Abstract
Human ether á go-go related gene (hERG) potassium channels play a central role in cardiac repolarization where channel closing (deactivation) regulates current density during action potentials. Consequently, mutations in hERG that perturb deactivation are linked to long QT syndrome (LQTS), a catastrophic cardiac arrhythmia. Interactions between an N-terminal domain and the pore-forming “core” of the channel were proposed to regulate deactivation, however, despite its central importance the mechanistic basis for deactivation is unclear. Here, to more directly examine the mechanism for regulation of deactivation, we genetically fused N-terminal domains to fluorescent proteins and tested channel function with electrophysiology and protein interactions with Förster resonance energy transfer (FRET) spectroscopy. Truncation of hERG N-terminal regions markedly sped deactivation, and here we report that reapplication of gene fragments encoding N-terminal residues 1–135 (the “eag domain”) was sufficient to restore regulation of deactivation. We show that fluorophore-tagged eag domains and N-truncated channels were in close proximity at the plasma membrane as determined with FRET. The eag domains with Y43A or R56Q (a LQTS locus) mutations showed less regulation of deactivation and less FRET, whereas eag domains restored regulation of deactivation gating to full-length Y43A or R56Q channels and showed FRET. This study demonstrates that direct, noncovalent interactions between the eag domain and the channel core were sufficient to regulate deactivation gating, that an LQTS mutation perturbed physical interactions between the eag domain and the channel, and that small molecules such as the eag domain represent a novel method for restoring function to channels with disease-causing mutations.
Keywords: eag domain, FRET, LQTS
Human ether á go-go related gene (hERG) potassium channels were originally cloned from a human hippocampus library based on homology to mammalian ether á go-go (eag) potassium channels (1). hERG forms the major subunits of the “rapid component of the cardiac delayed rectifier potassium current” (IKr) in the heart (2, 3). The physiological role of IKr is to repolarize the late phase of cardiac action potentials (4). The clinical significance of hERG channels and IKr in the heart is emphasized by mutations in the hERG gene, which are linked to long QT syndrome (LQTS), a cardiac arrhythmia (5). hERG channels are also the targets for inhibitory compounds that cause acquired arrhythmias (6).
hERG channels exhibit unusual kinetics that help to specialize them for their role in the heart. With membrane depolarization, hERG channels activate relatively slowly and inactivate rapidly, which limits current amplitude. With subsequent membrane repolarization, hERG channels rapidly recover from inactivation and then very slowly deactivate, which gives rise to a large tail current. The resurgent tail current helps to repolarize the late phase of cardiac action potentials (2–4, 7).
The molecular basis for the slow deactivation mechanism that helps to determine the critical hERG tail current is not understood. Like other members of the Kv family of potassium channels, hERG channels are activated by membrane voltage by a mechanism that involves the voltage-sensor domains (8–12). In hERG channels, voltage-dependent gating is regulated by the large, cytoplasmic N-terminal domain (13–17). The N terminus is involved in the regulation of activation and inactivation gating (16, 17) and regulates current amplitude during the action potential (7). Previous work identified a conserved “eag domain” comprising residues 1–135 in the N-terminal region of hERG channels (13, 18) that regulates deactivation gating (13). Channels with a deletion of most of the N-terminal region (14, 15, 17) or a deletion of the eag domain (13) have deactivation kinetics that are ≈5-fold faster than the deactivation kinetics in wild-type channels, indicating that the eag domain is necessary for regulation of slow deactivation. Previous work proposed that the eag domain interacts with the “core” of the channel (13), possibly at the S4–S5 linker region (13, 17), to regulate deactivation. The critical role of the eag domain was emphasized by mutations within this domain that perturb deactivation gating and are linked to LQTS (19).
In this study, we sought to more directly determine whether the N-terminal eag domain made an interaction with the core of hERG channels using fluorescence methods that offer the advantage of determining physical proximity. We found that hERG eag domains tagged with fluorescent proteins regulated deactivation in N-truncated hERG channels with kinetics that were not measurably different from those of hERG channels with an intact N terminus. Using FRET spectroscopy, we observed that the eag domains were in close proximity to the core of hERG channels at the plasma membrane. The eag domain fragments with point mutations Y43A or R56Q had perturbed regulation of gating and decreased FRET and eag domains restored regulation of deactivation gating to full-length hERG Y43A or R56Q channels and showed FRET. Our results show that the mutations weakened an interaction between the eag domain and the rest of the channel, suggesting that a weakened interaction is a mechanistic determinant of cardiac arrhythmias. Furthermore, soluble eag domains can rescue deficient gating in disease-related (hERG R56Q) mutant channels.
Results
A Recombinant N-Terminal eag Domain Fragment Restored Slow Deactivation Gating to N-Truncated hERG Channels.
