Abstract
The actions of Escherichia coli DNA Polymerase IV (Pol IV) in mutagenesis are managed by its interaction with the β sliding clamp. In the structure reported by Bunting et al. [EMBO J (2003) 22:5883–5892], the C-tail of Pol IV contacts a hydrophobic cleft on the clamp, while residues V303–P305 reach over the dimer interface to contact the rim of the adjacent clamp protomer. Using mutant forms of these proteins impaired for either the rim or the cleft contacts, we determined that the rim contact was dispensable for Pol IV replication in vitro, while the cleft contact was absolutely required. Using an in vitro assay to monitor Pol III*-Pol IV switching, we determined that a single cleft on the clamp was sufficient to support the switch, and that both the rim and cleft contacts were required. Results from genetic experiments support a role for the cleft and rim contacts in Pol IV function in vivo. Taken together, our findings challenge the toolbelt model and suggest instead that Pol IV contacts the rim of the clamp adjacent to the cleft that is bound by Pol III* before gaining control of the same cleft that is bound by Pol III*.
Keywords: mutagenesis, toolbelt, translesion DNA synthesis, Pol IV, DinB
Endogenous and exogenous agents continuously damage cellular DNA. As a result, faithful duplication of an organism's genetic information requires both high fidelity DNA polymerases (Pols), as well as a multitude of DNA repair and DNA damage tolerance mechanisms (reviewed in ref. 1). Generally speaking, repair functions act to maintain the fidelity of the genome by removing lesions in the DNA before its replication and by correcting replication errors (reviewed in ref. 1). In contrast, damage tolerance pathways, such as translesion DNA synthesis (TLS), do not repair DNA damage, but rather catalyze replication over damaged bases that cannot for whatever reason be repaired. Due to their high fidelity, replicative Pols are usually unable to catalyze TLS. Therefore, specialized Pols (TLS Pols) capable of replicating imperfect DNA substrates are required for most TLS (2, 3). Because most TLS Pols possess an open active site, and lack a detectable proofreading activity, they display relatively low fidelity. Thus, the actions of these Pols must be tightly regulated to guard against introducing gratuitous mutations.
Although multiple mechanisms likely act to coordinate the actions of replicative and TLS Pols, 1 mechanism that has received considerable attention in recent years pertains to the role played by sliding clamp proteins (4–6). The Esherichia coli β sliding clamp, which is encoded by the dnaN gene, is the founding member of the ubiquitous sliding clamp family of proteins (7). The E. coli β clamp was originally discovered based on its ability to tether the replicative Pol, Pol III holoenzyme (Pol III HE) to the DNA, thereby increasing its processivity (7). Pol III HE is comprised of 3 subassemblies: core (αεθ), DnaX clamp loader (τ2γδδ'ψχ), and β (reviewed in ref. 8). The α subunit of core tethers Pol III HE to the clamp and catalyzes polymerization, while ε acts as the 3′→5′ exonuclease proofreader. DnaX acts to load β clamps onto primed DNA. In addition, the τ2 subunit of DnaX tethers 2 cores for simultaneous replication of leading and lagging strands. The form of Pol III HE lacking β clamp is called Pol III*. In addition to interaction with Pol III*, it is now clear that the β clamp also interacts with each of the other 4 E. coli Pols, as well as several other proteins involved in DNA replication, DNA repair, and cell cycle progression (9). A similar situation is true for many eukaryotic Pols (10). Taken together, these findings suggest that sliding clamps act like traffic cops to coordinate the actions of different partners on DNA.
