Abstract
Hepatitis E virus (HEV) is a causative agent of acute hepatitis. The crystal structure of HEV-like particles (HEV-LP) consisting of capsid protein was determined at 3.5-Å resolution. The capsid protein exhibited a quite different folding at the protruding and middle domains from the members of the families of Caliciviridae and Tombusviridae, while the shell domain shared the common folding. Tyr-288 at the 5-fold axis plays key roles in the assembly of HEV-LP, and aromatic amino acid residues are well conserved among the structurally related viruses. Mutational analyses indicated that the protruding domain is involved in the binding to the cells susceptive to HEV infection and has some neutralization epitopes. These structural and biological findings are important for understanding the molecular mechanisms of assembly and entry of HEV and also provide clues in the development of preventive and prophylactic measures for hepatitis E.
Keywords: capsid, HEV, VLP
Hepatitis E is an acute viral hepatitis caused by infection with hepatitis E virus (HEV) that is transmitted primarily by a fecal-oral route (1, 2). Numerous epidemic and sporadic cases have occurred in developing countries of Asia, the Middle East, and North Africa, where sanitary conditions are not well-maintained. Hepatitis E affects predominantly young adults, and HEV infection in pregnancy is one of the risk factors for severe disease and death (3). Recent epidemiological studies show that significant prevalence of HEV and anti-HEV antibody is found in humans and several animals worldwide, even in developed countries (4–8).
HEV is the sole member of the genus Hepevirus within the family Hepeviridae and has a 7.2-kb positive-sense RNA genome (9). Five major genotypes have been identified so far (2). The viruses in the genotypes 1 and 2 are maintained among only humans, while those in the genotypes 3 and 4 are found in pigs or wild animals (4–6). However, infections of human with genotypes 3 and 4 via zoonotic transmission or blood transfusion were reported in the developed countries, such as Japan and the United States (7, 8, 10), suggesting that hepatitis E caused by infection with genotypes 3 and 4 of HEV is an important emerging infectious disease. The viruses in the genotype 5 are of avian origin and are thought to be uninfectious to humans (11). The genomic RNA contains three ORFs (ORFs) encoding nonstructural proteins (ORF1), the viral capsid protein composed of 660 amino acids (ORF2) and a small phosphorylated protein of unidentified function (ORF3) (1, 9). The viral capsid protein induces neutralizing antibodies by its immunization (12–15) or during the course of infection (16, 17). A typical signal sequence at the N terminus and 3 potential N-glycosylation sites (Asn-X-Ser/Thr) are well-conserved in the capsid protein derived from all mammalian genotypes (18, 19), but the glycosylation status of this protein is still controversial and the biological significance of the modification in the viral life cycle remains unknown. Although propagation of HEV in the cell culture systems reported in earlier studies was not efficient (20–23), Tanaka et al. succeeded in the establishment of a persistent infection system of HEV genotype 3 in human hepatoma (PLC/PRF/5) and human carcinomic alveolar epithelial (A549) cell lines (24). However, sufficient amounts of viral particles cannot be obtained for studies of the structure, life cycle, and pathogenesis of HEV.
Electron microscopy of human stool specimens showed that HEV is a nonenveloped spherical particle with a diameter of approximately 320 Å (25). As an alternative to in vitro propagation of HEV, the baculovirus expression system opens the prospect of studying HEV capsid assembly, since HEV-like particles (HEV-LP) with protruding spikes on the surface can be formed in insect cells infected with a recombinant baculovirus expressing the capsid protein of a genotype 1 strain (26–28). Cryo-electron microscopic (cryoEM) analysis has revealed that HEV-LP is a T = 1 icosahedral particle composed of 60 copies of truncated products of ORF2 (27, 28). The HEV-LP appeared to be empty due to a lack of significant density containing RNA inside and was 270 Å in diameter (26–28), which is smaller than the diameter of the native virions. However, the HEV-LP retained the antigenicity and capsid formation of the native HEV particles.
The crystal structures of the recombinant or native T = 3 viral particles derived from structurally related mammalian and plant viruses, such as recombinant Norwalk virus (rNV; PDB accession code 1IHM) (29), San Miguel sea lion virus (SMSV; PDB accession code 2GH8) (30), the members of the family Caliciviridae, and Carnation mottle virus (CARMV; PDB accession code 1OPO) (31), a member of the family Tombusviridae, have been determined at resolutions of 3.4 Å, 3.2 Å, and 3.2 Å, respectively. In this study, to understand the structural basis on HEV, we solved the crystal structure of HEV-LP derived from a genotype 3 strain at 3.5-Å resolution and found differences in the folding of the capsid protein among these viruses. On the other hand, we found several structural similarities of shell formation. In particular, it was revealed that aromatic amino acids (Tyr-288 in the case of HEV-LP) at the 5-fold axis play a crucial role in the hydrophobic interaction required for particle formation and are well conserved among these viruses. Furthermore, mutational analyses depicted the putative cellular receptor-binding regions and epitopes for neutralizing of binding (NOB) antibodies on the 3D structure of HEV-LP. The availability of the 3D structure of HEV-LP at high resolution will provide valuable information not only for analyses of the entry and assembly of HEV, but also for the development of a vaccine for hepatitis E.
Results
Preparation of HEV-LP of a Genotype 3.