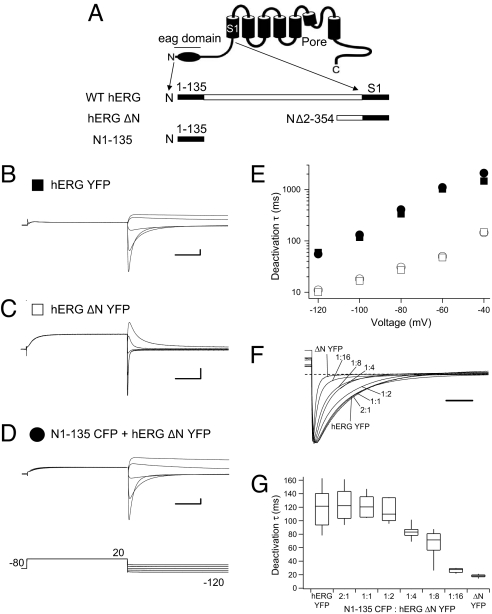
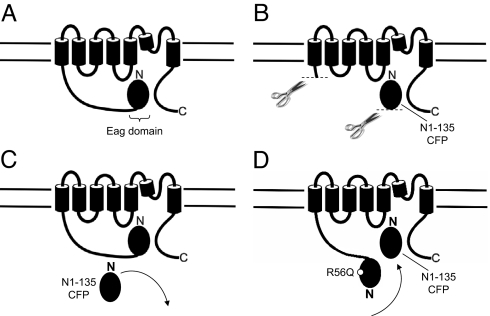
To investigate the mechanism for regulation of gating in hERG channels by the eag domain (the first 135 residues of hERG) we generated a recombinant eag domain (Fig. 1A) that we genetically fused to CFP (N1–135 CFP). We coexpressed the eag domain fragment (N1–135 CFP) with hERG channels encoding the “core” of the channel (hERG ΔN YFP) at a 2:1 ratio of RNAs in Xenopus oocytes and performed two-electrode voltage-clamp recordings. To elicit a family of deactivating traces, channels were activated by a voltage pulse to 20 mV, followed by a series of pulses to voltages from −120 mV to −40 mV in 20 mV steps (Fig. 1). To compare deactivation kinetics, currents were fit with a single exponential function and the time constant was derived (see Materials and Methods). As anticipated, currents from hERG YFP channels (Fig. 1 B and E) had deactivation kinetics that were ≈5-fold slower than the deactivation kinetics in hERG ΔN YFP channels, which have a truncated N-terminal region (Fig. 1 C and E). In cells coexpressing hERG N1–135 CFP with N-truncated hERG ΔN YFP channels, the kinetics of deactivation were markedly slower (Fig. 1D), that is, the time constants of deactivation were larger, than those for hERG ΔN YFP channels and were not measurably different from those for hERG YFP channels with an intact N-terminal region (Fig. 1 D and E and Table S1). As a negative control, we coexpressed CFP with hERG ΔN YFP channels and found that the kinetics of deactivation gating were not different from the kinetics of hERG ΔN YFP channels alone (open circles, Fig. 1E). These results indicate that a recombinant eag domain, coexpressed as a separate piece, was sufficient to recapitulate regulation of deactivation in hERG ΔN channels.
Fig. 1.
Regulation of deactivation gating by a recombinant N-terminal eag domain. (A) Schematic of hERG channel constructs. Two-electrode voltage-clamp recordings of a family of tail currents from (B) hERG YFP (closed squares), (C) hERG ΔN YFP (open squares) and (D) N1–135 CFP + hERG ΔN YFP (closed circles). Currents were elicited by the voltage pulse protocol indicated. Calibration bars, 1 μA and 250 ms. (E) Plots of the time constants (τ) derived from a single exponential fit (see Materials and Methods) to the tail currents in (B–D). Plots also include CFP + hERG ΔN YFP (open circles). Error bars are the SEM and are inside the points when not visible. n ≥ 6 for each. (F) Current traces from N-truncated hERG channels, eag domains: N-truncated hERG channels at the indicated ratios and wild-type hERG scaled to the peak at −100 mV for comparison. Zero current given by dotted line. Scale bar, 100 ms. (G) Box plot of time constants of deactivation, in which the middle line of the box is the mean, the top and bottom are the 75th and 25th percentiles, and the straight lines are the 90th and 10th percentiles. n ≥ 5 for each.
Progressive Recovery of Deactivation Gating by a Recombinant eag Domain.
We next asked whether the regulatory effect of the eag domain fragment was due the maximal association of eag domains with N-truncated hERG channels. As a first test of this we coexpressed different input ratios of hERG N1–135 CFP relative to that of hERG ΔN YFP channels (at mRNA ratios of 2:1, 1:1, 1:2, 1:4, 1:8, and 1:16) as indicated (Fig. 1 F and G). We recorded inward tail currents at −100 mV, scaled the currents for comparison, and measured the rate of decay of the tail currents using an exponential fit. We found that deactivation was slower (the time constant of the relaxation was larger) with larger input amounts of N1–135 CFP relative to hERG ΔN YFP channels (Fig. 1F). Notably, the kinetics of the tail currents reached a plateau starting at approximately a 1:2 RNA ratio of N1–135 CFP to hERG ΔN YFP and, with increased input of N1–135 CFP, the deactivation time constant did not exceed the kinetics of wild-type hERG (Fig. 1G). These results show that regulation of deactivation was a function of the ratio of the eag domain to the pore-forming core regions of the channel. Moreover, these results indicate that the regulatory effect of the eag domain was not a nonspecific slowing effect but rather was consistent with a specific interaction of the eag domain with the core of the hERG channel.
FRET Spectroscopy Showed That Fragments Encoding the eag Domain Were in Close Physical Proximity to the Core of hERG Channels at the Plasma Membrane.
To directly test for the physical proximity between the eag domain and the core of hERG channels at the plasma membrane, we used FRET spectroscopy. FRET is the transfer of light energy between a donor and acceptor fluorophore that are within a characteristic distance. For fluorescent proteins based on GFPs, FRET efficiency is steeply distance dependent and becomes negligible at distances greater than 80 Å (20, 21). Therefore, FRET reports proteins that are within close proximity to one another.
