of hepatocellular carcinoma to systemic and hepatic arterial administration of platinol, recombinant interferon alpha 2B, doxorubicin, and 5-fluorouracil: A communication. Am J Clin Oncol. 1999;22:209–213. doi: 10.1097/00000421-199904000-00024. [DOI] [PubMed] [Google Scholar]
- 12.Abou-Alfa GK, et al. Phase II study of sorafenib in patients with advanced hepatocellular carcinoma. J Clin Oncol. 2006;24:4293–4300. doi: 10.1200/JCO.2005.01.3441. [DOI] [PubMed] [Google Scholar]
- 13.Simpson D, Keating GM. Sorafenib: In hepatocellular carcinoma. Drugs. 2008;68:251–258. doi: 10.2165/00003495-200868020-00007. [DOI] [PubMed] [Google Scholar]
- 14.Zhu AX, et al. Phase II study of gemcitabine and oxaliplatin in combination with bevacizumab in patients with advanced hepatocellular carcinoma. J Clin Oncol. 2006;24:1898–1903. doi: 10.1200/JCO.2005.04.9130. [DOI] [PubMed] [Google Scholar]
- 15.Su ZZ, et al. Identification and cloning of human astrocyte genes displaying elevated expression after infection with HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid subtraction hybridization, RaSH. Oncogene. 2002;21:3592–3602. doi: 10.1038/sj.onc.1205445. [DOI] [PubMed] [Google Scholar]
- 16.Kang DC, et al. Cloning and characterization of HIV-1-inducible astrocyte elevated gene-1, AEG-1. Gene. 2005;353:8–15. doi: 10.1016/j.gene.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Kikuno N, et al. Knockdown of astrocyte-elevated gene-1 inhibits prostate cancer progression through upregulation of FOXO3a activity. Oncogene. 2007;26:7647–7655. doi: 10.1038/sj.onc.1210572. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar D, et al. Molecular basis of nuclear factor-kappaB activation by astrocyte elevated gene-1. Cancer Res. 2008;68:1478–1484. doi: 10.1158/0008-5472.CAN-07-6164. [DOI] [PubMed] [Google Scholar]
- 19.Emdad L, et al. Astrocyte elevated gene-1: Recent insights into a novel gene involved in tumor progression, metastasis and neurodegeneration. Pharmacol Ther. 2007;114:155–170. doi: 10.1016/j.pharmthera.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown DM, Ruoslahti E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer cell. 2004;5:365–374. doi: 10.1016/s1535-6108(04)00079-0. [DOI] [PubMed] [Google Scholar]
- 21.Li J, et al. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res. 2008;14:3319–3326. doi: 10.1158/1078-0432.CCR-07-4054. [DOI] [PubMed] [Google Scholar]
- 22.Yoo BK, et al. Astrocyte elevated gene 1 regulates hepatocellular carcinoma development and progression. J Clin Invest. 2009;119:465–477. doi: 10.1172/JCI36460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim CH, Heath C, Bertuch A, Hansen U. Specific stimulation of simian virus 40 late transcription in vitro by a cellular factor binding the simian virus 40 21-base-pair repeat promoter element. Proc Natl Acad Sci USA. 1987;84:6025–6029. doi: 10.1073/pnas.84.17.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veljkovic J, Hansen U. Lineage-specific and ubiquitous biological roles of the mammalian transcription factor LSF. Gene. 2004;343:23–40. doi: 10.1016/j.gene.2004.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim LC, Swendeman SL, Sheffery M. Molecular cloning of the alpha-globin transcription factor CP2. Mol Cell Biol. 1992;12:828–835. doi: 10.1128/mcb.12.2.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones KA, Luciw PA, Duchange N. Structural arrangements of transcription control domains within the 5′-untranslated leader regions of the HIV-1 and HIV-2 promoters. Genes Dev. 1988;2:1101–1114. doi: 10.1101/gad.2.9.1101. [DOI] [PubMed] [Google Scholar]
- 27.Wu FK, Garcia JA, Harrich D, Gaynor RB. Purification of the human immunodeficiency virus type 1 enhancer and TAR binding proteins EBP-1 and UBP-1. EMBO J. 1988;7:2117–2130. doi: 10.1002/j.1460-2075.1988.tb03051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim CG, Barnhart KM, Sheffery M. Purification of multiple erythroid cell proteins that bind the promoter of the alpha-globin gene. Mol Cell Biol. 1988;8:4270–4281. doi: 10.1128/mcb.8.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang JH, Liao WS. Induction of the mouse serum amyloid A3 gene by cytokines requires both C/EBP family proteins and a novel constitutive nuclear factor. Mol Cell Biol. 1994;14:4475–4484. doi: 10.1128/mcb.14.7.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powell CM, Rudge TL, Zhu Q, Johnson LF, Hansen U. Inhibition of the mammalian transcription factor LSF induces S-phase-dependent apoptosis by downregulating thymidylate synthase expression. EMBO J. 2000;19:4665–4675. doi: 10.1093/emboj/19.17.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casolaro V, et al. Identification and characterization of a critical CP2-binding element in the human interleukin-4 promoter. J Biol Chem. 2000;275:36605–36611. doi: 10.1074/jbc.M007086200. [DOI] [PubMed] [Google Scholar]