Upon infection with a recombinant baculovirus possessing a genome of the truncated capsid protein (amino acids 112–608) from a genotype 3 strain under the control of polyhedrin promoter, a large amount of HEV-LP was secreted into the culture supernatant as described in the case of HEV-LP of genotype 1 strain (26–28). The purified HEV-LP of genotype 3 was used for further structural and biological analyses.
Overall Structure of HEV-LP.
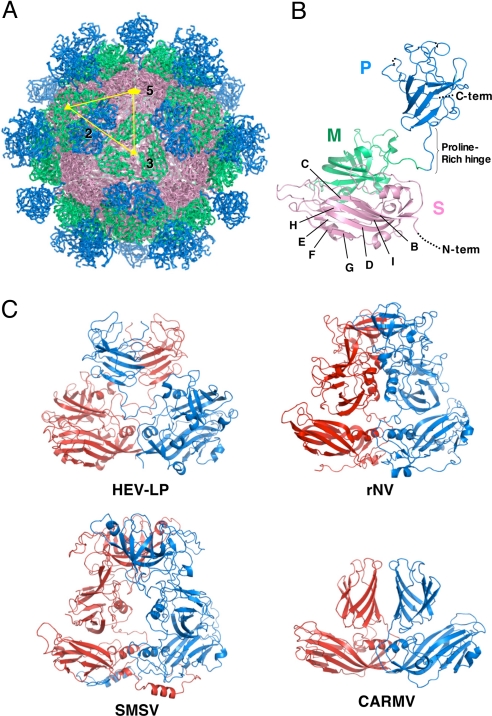
The crystal structure of HEV-LP derived from the genotype 3 strain was determined at 3.5-Å resolution by the molecular replacement method by using a cryoEM map of HEV-LP derived from the genotype 1 strain (27, 28) as an initial phasing model (Fig. 1A). As shown in the previous papers (27, 28), HEV-LP shows a T = 1 icosahedral symmetry with an external diameter of 270 Å. This particle is composed of 60 subunits of the truncated capsid proteins, forming the icosahedral 2-, 3-, and 5-fold axes. It has 30 protrusions at the 2-fold axis of the surface with large depressions at the 3- and 5-fold axes.
Fig. 1.
Crystal structure of HEV-LP and comparison of capsid protein dimers of HEV-LP, rNV, SMSV, and CARMV. The S, M, and P domains of the HEV capsid protein are indicated by pink, green, and blue, respectively. (A) HEV-LP is composed of sixty capsid subunits forming icosahedral 2-, 3-, and 5-fold axes and indicating a T = 1 symmetry. (B) The ribbon diagram of a capsid subunit of HEV-LP (PDB accession code: 2ZTN) shows P, M, and S domains at the top, middle, and bottom, respectively. The disordered regions are shown with dashed lines. The S domain shows a jerry roll-like β-barrel structure conserved in some viruses. The conserved anti-parallel β-strands are indicated (B to I). (C) The ribbon diagrams of crystal structures of capsid protein dimers of HEV-LP and those of rNV (PDB accession code 1IHM), SMSV (PDB accession code 2GH8), and CARMV (PDB accession code 1OPO) are indicated. Each capsid protein monomer is colored in red and blue.
Structure of the HEV Capsid Protein.
The truncated HEV capsid protein has 3 definite domains designated as S (shell), M (middle), and P (protruding) composed of the amino acid residues 129–319, 320–455, and 456–606, respectively (Fig. 1B). Because the N- and C-terminally truncated capsid proteins were used for the characterization, the typical signal sequence (amino acids 1–22) and following arginine-rich domain (amino acids 23–111) and the C-terminal domain removed by cleavage in insect cells (amino acids 609–660) were not determined in this study. Additionally, the amino acid residues 112–128, 486–487, 555–560, and 607–608 were disordered in this study. The S domain, which forms an internal scaffold structure of the particle, folds into a classical anti-parallel jelly roll-like β-sandwich structure with 8 β-strands (designated as B to I) and 4 short α-helices that are conserved among many viral capsids (Fig. 1B and Fig. S1) (29–33). The M domain, which is one of the characteristic domains, has a twisted anti-parallel β-barrel structure composed of 6 β-strands and 4 short α-helices. This domain is tightly associated with the S domain and located on the surface around the icosahedral 3-fold axis (Fig. 1 A and B). The M and P domains are linked with a long proline-rich hinge (amino acids 445–467). Previous studies on the structures of rNV (29) and SMSV (30) revealed that the P domains of the viruses are composed of 2 subdomains, P1 and P2, and the P2 subdomain is located as a large protrusion of the P1 subdomain (Fig. S1). In contrast, the P domain of HEV-LP is composed of a single individual domain forming a twisted anti-parallel β-sheets structure (Fig. 1B and Fig. S1), demonstrating that the capsid protein of HEV-LP has a significantly different fold from those of caliciviruses, except for the S domain. Although we have no evidence of glycosylation of HEV-LP prepared in insect cells, the HEV capsid protein has 3 potential N-glycosylation sites, Asn-137-Leu-Ser, Asn-310-Leu-Thr and Asn-562-Thr-Thr (19). In the dimer structure, the former 2 sites are mapped on the horizontal surface of the S domain, as shown in Fig. S2A. However, Asn-137 and Asn-310 are located in the interfaces of the pentamer and trimer structures, respectively (Fig. S2B and C), suggesting that, if it occurs at all, N-glycosylation in these sites may inhibit assembly of HEV-LP. Indeed, Graff et al. (18) reported that HEV carrying mutations in Asn-137 or Asn-310 to Glu lost infectivity to cells or rhesus macaques due to a defect in the virion assembly. On the other hand, Asn-562 is mapped in the central region in the top of the P dimer and exposed in the surface of HEV-LP.