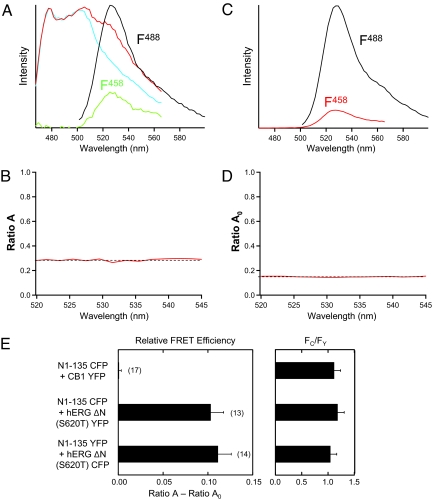
To enhance expression necessary to measure FRET in hERG channels, we introduced a well-characterized mutation in the pore region, serine 620 to threonine (S620T), which we previously showed markedly increased ionic currents and attenuated inactivation but, importantly, left deactivation intact (16, 22–24). To measure FRET, we recorded the stimulated emission of YFP (the acceptor) by proximate CFP (the donor) molecules using spectral separation analysis (see Materials and Methods). We coexpressed eag domains fused to CFP (N1–135 CFP) with hERG ΔN S620T YFP, and to examine potential effects of fluorescent protein placement we reversed the fluorophores and coexpressed eag domains fused to YFP (N1–135 YFP) with hERG ΔN S620T CFP. We confirmed that these channels were regulated by eag domains (Fig. S1 and Table S1). We recorded spectra using laser excitation and scanning confocal microscopy from the surface of oocytes. To excite CFP we used a 458-nm laser line. The emission spectra contained emission from CFP and YFP (Fig. 2A, red trace). The spectral component due to CFP was subtracted using a scaled spectrum from a separate control experiment (Fig. 2A, cyan trace), which revealed the spectral component due to YFP emission (Fig. 2A, F458, green trace). F458 contained a FRET component and a component due to direct excitation of the YFP by the 458 laser line. To solve the FRET component, we normalized F458 to the emission from direct excitation of the YFP fluorophore by the 488 laser line (Fig. 2A, F488, black trace) and calculated Ratio A (Fig. 2B). The Ratio A component caused by direct excitation of YFP (Ratio A0) was measured directly from control oocytes expressing YFP fused to hERG channels (Fig. 2 C and D). Ratio A0 was determined by measuring the direct excitation of YFP by the 458 laser line (Fig. 2C, F458, red trace) and normalizing this to the emission of YFP by the 488 laser line (Fig. 2C, F488, black trace). The FRET component of Ratio A was solved by subtracting the direct component, Ratio A0 (Fig. 2E). Values of Ratio A − Ratio A0 that were greater than zero indicate FRET.
Fig. 2.
Proximity of the soluble hERG N-terminal eag domain and the core of the hERG channel measured using FRET. (A) Spectral method for measuring FRET and determining Ratio A. Spectra (red trace) were measured from cells coexpressing N1–135 CFP + hERG ΔN S620T YFP by excitation at 458 nm. Emission spectra of CFP (cyan) measured in a control experiment from oocytes expressing N1–135 CFP. Extracted spectra (F458, green trace) is the cyan spectra subtracted from the red spectra and contains the emission of YFP. Spectra (F488, black trace) from excitation of YFP by a 488-nm laser line. Ratio A is the F458 spectra normalized to F488 spectra. (B) Plot of Ratio A (red line) versus wavelength. (C) Determination of Ratio A0. As a control, the emission spectra from oocytes expressing hERG ΔN S620T YFP channels was determined with excitation with a 458 laser line (F458, red trace) and with excitation of a 488 laser line (F488, black trace). Ratio A0 is the F458 spectra normalized to F488 spectra. (D) Plot of Ratio A0 (red line) versus emission wavelength. (E) Bar graph of Ratio A − Ratio A0, a value directly related to FRET efficiency, for the indicated channels and bar graph of ratio of CFP intensity to YFP intensity (Fc/Fy). SEM is denoted by error bars; the number of experiments (n) is indicated in parentheses.
We detected FRET in cells coexpressing N1–135 CFP with hERG ΔN S620T YFP and in cells coexpressing N1–135 YFP with hERG ΔN S620T CFP (Fig. 2E). These findings show that the eag domains were in physical proximity to N-truncated hERG channels at the plasma membrane.
In a negative control experiment coexpressing N1–135 CFP domains with cannabinoid-1 receptor YFP (CB1 YFP), a membrane protein unrelated to hERG, we did not detect FRET (Fig. 2E). The similar ratio of donor CFP to acceptor YFP (Fc/Fy) in these experiments indicates that the lack of FRET in negative controls was not simply caused by a low ratio of donor to acceptor. These results indicate that FRET was not caused by nonspecific interactions between donors and acceptors at the membrane surface; rather FRET was caused by a specific interaction between eag domains and hERG channels (Fig. 2E). Taken together, the functional restoration of regulation of deactivation by soluble N-terminal eag domains and the structural proximity of N-terminal eag domains and N-truncated hERG channels measured with FRET indicate that the N-terminal domain made a noncovalent interaction with hERG channels at the membrane surface to regulate gating. These results suggest that in full-length channels, the eag domain is likely in close physical proximity to the core of hERG channels at the membrane.
Mutations Perturb a Physical Interaction Between the N-Terminal Domain and the Channel.