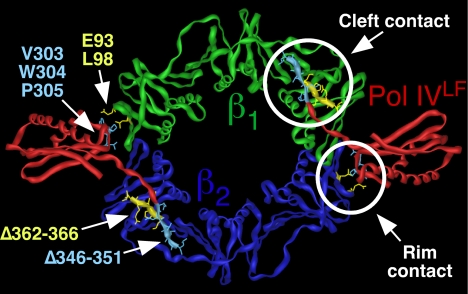
One contact site between these partners and the clamp involves a clamp-binding motif (CBM) present in the partners and a hydrophobic cleft located near the C-terminal tail of each clamp protomer (11, 12). Based on a crystal structure of the β clamp-Pol IV little finger domain (Pol IVLF; residues 243–351) complex (13), Pol IV contacts at least 2 distinct surfaces of the clamp: The Pol IV CBM (residues 346–351) interacts with the hydrophobic cleft of β (cleft, or C contact), while residues 303VWP305 of Pol IV reach over the dimer interface to contact residues E93 and L98 on the rim (rim, or R contact) of the adjacent clamp protomer (see Fig. 1). Likewise, Pol II, Pol III, and Pol V also contact noncleft surfaces (14–16). However, the roles of these contacts in replication and Pol switching have thus far received very little attention.
Fig. 1.
Structure of the Pol IVLF-β clamp complex depicting the cleft and rim contacts. The Pol IVLF domain is shown in red, and the clamp is shown in green and blue. Positions of mutations affecting the rim or the cleft contacts between the clamp and Pol IVLF are indicated in yellow and blue, respectively. This figure was generated using imol and PDB coordinates 1UNN (13).
A centrally important yet unanswered question in the field pertains to how different partners exchange with each other, or switch, on a clamp assembled on DNA. This is a particularly important question in light of the fact that clamp proteins, and the events that they manage, are so widely conserved throughout evolution. Indeed, mismanagement of Pol function is widely believed to contribute to mutations that lead to human diseases such as cancer (17, 18). Many models propose that multiple partners interact simultaneously with the same clamp during switching. One such model, referred to as the “toolbelt” model, postulates that 2 different Pols simultaneously bind to the same β clamp, with each Pol contacting a separate cleft within the clamp dimer (19). The purpose of the work discussed in this report was to directly test several key aspects of the toolbelt model. Our results indicate that Pol IV can switch with a stalled Pol III*, and this switch relies on both the cleft and the rim contacts. In contrast, processive replication by Pol IV required only the cleft contact. Results from genetic experiments support roles for both the rim and cleft contacts in Pol IV function in vivo. Finally, using a heterodimeric clamp bearing a single cleft (14), we demonstrate that the Pol III*/Pol IV switch operates at a single cleft. Taken together, our results support a variation of the toolbelt model in which switching involves contact of the incoming Pol IV with the rim of the β clamp adjacent to the cleft that is bound by a stalled Pol III*.
Results
Pol IV Switches More Efficiently with a Stalled Pol III* Than It Does with an Actively Replicating Pol III*.
To begin to define the features of the β clamp required for a Pol III*/Pol IV switch, we exploited conditions under which Pol III HE was stalled at the 3′-end of a primer annealed to an SSB-coated ssDNA template by virtue of the omission of 2 nucleotides. The resulting stalled Pol III HE complex resembles a Pol III HE stalled at a replication-blocking lesion in at least 3 ways, including: (i) an inability to extend the 3′-primer end; (ii) repeated cycles of polymerization and exonuclease proofreading of a mismatched 3′-primer end; and (iii) the presence of an SSB-coated ssDNA template downstream from the stall site, reminiscent of a ssDNA gap. Several labs have reported that this stalled Pol III* complex is stable at the 3′ terminus for several minutes (20, 21). Using a template challenge assay (Fig. S1), we determined that in the presence of wild-type β clamp the stalled Pol III* had a half-life on the template of well over 2 min under our assay conditions. This stalled complex was rapidly rescued by addition of dATP and [3H]dTTP, yielding >10 pmol on average of nascent product within a 15-s burst of replication (Fig. 2). Importantly, replication under these conditions is dependent upon β clamp (7, 14), indicating that replication products result from processive Pol III HE synthesis. Taken together, these results confirm that Pol III was merely stalled at the 3′-primer terminus and was not dissociated from the β clamp/primed template.