The Dimer Structure at the 2-Fold Axis.
It is noteworthy that the HEV-LP dimer at the icosahedral 2-fold axis shows a crossing topology of the P versus M and S domains, while that of the other viruses with protrusions at the 2-fold axis, containing rNV, SMSV, and CARMV, exhibits a parallel topology of each domain (Fig. 1C). The flexibility of the long proline-rich hinge region between the M and P domains allows this unique topology of HEV-LP. The P domain of HEV-LP interacts with not only the P domain but also the M domain of the counterpart to stabilize the dimer structure. Despite these topological differences, the overall structure of the protrusion dimeric structure at the 2-fold axis is similar to that of rNV and SMSV. The disordered residues 486–487 and 555–560 are located in the apical region of the protrusion, suggesting that this region is flexible to take advantage of the interaction with other molecules.
Five-Fold Axis Packaging.
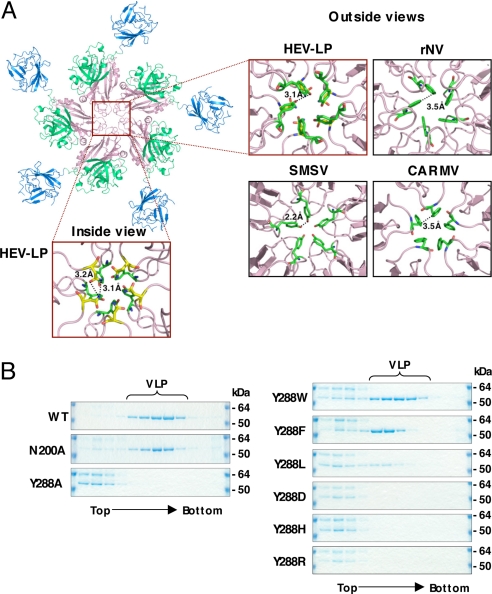
The inter-molecule-interface of the capsid pentamer at the icosahedral 5-fold axis is composed of only S domains, and these interaction regions are narrower than those of the dimer and trimer at the 2-fold and 3-fold axes, respectively (Fig. 2A), suggesting that the pentamer formation is a key step of HEV-LP assembly. There are 4 loops between the β-sheets in the S domain, designated as loops B–C (amino acids 139–152), D–E (amino acids 196–206), F–G (amino acids 236–241), and H–I (amino acids 281–296), around the center of the pentamer structure. Among them, the loops B–C and F–G are not in close proximity to the next subunits, suggesting they are not implicated in the inter-molecular interaction. In contrast, loops D–E and H–I do interact with the next subunits. In particular, the side chains of Asn-200 and Tyr-288 in loops D–E and H–I, respectively, interact with those of the next subunits, from which they are separated by a distance of approximately 3.2 Å, filling in the central pore (Fig. 2A). These observations led us to hypothesize that these amino acid residues are important for assembly and stability of the particles. To examine this hypothesis, we constructed 2 mutant capsid proteins in which Asn-200 was replaced with alanine (N200A) or Tyr-288 was replaced with alanine (Y288A), and the effect of these mutations on the particle formation was determined by a density-fractionation assay (Fig. 2B). Comparative amounts of the mutant proteins to the wild-type capsid were expressed and released into the supernatants of cells infected with the recombinant baculoviruses. N200A but not Y288A formed VLP as the wild-type, indicating that Tyr-288 plays a more crucial role in particle formation than Asn-200. The aromatic amino acids, Phe-118, Tyr-330, and Phe-145, are also found in the icosahedral 5-fold axis of rNV, SMSV, and CARMV, respectively (Fig. 2A). To examine the role of the aromatic side chain in Tyr-288 in the particle formation, a series of mutants in which Tyr-288 was replaced with tryptophan, phenylalanine, leucine, asparatic acid, histidine, or arginine (Y288W, Y288F, Y288L, Y288D, Y288H, or Y288R) were generated. All of them were expressed and released into the culture medium, as well as was the wild type. The mutants with aromatic amino acids, Y288W and Y288F, were able to form HEV-LP, whereas other mutants produced no or very few particles (Fig. 2B). These results suggest that the aromatic side chain of Tyr-288 plays a crucial role in the HEV-LP formation by shutting off the central pore of the pentamer, and that the aromatic amino acids in the positions corresponding to Tyr-288 of HEV are functionally conserved among the structurally related viruses.
Fig. 2.
Interaction of capsid protein subunits of HEV-LP around the 5-fold axis. (A) The pentamer of the capsid protein of HEV-LP. The close-up surface diagram of the 5-fold axis showed from outside and inside of HEV-LP. Amino acid residues Asn-200 and Tyr-288 are shown in yellow and green, respectively. The close-up surface diagram of the 5-fold axis showed from outside of rNV, SMSV, and CARMV. The aromatic amino acids Phe-118 of rNV, Tyr-330 of SMSV, and Phe-145 of CARMV are indicated in green. The deduced interacting atoms are connected with dashed lines, and the distances are indicated. (B) Sucrose density fractionation assay using the wild-type or mutant capsid proteins (53 kDa) in which the amino acids composing the 5-fold axis were substituted. The capsid protein composing HEV-LP was found in the 5–9th fractions from the top, while that which failed to form particles was found in the top fractions. The molecular mass of approximately 64 kDa was a non-specific protein.