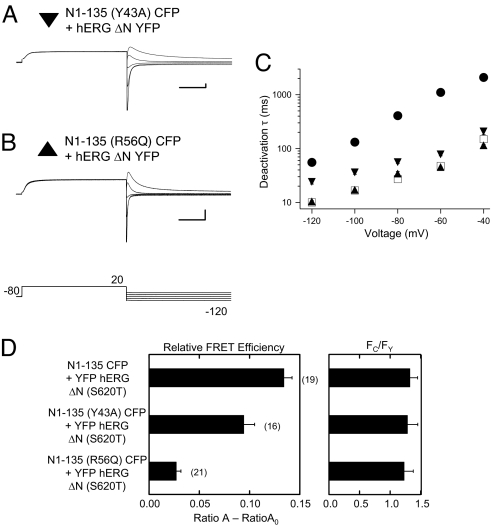
Like hERG channels with truncations of the N-terminal region, hERG channels with point mutations within the N-terminal eag domain had deactivation kinetics that were markedly faster than those in wild-type hERG channels (13, 19). To examine the mechanism for dysregulation of deactivation, we chose to investigate two hERG channels bearing mutations at single sites in the eag domain that were reported to have the most prominent speeding effect on deactivation gating; a tyrosine to alanine change at position 43 (Y43A) and an arginine to asparagine change at position 56 (R56Q). We coexpressed soluble eag domains containing either Y43A or R56Q mutations with N-truncated hERG channels at a 2:1 RNA ratio. We found that eag domain fragments with Y43A mutations partially regulated deactivation (Fig. 3 A and C) and that eag domain fragments with R56Q mutations did not measurably alter deactivation kinetics (Fig. 3 B and C). These findings indicate that the functional effects of the two mutations in the eag domain were not conferred to the channel via the covalent backbone but instead that the mutations perturbed the noncovalent interaction of the eag domain with the channel. We directly tested whether Y43A or R56Q mutations perturbed the physical proximity of the eag domain and the channel by examining FRET. To carry out these experiments, we coexpressed eag domains containing either Y43A or R56Q mutations with N-truncated hERG channels tagged with YFP at the N terminus (Fig. 3D). (These channels contained the S620T mutation and showed regulation by eag domains similar to those of their counterparts in Fig. 3 A–C and Table S1). Using similar amounts of donor to acceptor (Fc/Fy), we detected a reduction in relative FRET efficiency of 30% for eag domain fragments with Y43A (P < 0.01, analysis of variance [ANOVA]) and 80% for eag domain fragments with R56Q (P < 0.01, ANOVA) compared to that with wild-type eag domain fragments (Fig. 3D). A reduction in FRET in these experiments suggests that the mutations altered the orientation of the eag domain with respect to the channel and/or weakened an interaction of the eag domain with the channel.
Fig. 3.
Reduced regulation of deactivation gating by soluble eag domains containing point mutations. Two-electrode voltage-clamp recordings of ionic current from (A) N1–135 (Y43A) CFP fragment + hERG ΔN YFP channels (inverted, closed triangle) and (B) N1–135 (R56Q) CFP fragment + hERG ΔN YFP channels (closed triangle). Calibration bar is 2 μA and 250 ms. (C) Plot of time constant (τ) of deactivation versus voltage. Time constants for N1–135 CFP + hERG ΔN YFP (closed circles) and hERG ΔN YFP (open squares) included for comparison. Error bars are the SEM and the n ≥ 3 for each point. (D) Bar graph of relative FRET efficiency (Ratio A − Ratio A0) and ratio of CFP to YFP (Fc/Fy) for the indicated channels. SEM is denoted by error bars; the number of experiments (n) is indicated in parentheses.
Recombinant N-Terminal Domain Fragments Restore Deactivation Gating in a hERG Channel with an LQTS-Associated Mutation.
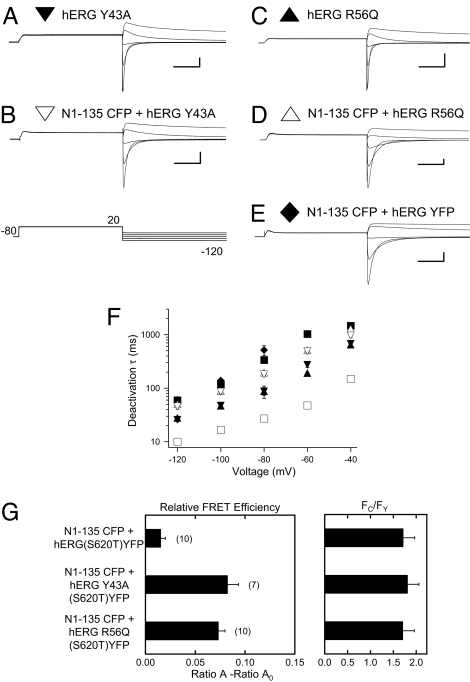
To test for a weakened interaction between the eag domain and the channel, we tested whether eag domain fragments could restore slow deactivation gating and make a physical interaction with full-length hERG Y43A or hERG R56Q channels. We first confirmed that full-length hERG channels with the Y43A or R56Q mutations had deactivation kinetics that were faster than those for wild-type hERG channels (Fig. 4A, C, and F and Table S1). When coexpressed with eag domain fragments (N1–135 CFP), we found that the deactivation kinetics of hERG Y43A (Fig. 4B) and hERG R56Q (Fig. 4D) channels were slowed and were similar to those measured for wild-type hERG channels (Fig. 4F and Table S1). As a negative control, we expressed N1–135 CFP with hERG YFP channels, and we did not detect a change in deactivation kinetics (Fig. 4 E and F) consistent with the lack of a noncovalent interaction with soluble eag domains. To directly examine the physical proximity of soluble eag domains and full-length hERG channels, we coexpressed eag domains (N1–135 CFP) with hERG YFP, hERG Y43A YFP, and hERG R56Q YFP (all channels contained the S620T mutation and showed regulation by eag domains similar to that that of their counterparts in Fig. 4 A–E and Table S1). Using similar donor-to-acceptor ratios (Fc/Fy), we detected robust FRET between N1–135 CFP and hERG Y43A YFP or N1–135 CFP and hERG R56Q YFP, but we detected very weak FRET between N1–135 CFP and hERG YFP (Fig. 4G). Together, the electrophysiological and FRET data suggest that the soluble eag domains physically supplanted the covalently attached eag domains that contained Y43A or R56Q mutations but did not supplant the wild-type eag domain. These results indicate that the Y43A or R56Q mutations weaken the noncovalent interaction between the eag domain and the channel.