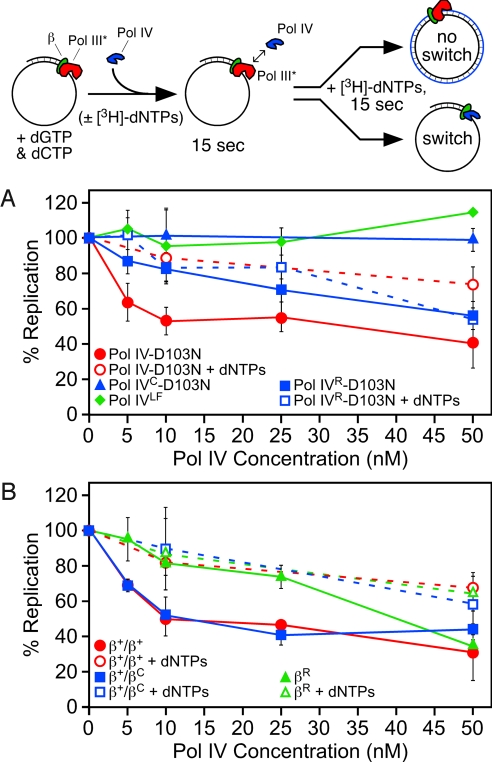
Fig. 2.
Assay to monitor switching between Pol III* and Pol IV in vitro. A cartoon depiction of the assay is shown at the top. (A) Abilities of Pol IV-D103N, Pol IVC-D103N, Pol IVLF, and Pol IVR-D103N to switch with Pol III*. (B) Abilities of β+/β+, β+/βC, and βR to support the Pol III*/Pol IV switch. Results shown represent the average of 4 or more independent determinations. Error bars represent the standard deviation. Replication activity is expressed relative to the level of nucleotide incorporation (ranging from 8–16.4 pmol) observed for Pol III* alone under identical conditions, which was set equal to 100% for each independent experiment.
Switching between the stalled Pol III* and Pol IV was measured by adding Pol IV to the stalled Pol III HE complex before addition of the missing dNTPs. For these experiments, we used a mutant form of Pol IV lacking catalytic activity. We hypothesized that if Pol IV switched with the stalled Pol III*, then we would observe a reduction in total DNA synthesis, as reported previously with similar assays (5, 6). Position D103 of Pol IV is absolutely required for catalytic activity, and its substitution with asparagine (D103N) resulted in an inactive enzyme (22) (Fig. S2 and Fig. 3A). Premixing the wild-type and Pol IV-D103N proteins at ratios of 1:1 or 1:5 resulted in an ∼50% and ∼80% inhibition of DNA synthesis in vitro, respectively (Fig. S2). These results indicate that Pol IV-D103N retains wild-type affinity for both the β clamp and the DNA template.
Titration of Pol IV-D103N into the reaction consisting of the stalled Pol III HE (5 nM) complex before addition of the missing dNTPs resulted in a pronounced inhibition of replication. A 2-fold molar excess of 10 nM Pol IV-D103N inhibited Pol III HE replication by ∼50% (Fig. 2A). Pol III HE replication was essentially completely inhibited by addition of 130 nM Pol IV-D103N (not shown). In striking contrast, simultaneous addition of Pol IV-D103N and the missing dNTPs failed to significantly inhibit replication, except at higher Pol IV-D103N levels, which resulted in modest inhibition (Fig. 2A). Taken together, these results indicate that Pol IV did not switch efficiently with a replicating Pol III*. Since the stalled Pol III* is stable at the 3′ terminus for well over the 15-s incubation used in our assay (Fig. S1), the ability of Pol IV to inhibit the stalled Pol III* replication suggests that Pol IV is not merely waiting for Pol III* to dissociate from the clamp, but is rather actively gaining access to both the clamp and the 3′-primer terminus via a coordinated switch with the stalled Pol III*. Two other groups recently reported similar conclusions (5, 6).
The Pol III*/Pol IV Switch Requires More Than Pol IV Simply Contacting the Cleft of the β Clamp.