Binding of HEV-LP to Cultured Cells.
The early steps of HEV entry remain unclear because of the lack of a robust cell culture system for HEV, despite recent progress in the in vitro propagation of HEV in the cell lines PLC/PRF/5 and A549 (24). HEV-LP was able to bind to several cell lines, including PLC/PRF/5 and A549 cells, but not to mouse myeloma P3 × 63Ag8U.1 (P3U1) cells (Fig. S3), suggesting that a binding assay using HEV-LP is useful to examine the first step of receptor-binding of HEV to the target cells. Among the cell lines examined, the human hepatoma cell line Huh7, exhibited a greater ability to bind to HEV-LP than the cell lines PLC/PRF/5 and A549. Therefore, Huh7 cells were used for the following binding experiments of HEV-LP.
Three-Dimensional Mapping of Epitopes for NOB Antibodies.
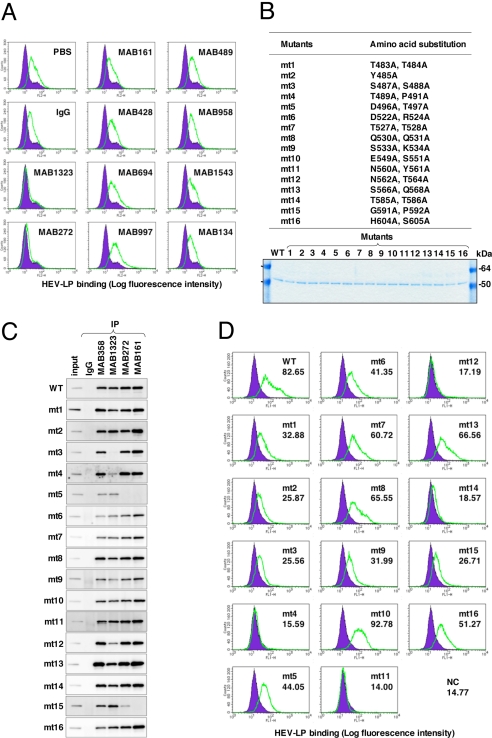
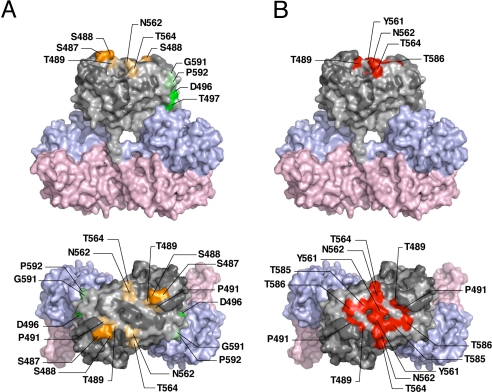
We examined the ability of the 10 newly produced anti-HEV-LP monoclonal antibodies to inhibit the binding of HEV-LP to Huh7 cells (Fig. 3A). Two of the monoclonal antibodies, MAB1323 and MAB272, exhibited NOB of HEV-LP to Huh7 cells and recognized the P domain by immunoblotting using the GST (GST)-fused HEV capsid proteins (Fig. S4). However, further truncation of the C-terminal 28 or N-terminal 24 amino acids from the GST-fused P domain abrogated the binding with the antibodies, indicating that it is difficult to determine the epitopes of the antibodies in more detail using a series of truncated mutants of the P domain. A competitive enzyme-linked immunosorbent assay (ELISA) suggested that MAB1323, MAB272, and MAB161, but not MAB358, which was used as a detector in the binding assay, recognized the same or adjacent epitopes (Fig. S5). The P domains of rNV and feline calicivirus were suggested to be involved in the binding to the receptor molecules (34–36), and we therefore hypothesized that the P domain of HEV-LP might also be involved in the cell binding. To examine this possibility, we prepared 16 HEV-LP mutants in which 1 or 2 amino acid residues at the surface of the P domain were substituted (Fig. 3B). The density fractionation assay indicated that all of the mutant proteins formed HEV-LP in the manner of the wild-type capsid protein. MAB358, which recognized an epitope on the M domain (Fig. S4), was capable of precipitating all of the mutants (Fig. 3C). MAB1323 exhibited no interaction with mt3 and a weak precipitation of mt4 and mt12. Both MAB272 and MAB161 exhibited no or weak precipitation of mt5 and mt15, whereas MAB272 but not MAB161 exhibited NOB of HEV-LP to Huh7 cells (Fig. 3 A and C). The substituted amino acids of these mutants are illustrated in the 3D structure of the capsid dimer (Fig. 4A), and these results suggest that the NOB antibodies MAB1323 and MAB272 recognize the peripheral region of the apical surface (orange) and the horizontal region (green) of the P domain above the M domain at the 3-fold axis, respectively.
Fig. 3.