Fig. 4.
Regulation of deactivation gating in a hERG channel with an LQTS mutation by soluble eag domains. Two-electrode voltage-clamp recordings of ionic currents from (A) hERG Y43A (closed inverted triangle), (B) N1–135 CFP + hERG Y43A (open inverted triangle), (C) hERG R56Q (closed triangle), (D) N1–135 CFP + hERG R56Q (open triangle), and (E) N1–135 CFP + hERG YFP (diamond) elicited with the voltage pulse as indicated. Calibration bars are 2 μA and 250 ms. (F) Plot of deactivation time constant (τ). hERG YFP (closed squares) and hERG ΔN YFP (open squares) are included as a reference. Error bars are the SEM and the n ≥ 3 for each point. (G) Bar graph of relative FRET efficiency (Ratio A − Ratio A0) and ratio of CFP to YFP (Fc/Fy) for the indicated channels. SEM is denoted by error bars; the number of experiments (n) is indicated in parentheses.
Because hERG R56Q is associated with LQTS, our findings show that a weakened interaction between the eag domain and the channel may be a determinant of heart disease. The restoration of deactivation gating in hERG R56Q channels by soluble eag domains points to one potential mechanism for restoring normal deactivation gating to a LQTS-associated mutant channel.
Discussion
Here we have shown evidence for an interaction between the eag domain (encoded by residues 1–135 of the amino terminus) of hERG channels and hERG channels lacking the N terminus. A model we propose to explain our results is that the soluble eag domains are in close physical proximity to N-truncated hERG channels at the membrane, that an interaction between these two proteins is formed, and that this interaction is likewise formed in intact hERG channels, such that the kinetics of deactivation gating are completely reconstituted in channels with N-truncated regions (Fig. 5 A and B). Mutations Y43A and R56Q in soluble eag domains partially or did not measurably alter deactivation and decreased FRET, indicating a perturbation of an interaction between the eag domain and the channel. Soluble eag domains reconstituted gating and made a physical interaction with full-length hERG channels bearing Y43A or R56Q point mutations in the eag domain. A model that fits with these findings is that the soluble eag domain can supplant the covalently attached dysfunctional eag domains but not the wild-type eag domains (Fig. 5 C and D). These results suggest that the eag domain makes a noncovalent interaction with intracellular regions of the hERG channel to directly regulate deactivation gating.
Fig. 5.
Schematic for function of eag domain. (A) Interaction of the N-terminal eag domain with other regions of the hERG channel. (B) Interaction of the eag domain with the channel is noncovalent and does not require the proximal N-terminal region. (C) Soluble eag domains do not supplant eag domain in wild-type channels. (D) Soluble eag domains supplant eag domains with point mutations, indicating that the mutation weakened an eag domain–channel interaction.
How might the eag domain regulate deactivation gating? Previous work shows that the slow deactivation kinetics in wild-type hERG channels are apparently influenced by several different regions of the protein. Mutations in transmembrane regions, including the voltage-sensor (S1–S4) domains (12, 25, 26), the S4–S5 linker region that joins the voltage sensor to the pore (17, 27), and the lower part of the S6 (28, 29) change the kinetics of deactivation. Mutations at the amino terminal end of the eag domain (13, 17), within the Per-Arnt-Sim (PAS) region of the eag domain (13, 19) and in the C-terminal regions (30, 31), accelerate deactivation gating. Can all of these observations be combined into a unifying model? The eag domain might regulate deactivation by directly interacting with voltage-sensing machinery; or, if we consider that gating in ion channels may be a series of conformational changes in different parts of the channel that are coupled to channel opening (32, 33), the eag domain might allosterically regulate deactivation by interacting with another part of the channel. (a combination of direct and allosteric mechanisms is also possible). Although speculative at this time, we propose a model in which the eag domain makes extensive interactions with intracellular S4–S5 linker and C-terminal regions of the channel to regulate gating (Fig. 5 A and B). Examining the interaction of the eag domain with the channel as a function of channel deactivation state or with channels with mutations in other regions that affect deactivation may clarify the role of the eag domain in regulation of deactivation gating.