- 32.Murata T, Nitta M, Yasuda K. Transcription factor CP2 is essential for lens-specific expression of the chicken alphaA-crystallin gene. Genes Cells. 1998;3:443–457. doi: 10.1046/j.1365-2443.1998.00204.x. [DOI] [PubMed] [Google Scholar]
- 33.Zheng JB, Zhou YH, Maity T, Liao WS, Saunders GF. Activation of the human PAX6 gene through the exon 1 enhancer by transcription factors SEF and Sp1. Nucleic Acids Res. 2001;29:4070–4078. doi: 10.1093/nar/29.19.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundseth R, Hansen U. Activation of RNA polymerase II transcription by the specific DNA-binding protein LSF. Increased rate of binding of the basal promoter factor TFIIB. J Biol Chem. 1992;267:7845–7855. [PubMed] [Google Scholar]
- 35.Romerio F, Gabriel MN, Margolis DM. Repression of human immunodeficiency virus type 1 through the novel cooperation of human factors YY1 and LSF. J Virol. 1997;71:9375–9382. doi: 10.1128/jvi.71.12.9375-9382.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez PM, Villanueva A, Llovet JM. Systematic review: Evidence-based management of hepatocellular carcinoma–an updated analysis of randomized controlled trials. Aliment Pharmacol Ther. 2006;23:1535–1547. doi: 10.1111/j.1365-2036.2006.02932.x. [DOI] [PubMed] [Google Scholar]
- 37.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: Mechanisms of action and clinical strategies. Nature Rev. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 38.Yoshinare K, et al. Gene expression in colorectal cancer and in vitro chemosensitivity to 5-fluorouracil: a study of 88 surgical specimens. Cancer Sci. 2003;94:633–638. doi: 10.1111/j.1349-7006.2003.tb01495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oguri T, et al. The role of thymidylate synthase and dihydropyrimidine dehydrogenase in resistance to 5-fluorouracil in human lung cancer cells. Lung Cancer. 2005;49:345–351. doi: 10.1016/j.lungcan.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 40.Shirra MK, Zhu Q, Huang HC, Pallas D, Hansen U. One exon of the human LSF gene includes conserved regions involved in novel DNA-binding and dimerization motifs. Mol Cell Biol. 1994;14:5076–5087. doi: 10.1128/mcb.14.8.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emdad L, et al. Activation of the nuclear factor kappaB pathway by astrocyte elevated gene-1: Implications for tumor progression and metastasis. Cancer Res. 2006;66:1509–1516. doi: 10.1158/0008-5472.CAN-05-3029. [DOI] [PubMed] [Google Scholar]
- 42.Lee SG, et al. Astrocyte elevated gene-1 activates cell survival pathways through PI3K-Akt signaling. Oncogene. 2008;27:1114–1121. doi: 10.1038/sj.onc.1210713. [DOI] [PubMed] [Google Scholar]
- 43.Lee SG, Su ZZ, Emdad L, Sarkar D, Fisher PB. Astrocyte elevated gene-1 (AEG-1) is a target gene of oncogenic Ha-ras requiring phosphatidylinositol 3-kinase and c-Myc. Proc Natl Acad Sci USA. 2006;103:17390–17395. doi: 10.1073/pnas.0608386103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Kuilenburg AB. Dihydropyrimidine dehydrogenase and the efficacy and toxicity of 5-fluorouracil. Eur J Cancer. 2004;40:939–950. doi: 10.1016/j.ejca.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 45.Maring JG, et al. Reduced 5-FU clearance in a patient with low DPD activity due to heterozygosity for a mutant allele of the DPYD gene. Br J Cancer. 2002;86:1028–1033. doi: 10.1038/sj.bjc.6600208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diasio RB, Beavers TL, Carpenter JT. Familial deficiency of dihydropyrimidine dehydrogenase. Biochemical basis for familial pyrimidinemia and severe 5-fluorouracil-induced toxicity. J Clin Invest. 1988;81:47–51. doi: 10.1172/JCI113308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang W, Lu Z, He Y, Diasio RB. Dihydropyrimidine dehydrogenase activity in hepatocellular carcinoma: Implication in 5-fluorouracil-based chemotherapy. Clin Cancer Res. 1997;3:395–399. [PubMed] [Google Scholar]
- 48.Johnston SJ, Ridge SA, Cassidy J, McLeod HL. Regulation of dihydropyrimidine dehydrogenase in colorectal cancer. Clin Cancer Res. 1999;5:2566–2570. [PubMed] [Google Scholar]
- 49.Takahashi T, et al. Profiling of fluorouracil-related genes by microdissection technique in hepatocellular carcinoma. Hepatogastroenterology. 2007;54:1612–1616. [PubMed] [Google Scholar]
- 50.Ukon K, et al. Activator protein accelerates dihydropyrimidine dehydrogenase gene transcription in cancer cells. Cancer Res. 2005;65:1055–1062. [PubMed] [Google Scholar]
- 51.Kock N, Kasmieh R, Weissleder R, Shah K. Tumor therapy mediated by lentiviral expression of shBcl-2 and S-TRAIL. Neoplasia. 2007;9(5):435–442. doi: 10.1593/neo.07223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.