Characterization of monoclonal antibodies and mutant HEV-LPs. (A) Neutralization of binding (NOB) of HEV-LP to Huh7 cells by monoclonal antibodies to HEV-LP. After preincubation of HEV-LP (10 μg/mL) with each of the monoclonal antibodies (20 μg/mL) for 1 h at 37°C, the mixture was inoculated into Huh7 cells and incubated for 1 h at 4°C. HEV-LP (lined area) bound to cells was detected by flow cytometry. The filled area indicates mock-incubated cells. (B) Construction of HEV-LP mutants. Sixteen HEV-LP mutants, in which the surface amino acid residues of the P domain were substituted, were constructed. The protein bands of 100 ng each of the purified wild-type and mutant HEV-LPs were visualized by Coomassie brilliant blue staining after SDS/PAGE. (C) Reactivities of NOB antibodies with the mutant HEV-LPs. Immunoprecipitation analyses of a series of HEV-LPs by NOB (MAB1323 and MAB272) or non-NOB antibodies (MAB358 and MAB161). Immunoprecipitated HEV-LPs were detected by an anti-HEV capsid rabbit polyclonal antibody. (D) Binding capability of the mutant HEV-LPs to Huh7 cells. Wild-type or mutant HEV-LPs (10 μg/mL) were incubated with Huh7 cells for 1 h at 4°C, and then HEV-LP (lined area) bound to cells was detected by flow cytometry. The filled area indicates mock-incubated cells. The MFI is shown in each panel.
Fig. 4.
Amino acid residues involved in the recognition by NOB antibodies and in the binding to Huh7 cells. Surface diagrams of the capsid protein dimer from a lateral (Upper) or top (Lower) view. (A) Amino acids in HEV-LP involved in the complete loss (deep color) or reduction (light color) of reactivity to MAB1323 and MAB272 are shown in orange and green, respectively. (B) Amino acids in HEV-LP responsible for binding to Huh7 cells are shown in red. Domains S, M, and P are colored pink, blue and gray, respectively. The substitutions in the P domain of HEV-LP that exhibited no effect on the reactivity with NOB antibodies or the binding to Huh7 cells are shown in dark gray.
Three-Dimensional Mapping of a Region Crucial for Binding to the Target Cells.
To determine the region important for binding to the cell surface, the mutant HEV-LPs substituted into the P domain were also used in the assay of binding to Huh7 cells (Fig. 3D). The wild-type HEV-LP bound to Huh7 cells with a geographic mean fluorescence intensity (MFI) of 82.65. Among the mutants examined, mt4, mt11, mt12, and mt14 exhibited significantly low MFI values of less than 20. Similar results were obtained using A549 cells (Fig. S6). The amino acid residues required for cell binding were mapped in the central flexible region of the apical surface as shown in Fig. 4B (red). This region is partially overlapped with epitopes of MAB1323 (Fig. 4A) and other neutralizing antibodies reported by Schofield et al. (16) as shown in Fig. S7. These results suggested that the apical center region of the P domain is involved in the association with not-yet-identified cellular receptor(s).
Discussion
The expression of the truncated HEV capsid protein (amino acids 112–608) in insect cells resulted in assembly of HEV-LP, which retains an antigenicity similar to that of the native HEV particles (26, 37). This particle with a T = 1 symmetry has a diameter of 270 Å, which is smaller than the 320-Å diameter of the native virion detected in the fecal specimens of patients (25). It has been reported that the interior cavity of HEV-LP is too small to accommodate a viral RNA of 7.8 kb in length (28) and that the particles show no evidence of nucleotide contents (26, 28). Therefore, native HEV particles are suggested to be composed of a larger number and/or a larger size of capsid proteins than HEV-LP. In some cases of plant viruses with a T = 3 symmetry, the capsid proteins assembled into particles with a T = 1 symmetry by deletion of the N-terminal basic region (38, 39) or amino acid substitutions either in the N-terminal region or in the linker domain between the N-terminal region and S domain (39), suggesting that the N-terminal basic region plays an important role in switching of the transition from T = 3 to T = 1 symmetry. In addition, expression of the NV capsid protein in insect cells resulted in production of not only T = 3 large particles but also small particles thought to have the T = 1 symmetry (40). Based on many similarities of the capsid structures and their packaging of structurally related viruses, the native HEV particles are suggested to possess a T = 3 surface lattice. The flexibility of the proline-rich hinge linking the M and P domains could allow the capsid protein dimer to switch conformations between the A/B and C/C subunits found in T = 3 viruses. Although structure of the native HEV may be slightly different from that of the HEV-LP, the data obtained in this study by using HEV-LP should provide useful information to understand the structure of viral particle, life cycle, and pathogenesis of HEV. The S domain shares the jellyroll fold with some other icosahedral viruses (29–33). It was found that the capsid proteins with substitutions of Tyr-288 positioned at the center of the pentamer structure built in interS domain-interaction failed to assemble into HEV-LP. Alignment analysis of amino acid sequences using data available in GeneBank showed that Tyr-288 is completely conserved within 5 genotypes of HEV. Furthermore, residues corresponding to Tyr-288 of the HEV capsid protein are found in the structures of rNV (Phe-118), SMSV (Tyr-330), and CARMV (Phe-145), although the positions of these aromatic residues are different. Tyr-288 of HEV and Tyr-330 of SMSV located in the H-I loop and Phe-110 of rNV in the D–E loop are exposed at the outside surface of the particles, whereas Phe-145 of CARMV located in the D–E loop is exposed at the interior of the particle. These data suggest that the aromatic side chains of these residues are involved in hydrophobic interactions with those of the next subunits, assuring stable assembly of the particles. During entry into cells, rearrangement of the virion structure is required for release of the genome from the shell. However, the entry and uncoating mechanisms of HEV remain unknown. Because the center of the pentamer is the thinnest region of the particle and takes a channel-like structure (28), this region might also be important for uncoating and release of the viral RNA. It has been proposed that the 5-fold axis of poliovirus is involved in the genomic RNA translocation via conformational change of the virion initiated by binding to the receptor molecules (41, 42).