Our data indicate that one region of hERG that is apparently not a key determinant for an interaction of the eag domain with the channel is the proximal N-terminal region. In channels reconstituted from eag domains and N-truncated channels, the proximal N-terminal region (residues 136–354) was not included (Fig. 5B). However, reconstituted channels had deactivation kinetics that were not measurably different from those in hERG channels with an intact N-terminal region. We conclude that the proximal N-terminal region is neither a molecular determinant for regulation of deactivation gating nor a determinant of the interaction of the eag domain with the internal regions of the hERG channel.
Our results here with a recombinant eag domain differ from those of a previous report where a genetically encoded eag domain fragment was reported to have no effect on channel gating (13). Instead, a purified fusion protein composed of the eag domain partially (≈50%) recovered deactivation in N-truncated hERG channels but did not change gating in hERG F29A channels. An explanation consistent with both results may be that fusion of the eag domain to a fluorescent protein in this study may have helped to stabilize the eag domain in the cellular milieu. An explanation for 50% (previous study) versus 100% (this study) regulation of deactivation may be due to the differences in methodology between the studies. One possibility is that recombinant eag domain fragments might have been expressed at a higher ratio relative to N-truncated hERG channels, or perhaps recombinant eag domains undergo key stabilizing interactions at earlier stages of biogenesis or undergo posttranslational processing important for gating that fusion protein eag domains did not undergo. The significance of full recovery of gating by the recombinant eag domain in this study is that it means that the polypeptide backbone that covalently attaches the eag domain to the channel does not appear to be required for regulation of deactivation gating and that, instead, the noncovalent interaction of the eag domain with the channel regulates deactivation gating.
Does information contained in the FRET signals address the number of eag domains necessary for gating or the relative location of the eag domain within the channel? In intact channels, there are four eag domains per channel, but the precise number of eag domains necessary to regulate gating is not known. In our experiments, eag domains with CFP or YFP coexpressed with N-truncated hERG channels with YFP or CFP, respectively, showed equal FRET efficiencies and equal regulation of gating (Fig. 2E and Fig. S1). These results are consistent with the interpretation that four eag domains interacted with N-truncated channels but are also consistent with fewer than four eag domains being sufficient to regulate gating. In experiments in which N-truncated channels were tagged with YFP at the N terminus, we detected a 24% larger relative FRET efficiency (P < 0.05, ANOVA) than when the channel was tagged with YFP at the C terminus (Figs. 2E and 3D). This result is consistent with the idea that the eag domain is more proximate to the transmembrane domains than to the C terminus; however, FRET efficiency is also dependent the relative orientation of fluorophores (34), and thus calculations of absolute distances are not meaningful.
hERG R56Q is a naturally occurring human mutation that was identified based on its link to LQTS (19), and the dysfunctional gating properties of hERG R56Q channels were shown in computational models to be pro-arrhythmogenic (35). Our data indicates that R56Q disrupts and weakens an interaction between the eag domain and the channel, indicating that perturbations of this interaction may underlie heart disease. Soluble eag domain fragments restored deactivation gating to hERG R56Q, indicating that soluble eag domains are a means for restoring the dysfunctional deactivation gating in LQTS-associated hERG channels containing mutations within the eag domain.
Materials and Methods
Molecular Biology.
The hERG expression clone was previously described (3, 36). hERG R56Q was a gift from M. Sanguinetti (University of Utah). Cannabinoid receptor YFP was a gift from W.N. Zagotta (University of Washington). hERG channels were fused directly at position 1159 (with no linker) to monomeric enhanced cyan fluorescent protein (eCFP) or monomeric citrine (gifts from R.Y. Tsien, University of California) using oligonucleotide-based mutagenesis and were sequenced using standard fluorescence-based sequencing. hERG ΔN channels (17) were also fused to monomeric citrine at position 356 for ease of cloning. Consistent with previous studies (37), fluorescent proteins fused at the C terminus of hERG did not affect the kinetics of channel gating. The eag domains were fused directly (with no linker) at amino acid 135 to mCFP or monomeric citrine. For clarity, fluorescent proteins are referred to as CFP or YFP in this study. For expression in Xenopus oocytes, mRNAs were made with the Message Machine kit (Ambion, Austin, TX).
Electrophysiology and Analysis.
For electrophysiological recordings, Xenopus oocytes were injected with the same volume of mRNA (50 nl) for each experiment and incubated for 3–14 days at 16 °C. Recordings from whole oocytes were performed with an OC-725C two-electrode voltage-clamp (Warner Instruments) connected to an ITC-18 AD converter (Instrutech, Great Neck, NY). Data were recorded using Patchmaster software (HEKA, Instrutech) and analyzed using Igor software (Wavemetrics, Lake Oswego, OR). Glass recording microelectrodes were filled with 3 M KCl. The bath (external) solution contained 4 mM KCl, 94 mM NaCl, 1 mM MgCl2, 0.3 mM CaCl2 5 mM Hepes, pH 7.4. Current relaxations with repolarizing voltage steps (deactivation) were fit with an exponential function (y = Ae−t/τ), where t is time and τ is the time constant.
Confocal Microscopy.