Processive synthesis by Pol IV requires that it contacts the cleft of the clamp (23). We therefore asked whether a Pol IV mutant impaired for interaction with the cleft was proficient for switching with Pol III*. The C-terminal 6 residues of Pol IV (346QLVLGL351) contain the CBM motif that interacts with the clamp cleft (23). Deletion of these residues in Pol IVC serves to severely impair interaction of Pol IV with the clamp (23) (Fig. S3). Addition of Pol IVC-D103N in place of Pol IV-D103N at levels up to 50 nM failed to inhibit Pol III HE replication in our assay (Fig. 2A). These results indicate that Pol IV must bind the cleft to switch with the stalled Pol III*.
To determine whether simply binding to the cleft of the β clamp was sufficient for Pol IV to switch with a stalled Pol III*, we asked whether the Pol IV little finger domain (Pol IVLF; residues 243–351) was proficient for switching. Pol IVLF contains both known β-binding regions (i.e., cleft and rim contacts), but lacks Pol activity (13). Therefore, if simply binding to the cleft and/or rim of the clamp was sufficient to promote a switch with a stalled Pol III*, then Pol IVLF should behave like Pol IV-D103N. That Pol IVLF was unable to inhibit Pol III HE replication at levels up to 50 nM (Fig. 2A) indicates that switching in our assay is not simply the result of Pol IV binding to the clamp cleft. Taken together, these findings indicate that Pol IV must contact the cleft of the β clamp to switch with Pol III, but this interaction is on its own insufficient for the switch.
A Single Hydrophobic Cleft on the β Clamp Is Sufficient to Support the Pol III*/Pol IV Switch.
The homodimeric β clamp possesses 2 identical hydrophobic clefts on the same face of the ring. This structural feature served as the basis for the toolbelt hypothesis, which postulates that 2 Pols simultaneously bind to the same β clamp, with each Pol contacting a separate cleft (19). To determine whether both clefts are required for the Pol III*/Pol IV switch, we used a heterodimeric clamp protein (his6-β+/myc-βC) comprised of 1 protomer containing a functional cleft (β+) and 1 bearing a mutant cleft (βC) by virtue of the fact that its C-terminal 5 residues (362MPMRL366) were deleted. Detailed biochemical characterization of both the βC mutant and the β+/βC heterodimer were recently discussed, as were the methods by which we verified purity of the β+/βC heterodimer (14). Based on gel filtration and SPR experiments, βC was severely impaired for interaction with Pol IIIα, displaying a more than 16-fold increase in KD (108 nM vs. 1.78 μM), consistent with deletion of the C-terminal 5 residues of the clamp severely impairing the ability of the cleft to interact with the CBM (14; see Table 1). Despite having only 1 functional cleft, the β+/βC clamp was comparable to β+ with respect to both interaction with Pol IIIα (108 vs. 172 nM), as well as stimulation of Pol III* replication (14). The β+/βC heterodimer was slightly less proficient than the wild-type β clamp at retaining the stalled Pol III* complex on a primed DNA, displaying a half-life of ∼70 s (Fig. S1). The his6-β+/myc-β+ “heterodimer” was indistinguishable from β+, indicating that the tags did not affect clamp function in vitro (Fig. S1). Based on in vitro primer extension assays, the β+/βC clamp was impaired for stimulation of Pol IV replication (Fig. 3B). This is in agreement with results from SPR experiments, which indicated that β+/βC was impaired for interaction with Pol IV (465 vs. 600 nM; see Table 1). Since this clamp bears only 1 functional cleft, 1 Pol IV may be bound to the β+ subunit of the β+/βC clamp in the appropriate manner for replication, while a second Pol IV may be bound to the rim of the βC subunit, perturbing replication. Importantly, this defect of β+/βC in Pol IV replication did not translate into a Pol III*/Pol IV switching defect; the β+/βC heterodimer was indistinguishable from β+ with respect to the Pol III*/Pol IV switch (Fig. 2B). These findings indicate that a single cleft on the β clamp is capable of coordinating the actions of Pol III* with Pol IV in vitro.
Table 1.