The first step in viral entry into a target cell is binding to the cellular receptors. The human hepatoma PLC/PRF/5 and lung epithelial A549 cell lines, which are highly susceptible to persistent HEV-infection (24), are likely to express functional HEV receptors on the cell surface. However, HEV-LP had reduced binding to these cells compared to the other cell lines examined. Therefore, the human hepatoma cell line Huh7, which also exhibited a susceptibility to HEV infection (13, 18) and readily bound to HEV-LP, was mainly used in this study. It has been reported that the P domains of noroviruses and the feline calicivirus were involved in the binding to the putative receptors, histo-blood antigens (35, 36) and the feline junctional adhesion molecule (34), respectively. The peptide of the HEV capsid protein (amino acids 368–606), which consists of a part of the M and an entire P domain, was shown to be capable of binding to several cell lines (13), suggesting that the P domain of HEV is also involved in the binding to the cell receptors. Indeed, the mutational analyses in this study indicated that the central flexible region of the top of the P domain of HEV-LP plays a crucial role for binding to Huh7 and A549 cells. This is consistent with a recent study by Graff et al. in which an N562Q mutant of HEV lost infectivity to culture cells and rhesus macaques despite the production of viral particles (18). Interestingly, a possible N-glycosylation site, Asn-562-Thr-Thr, is mapped in this region. N-glycosylation is an unusual posttranslational modification for nonenveloped viruses, except for rotaviruses (43). The mutant capsid mt12, which has substitutions of Asn-562 and Thr-564 to alanine, exhibited the same migration as the wild-type protein in SDS/PAGE, suggesting that the HEV-LP produced in insect cells was not glycosylated at Asn-562. Lack of N-glycosylation in the capsid protein has also been reported in mammalian cells infected with HEV (18), whereas some portion of the capsid protein was glycosylated and transported to the cell surface upon overexpression in mammalian cells (19). N-glycosylation of the HEV capsid at Asn-562 may have a negative effect on the receptor-binding, whereas it may play a positive role in other functions, including pathogenesis. The biological significance of the glycosylation of HEV capsid protein remains to be studied.
Although there is currently a lack of sensitive and reliable assays to determine the neutralizing activity of anti-HEV antibodies, the assay of NOB of HEV-LP binding to the target cells is thought to be a suitable alternative method. Measurement of the reactivity of a panel of mutant HEV-LPs revealed that the epitopes of MAB1323 and MAB272 antibodies are mapped in the peripheral region of the apical surface and the horizontal region of the P domain dimer, respectively. These results further support the notion that the P domain of HEV-LP is important for the binding to cells. MAB1323 is suggested to directly inhibit the interaction between HEV-LP and cellular receptors through binding to the apical surface, whereas MAB272 may have an allosteric effect, inducing conformational change of the P domain through binding to the horizontal region. A number of monoclonal antibodies are capable of neutralizing in vitro and in vivo infection of HEV (12–17), and many of them recognize conformational epitopes of the capsid protein, as seen in the MAB1323 and MAB272 antibodies prepared in this study. Monoclonal antibodies against linear epitopes located in amino acids 578–607 of a genotype 1 capsid protein (16) were overlapped with a part of the putative receptor-binding domain and the epitope of MAB272, supporting the data of the present study. On the other hand, monoclonal antibodies against the linear epitopes located in amino acids 423–438 and amino acids 423–443 in the M domain of a genotype 1 capsid protein neutralized binding of a peptide derived from the capsid protein to cells and HEV-infection (13), suggesting the importance of the M domain in the binding step.
In summary, we have determined the crystal structure of HEV-LP produced in insect cells and demonstrated its structural characteristics in comparison with the structurally related animal and plant viruses. This study will provide useful information for elucidation of the molecular mechanisms of HEV-life cycles and for the development of prophylactic and therapeutic measures for hepatitis E.
Materials and Methods
Expression, Purification, and Crystallization of HEV-LP.
The recombinant baculovirus encoding the ORF2 of the HEV genotype 3, 2712 strain was expressed in insect cells. HEV-LP was purified as described previously (28) and crystallized by the hanging-drop vapor-diffusion method. Details are reported in SI Materials and Methods.
Data Collection and Phase Determination.
x-ray diffraction data were collected at 100 K on beamlines BL17A at the Photon Factory (KEK). The statistics of X-ray diffraction data collection are summarized in Table 1. The solved 3D structure of HEV-LP was submitted to the Protein Data Bank under the PDB accession code of 2ZTN. Details are reported in SI Materials and Methods.
Table 1.