To measure FRET, we used a confocal microscope (Leica TCS SP2) at the National Institutes of Health (Bethesda, MD) or, in preliminary experiments, at the University of Washington Keck Center. For these experiments, cells were visualized using a Leica DMIRE 2 inverted microscope with a 10× objective with 0.3 NA. A tunable acousto-optical system (AOBS) was used to measure spectra in 5-nm emission windows. Fluorophores were excited using the 458 nm or 488 nm lines of an Ar/Kr laser. Fluorescence signals were collected from the animal pole of Xenopus oocytes after laser excitation. Background was determined from a blank area of the image and was subtracted. We used spectral separation analysis to calculate Ratio A − Ratio A0, a value proportional to FRET efficiency (34, 38, 39). Some of the measured CFP intensities in Fc/Fy ratios were reduced because of energy transfer to YFP, and were corrected using a method (40) that uses the extinction coefficients for mCFP and monomeric citrine (41, 42). Corrected Fc values were reported here (Figs. 2–4). Equations for FRET and Fc/Fy are in the SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. J. Lippincott-Schwartz for the use of a Leica confocal microscope at the National Institutes of Health. We thank N. Shuart and Dr. W. N. Zagotta for assistance with microscopy. We thank Dr. M. Rizzo, E. Gianulis and Dr. G. Patterson for helpful discussions, Dr. A. Meredith for comments on the manuscript, and L. Leung for technical assistance. This work was supported by a Scientist Development Grant, National American Heart Association (to M.C.T.), training grant T32-GM 008181 (to A.S.G.), and a gift from the Helen Pumphrey Denit Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900180106/DCSupplemental.
References
- 1.Warmke JW, Ganetzky B. A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci USA. 1994;91:3438–3442. doi: 10.1073/pnas.91.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanguinetti MC, Jiang C, Curran ME, Keating MT. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995;81:299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 3.Trudeau MC, Warmke JW, Ganetzky B, Robertson GA. HERG, a human inward rectifier in the voltage-gated potassium channel family. Science. 1995;269:92–95. doi: 10.1126/science.7604285. [DOI] [PubMed] [Google Scholar]
- 4.Sanguinetti MC, Jurkiewicz NK. Two components of cardiac delayed rectifier K+ current. Differential sensitivity to block by class III antiarrhythmic agents. J Gen Physiol. 1990;96:195–215. doi: 10.1085/jgp.96.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curran ME, et al. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 6.Roden DM. Long QT syndrome: Reduced repolrizaiton reserve and the genetic link. J Intern Med. 2006;259:59–69. doi: 10.1111/j.1365-2796.2005.01589.x. [DOI] [PubMed] [Google Scholar]
- 7.Sale H, et al. Physiological properties of hERG 1a/1b heteromeric currents and a hERG 1b-specific mutation associated with long-QT syndrome. Circ Res. 2008;103:81–95. doi: 10.1161/CIRCRESAHA.108.185249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrer T, Rupp J, Piper DR, Tristani-Firouzi M. The S4–S5 linker directly couples voltage sensor movement to the activation gate in the human ether-a-go-go-related gene (hERG) K+ channel. J Biol Chem. 2006;281:12858–12864. doi: 10.1074/jbc.M513518200. [DOI] [PubMed] [Google Scholar]
- 9.Nakajima T, et al. Voltage-shift of the current activation in HERG S4 mutation (R534C) in LQT2. Cardiovasc Res. 1999;44:283–293. doi: 10.1016/s0008-6363(99)00195-9. [DOI] [PubMed] [Google Scholar]
- 10.Piper DR, Varghese A, Sanguinetti MC, Tristani-Firouzi M. Gating currents associated with intramembrane charge displacement in HERG potassium channels. Proc Natl Acad Sci USA. 2003;100:10534–10539. doi: 10.1073/pnas.1832721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith PL, Yellen G. Fast and slow voltage sensor movements in HERG potassium channels. J Gen Physiol. 2002;119:275–293. doi: 10.1085/jgp.20028534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subbiah R, et al. Molecular basis of slow activation of the human ether-a-go-go related gene potassium channel. J Physiol. 2004;558:417–431. doi: 10.1113/jphysiol.2004.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morais Cabral JH, et al. Crystal structure and functional analysis of the HERG potassium channel N terminus: A eukaryotic PAS domain. Cell. 1998;95:649–655. doi: 10.1016/s0092-8674(00)81635-9. [DOI] [PubMed] [Google Scholar]
- 14.Schonherr R, Heinemann SH. Molecular determinants for activation and inactivation of HERG, a human inward rectifier potassium channel. J Physiol. 1996;493:635–642. doi: 10.1113/jphysiol.1996.sp021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spector PS, Curran ME, Zou A, Keating MT, Sanguinetti MC. Fast inactivation causes rectification of the IKr channel. J Gen Physiol. 1996;107:611–619. doi: 10.1085/jgp.107.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viloria CG, Barros F, Giraldez T, Gomez-Varela D, de la Pena P. Differential effects of amino-terminal distal and proximal domains in the regulation of human erg K+ channel gating. Biophys J. 2000;79:231–246. doi: 10.1016/S0006-3495(00)76286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Trudeau MC, Zappia AM, Robertson GA. Regulation of deactivation by an amino terminal domain in human ether-a-go-go-related gene potassium channels. J Gen Physiol. 1998;112:637–647. doi: 10.1085/jgp.112.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Xu J, Li M. The human delta1261 mutation of the HERG potassium channel results in a truncated protein that contains a subunit interaction domain and decreases the channel expression. J Biol Chem. 1997;272:705–708. doi: 10.1074/jbc.272.2.705. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Zou A, Splawski I, Keating MT, Sanguinetti MC. Long QT syndrome-associated mutations in the Per-Arnt-Sim (PAS) domain of HERG potassium channels accelerate channel deactivation. J Biol Chem. 1999;274:10113–10118. doi: 10.1074/jbc.274.15.10113. [DOI] [PubMed] [Google Scholar]