Data collection and processing statistics for HEV-LP
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c, Å | 336.8, 349.4, 359.5 |
| X-ray wavelength, Å | 1.0000 |
| Resolution, Å | 50–3.55 (3.68–3.55) |
| Rmerge* | 0.131 (0.498) |
| I/σI | 9.8 (3.2) |
| Completeness, % | 99.9 (99.8) |
| Redundancy | 5.6 (5.2) |
| Refinement | |
| Resolution range, Å | 20–3.56 |
| No. reflections | 494,466 |
| Rwork/Rfree | 30.5/30.9 |
| No. atoms | |
| Protein | 215,400 |
| B factors | |
| Protein | 94.9 |
| rmsd | |
| Bond length, Å | 0.009 |
| Bond angle, ° | 1.355 |
Values in square brackets refer to the highest-resolution shell.
*Rmerge = ΣhklΣi|I(hkl)i − 〈I(hkl)〉|/ΣhklI(hkl), where I(hkl)i is the ith measurement of the intensity of reflection hkl and 〈I(hkl)〉 is the mean intensity of reflection hkl.
Supplementary Material
Acknowledgments.
We thank H. Murase for her secretarial work and the staff of SPring-8 BL44XU beamline and synchrotron beamline NW-17A of the Photon Factory, High Energy Accelerator Research Organization for their assistance with the data collection. This work was supported in part by grants-in-aid from the Research and Development Program for New Bio-industry Initiatives of Bio-oriented Technology Research Advancement Institution (BRAIN) and the Foundation for Research Collaboration Center on Emerging and Re-emerging Infections.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2ZTN).
This article contains supporting information online at www.pnas.org/cgi/content/full/0903699106/DCSupplemental.
References
- 1.Panda SK, Thakral D, Rehman S. Hepatitis E virus. Rev Med Virol. 2007;17:151–180. doi: 10.1002/rmv.522. [DOI] [PubMed] [Google Scholar]
- 2.Purcell RH, Emerson SU. Hepatitis E: An emerging awareness of an old disease. J Hepatol. 2008;48:494–503. doi: 10.1016/j.jhep.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Navaneethan U, Al Mohajer M, Shata MT. Hepatitis E and pregnancy: Understanding the pathogenesis. Liver Int. 2008;28:1190–1199. doi: 10.1111/j.1478-3231.2008.01840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng XJ, et al. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci USA. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonoda H, et al. Prevalence of hepatitis E virus (HEV) Infection in wild boars and deer and genetic identification of a genotype 3 HEV from a boar in Japan. J Clin Microbiol. 2004;42:5371–5374. doi: 10.1128/JCM.42.11.5371-5374.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamoto H. Genetic variability and evolution of hepatitis E virus. Virus Res. 2007;127:216–228. doi: 10.1016/j.virusres.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Li TC, et al. Hepatitis E virus transmission from wild boar meat. Emerg Infect Dis. 2005;11:1958–1960. doi: 10.3201/eid1112.051041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yazaki Y, et al. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J Gen Virol. 2003;84:2351–2357. doi: 10.1099/vir.0.19242-0. [DOI] [PubMed] [Google Scholar]
- 9.Tam AW, et al. Hepatitis E virus (HEV): Molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsubayashi K, et al. Transfusion-transmitted hepatitis E caused by apparently indigenous hepatitis E virus strain in Hokkaido, Japan. Transfusion. 2004;44:934–940. doi: 10.1111/j.1537-2995.2004.03300.x. [DOI] [PubMed] [Google Scholar]
- 11.Huang FF, et al. Determination and analysis of the complete genomic sequence of avian hepatitis E virus (avian HEV) and attempts to infect rhesus monkeys with avian HEV. J Gen Virol. 2004;85:1609–1618. doi: 10.1099/vir.0.79841-0. [DOI] [PubMed] [Google Scholar]
- 12.Emerson SU, et al. Putative neutralization epitopes and broad cross-genotype neutralization of Hepatitis E virus confirmed by a quantitative cell-culture assay. J Gen Virol. 2006;87:697–704. doi: 10.1099/vir.0.81545-0. [DOI] [PubMed] [Google Scholar]
- 13.He S, et al. Putative receptor-binding sites of hepatitis E virus. J Gen Virol. 2008;89:245–249. doi: 10.1099/vir.0.83308-0. [DOI] [PubMed] [Google Scholar]
- 14.Meng J, et al. Identification and characterization of the neutralization epitope(s) of the hepatitis E virus. Virology. 2001;288:203–211. doi: 10.1006/viro.2001.1093. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi M, et al. Production of monoclonal antibodies against hepatitis E virus capsid protein and evaluation of their neutralizing activity in a cell culture system. Arch Virol. 2008;153:657–666. doi: 10.1007/s00705-008-0045-6. [DOI] [PubMed] [Google Scholar]
- 16.Schofield DJ, Glamann J, Emerson SU, Purcell RH. Identification by phage display and characterization of two neutralizing chimpanzee monoclonal antibodies to the hepatitis E virus capsid protein. J Virol. 2000;74:5548–5555. doi: 10.1128/jvi.74.12.5548-5555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schofield DJ, Purcell RH, Nguyen HT, Emerson SU. Monoclonal antibodies that neutralize HEV recognize an antigenic site at the carboxyterminus of an ORF2 protein vaccine. Vaccine. 2003;22:257–267. doi: 10.1016/j.vaccine.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Graff J, et al. Mutations within potential glycosylation sites in the capsid protein of hepatitis E virus prevent the formation of infectious virus particles. J Virol. 2008;82:1185–1194. doi: 10.1128/JVI.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zafrullah M, Ozdener MH, Kumar R, Panda SK, Jameel S. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J Virol. 1999;73:4074–4082. doi: 10.1128/jvi.73.5.4074-4082.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang R, et al. Cell culture of sporadic hepatitis E virus in China. Clin Diagn Lab Immunol. 1999;6:729–733. doi: 10.1128/cdli.6.5.729-733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kazachkov Yu A, et al. Hepatitis E virus in cultivated cells. Arch Virol. 1992;127:399–402. doi: 10.1007/BF01309603. [DOI] [PubMed] [Google Scholar]