- 20.Miyawaki A, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 21.Patterson GH, Piston DW, Barisas BG. Förster distances between green fluorescent protein pairs. Anal Biochem. 2000;284:438–440. doi: 10.1006/abio.2000.4708. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Trudeau MC, Zappia AM, Robertson GA. Regulation of deactivation by an amino terminal domain in human ether-a-go-go-related gene potassium channels. J Gen Physiol. 1998;112:637–647. doi: 10.1085/jgp.112.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herzberg IM, Trudeau MC, Robertson GA. Transfer of rapid inactivation and E-4031 sensitivity from HERG to M-EAG Channels. J Physiol. 1998;511:3–14. doi: 10.1111/j.1469-7793.1998.003bi.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ficker E, Jarolimek W, Kiehn J, Baumann A, Brown AM. Molecular determinants of dofetilide block of HERG K+ channels. Circ Res. 1998;82:386–395. doi: 10.1161/01.res.82.3.386. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Zhang M, Jiang M, Tseng GN. Negative charges in the transmembrane domains of the HERG K channel are involved in the activation- and deactivation-gating processes. J Gen Physiol. 2003;121:599–614. doi: 10.1085/jgp.200308788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piper D, Hinz W, Tallurri C, Sanguinetti M, Tristani-Firouzi M. Regional specificity of human ether-a-go-go-related gene channel activation and inactivation gating. J Biol Chem. 2005;280:7206–7217. doi: 10.1074/jbc.M411042200. [DOI] [PubMed] [Google Scholar]
- 27.Sanguinetti MC, Xu QP. Mutations of the S4–S5 linker alter activation properties of HERG potassium channels expressed in Xenopus oocytes. J Physiol. 1999;514:667–675. doi: 10.1111/j.1469-7793.1999.667ad.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tristani-Firouzi M, Chen J, Sanguinetti MC. Interactions between S4–S5 linker and S6 transmembrane domain modulate gating of HERG K+ channels. J Biol Chem. 2002;277:18994–9000. doi: 10.1074/jbc.M200410200. [DOI] [PubMed] [Google Scholar]
- 29.Wynia-Smith SL, Gillian-Daniel AL, Satyshur KA, Robertson GA. hERG gating microdomains defined by S6 mutagenesis and molecular modeling. J Gen Physiol. 2008;132:507–520. doi: 10.1085/jgp.200810083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Owais M, Bracey K, Wray D. Role of intracellular domains in the function of the herg potassium channel. Eur Biophys J. 2009;38:569–576. doi: 10.1007/s00249-009-0408-2. [DOI] [PubMed] [Google Scholar]
- 31.Aydar E, Palmer C. Functional characterization of the C-terminus of the human ether-a-go-go-related gene K(+) channel (HERG) J Physiol. 2001;534:1–14. doi: 10.1111/j.1469-7793.2001.t01-3-00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Craven KB, Zagotta WN. Salt bridges and gating in the COOH-terminal region of HCN2 and CNGA1 channels. J Gen Physiol. 2004;124:663–677. doi: 10.1085/jgp.200409178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horrigan FT, Aldrich RW. Coupling between voltage sensor activation, Ca2+ binding and channel opening in large conductance (BK) potassium channels. J Gen Physiol. 2002;120:267–305. doi: 10.1085/jgp.20028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selvin PR. Fluorescence resonance energy transfer. Methods Enzymol. 1995;246:300–334. doi: 10.1016/0076-6879(95)46015-2. [DOI] [PubMed] [Google Scholar]
- 35.Clancy CE, Rudy Y. Cellular consequences of HERG mutations in the long QT syndrome: Precursors to sudden cardiac death. Cardiovasc Res. 2001;50:301–313. doi: 10.1016/s0008-6363(00)00293-5. [DOI] [PubMed] [Google Scholar]
- 36.Trudeau MC, Titus SA, Branchaw JL, Ganetzky B, Robertson GA. Functional analysis of a mouse brain Elk-type K+ channel. J Neurosci. 1999;19:2906–2918. doi: 10.1523/JNEUROSCI.19-08-02906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miranda P, et al. FRET with multiply labeled HERG K(+) channels as a reporter of the in vivo coarse architecture of the cytoplasmic domains. Biochim Biophys Acta. 2008;1783:1681–1699. doi: 10.1016/j.bbamcr.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Trudeau MC, Zagotta WN. Dynamics of Ca2+-calmodulin-dependent inhibition of rod cyclic nucleotide-gated channels measured by patch-clamp fluorometry. J Gen Physiol. 2004;124:211–223. doi: 10.1085/jgp.200409101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng J, Trudeau MC, Zagotta WN. Rod cyclic nucleotide-gated channels have a stoichiometry of three CNGA1 subunits and one CNGB1 subunit. Neuron. 2002;36:891–896. doi: 10.1016/s0896-6273(02)01099-1. [DOI] [PubMed] [Google Scholar]
- 40.Erickson MG, Alseikhan BA, Peterson BZ, Yue DT. Preassociation of calmodulin with voltage-gated Ca(2+) channels revealed by FRET in single living cells. Neuron. 2001;31:973–985. doi: 10.1016/s0896-6273(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 41.Rizzo MA, Springer G, Segawa K, Zipfel WR, Piston DW. Optimization of pairings and detection conditions for measurement of FRET between cyan and yellow fluorescent proteins. Microsc Micro. 2006;12:238–254. doi: 10.1017/S1431927606060235. [DOI] [PubMed] [Google Scholar]
- 42.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.