- 22.Meng J, Dubreuil P, Pillot J. A new PCR-based seroneutralization assay in cell culture for diagnosis of hepatitis E. J Clin Microbiol. 1997;35:1373–1377. doi: 10.1128/jcm.35.6.1373-1377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tam AW, et al. In vitro infection and replication of hepatitis E virus in primary cynomolgus macaque hepatocytes. Virology. 1997;238:94–102. doi: 10.1006/viro.1997.8817. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka T, Takahashi M, Kusano E, Okamoto H. Development and evaluation of an efficient cell-culture system for Hepatitis E virus. J Gen Virol. 2007;88:903–911. doi: 10.1099/vir.0.82535-0. [DOI] [PubMed] [Google Scholar]
- 25.Bradley D, et al. Aetiological agent of enterically transmitted non-A, non-B hepatitis. J Gen Virol. 1988;69:731–738. doi: 10.1099/0022-1317-69-3-731. [DOI] [PubMed] [Google Scholar]
- 26.Li TC, et al. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J Virol. 1997;71:7207–7213. doi: 10.1128/jvi.71.10.7207-7213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li TC, et al. Essential elements of the capsid protein for self-assembly into empty virus-like particles of hepatitis E virus. J Virol. 2005;79:12999–13006. doi: 10.1128/JVI.79.20.12999-13006.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing L, et al. Recombinant hepatitis E capsid protein self-assembles into a dual-domain T = 1 particle presenting native virus epitopes. Virology. 1999;265:35–45. doi: 10.1006/viro.1999.0005. [DOI] [PubMed] [Google Scholar]
- 29.Prasad BV, et al. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286:287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- 30.Chen R, Neill JD, Estes MK, Prasad BV. X-ray structure of a native calicivirus: Structural insights into antigenic diversity and host specificity. Proc Natl Acad Sci USA. 2006;103:8048–8053. doi: 10.1073/pnas.0600421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgunova E, et al. The atomic structure of Carnation Mottle Virus capsid protein. FEBS Lett. 1994;338:267–271. doi: 10.1016/0014-5793(94)80281-5. [DOI] [PubMed] [Google Scholar]
- 32.Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 33.Tsao J, et al. The three-dimensional structure of canine parvovirus and its functional implications. Science. 1991;251:1456–1464. doi: 10.1126/science.2006420. [DOI] [PubMed] [Google Scholar]
- 34.Bhella D, Gatherer D, Chaudhry Y, Pink R, Goodfellow IG. Structural insights into calicivirus attachment and uncoating. J Virol. 2008;82:8051–8058. doi: 10.1128/JVI.00550-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bu W, et al. Structural basis for the receptor binding specificity of Norwalk virus. J Virol. 2008;82:5340–5347. doi: 10.1128/JVI.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi JM, Hutson AM, Estes MK, Prasad BV. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc Natl Acad Sci USA. 2008;105:9175–9180. doi: 10.1073/pnas.0803275105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li TC, et al. Protection of cynomolgus monkeys against HEV infection by oral administration of recombinant hepatitis E virus-like particles. Vaccine. 2004;22:370–377. doi: 10.1016/j.vaccine.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Hsu C, et al. Characterization of polymorphism displayed by the coat protein mutants of tomato bushy stunt virus. Virology. 2006;349:222–229. doi: 10.1016/j.virol.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 39.Kakani K, Reade R, Katpally U, Smith T, Rochon D. Induction of particle polymorphism by cucumber necrosis virus coat protein mutants in vivo. J Virol. 2008;82:1547–1557. doi: 10.1128/JVI.01976-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White LJ, Hardy ME, Estes MK. Biochemical characterization of a smaller form of recombinant Norwalk virus capsids assembled in insect cells. J Virol. 1997;71:8066–8072. doi: 10.1128/jvi.71.10.8066-8072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belnap DM, et al. Molecular tectonic model of virus structural transitions: The putative cell entry states of poliovirus. J Virol. 2000;74:1342–1354. doi: 10.1128/jvi.74.3.1342-1354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bubeck D, Filman DJ, Hogle JM. Cryo-electron microscopy reconstruction of a poliovirus-receptor-membrane complex. Nat Struct Mol Biol. 2005;12:615–618. doi: 10.1038/nsmb955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jayaram H, Estes MK, Prasad BV. Emerging themes in rotavirus cell entry, genome organization, transcription and replication. Virus Res. 2004;101:67–81. doi: 10.1016/j.virusres.2003.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.