Interactions of mutant β clamp proteins with Pol IIIα and Pol IV
| β clamp | Pol IIIα* |
Pol IV† |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| KD, nM | ka, M−1s−1 | kd, s−1 | n | χ2 | KD, nM | ka, M−1s−1 | kd, s−1 | n | χ2 | |
| β+ | 108 | 6.75 × 104 | 7.26 × 10−3 | 1 | 4.2 | 465 | 1.53 × 104 | 7.13 × 10−3 | 2 | 10.7 |
| βC | 1,780 | 2.38 × 103 | 4.23 × 10−3 | 1 | 7.3 | 1,290 | 3.77 × 103 | 4.84 × 10−3 | 2 | 10.1 |
| β+/βC | 172 | 7.27 × 104 | 12.5 × 10−3 | 1 | 3.0 | 600 | 4.89 × 103 | 2.94 × 10−3 | 2 | 9.3 |
| βR | 64 | 9.45 × 104 | 6.00 × 10−3 | 1 | 5.4 | 657 | 4.87 × 103 | 3.20 × 10−3 | 2 | 8.7 |
Kinetic constants describing the interaction of β+, βC, and β+/βC with Pol IIIα (14), and β + with Pol IV (40) were reported previously, and are included here for comparison to results observed with other mutant clamp proteins. n refers to the stoichiometry between Pol IIIα or Pol IV and the dimeric clamp, and was calculated under conditions of saturating analyte. χ2 values describe the fit of the raw data to the 1:1 Langmuir model.
*Approximately 100 RU of each clamp protein was captured using BSA free anti·Penta-His antibody (Qiagen) conjugated to a C1 chip (GE Healthcare), followed by injection of 1–1,000 nM Pol IIIα, as described (14).
†Approximately 250 RU of the indicated clamp protein was captured using BSA free anti·Penta-His antibody (Qiagen) conjugated to a CM5 chip (GE Healthcare), followed by injection of 15–1,500 nM Pol IV as described (40).
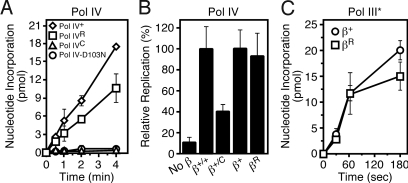
Fig. 3.
Importance of the cleft and rim contacts in stimulating Pol III* and Pol IV replication. (A) Replication activity of the indicated Pol IV mutant proteins was measured as described previously using a filter-binding assay (15, 40). Assays were incubated at 37 °C for the indicated time and contained 10 nM of the indicated Pol IV protein. Results shown represent the average of 2 independent experiments. Error bars represent the range. (B) The ability of the indicated mutant β clamp proteins to stimulate Pol IV+ replication was measured as described above. Replication activity observed with β+ was set equal to 100%. Results shown represent the average of 3 or more independent experiments. Error bars represent the standard deviation. (C) The ability of βR to stimulate Pol III* (5 nM) replication was measured as described above. Results shown represent the average of 3 or more independent experiments. Error bars represent the standard deviation.
Interaction of Pol IV with the Rim of the β Clamp Is Dispensable for Replication, but Is Absolutely Required for the Pol III*/Pol IV Switch.
To determine whether the rim contact contributed to either Pol IV replication, or to the Pol III*/Pol IV switch, we purified mutant forms of the clamp and Pol IV bearing substitutions designed to impair the Pol IV-clamp rim contact. βR bears E93K–L98K substitutions, while Pol IVR contains 303VWP305→303AGA305 mutations. Based on results of SPR experiments, the βR mutant was impaired for interaction with Pol IV, as expected (465 vs. 657 nM; Table 1). In contrast, βR interacted with Pol IIIα ∼2-fold more tightly than did β+ (63.5 vs. 108 nM; Table 1). We were unable to characterize Pol IVR using the same SPR technique due to nonspecific interactions of Pol IVR with the SPR chip surface. However, the Pol IVR little finger was impaired for interaction with the clamp as measured by gel filtration, forming only ∼50% as much complex as the wild-type Pol IV little finger (Fig. S3).
We next examined functional interactions of βR with Pol III* and Pol IV using a primer extension assay. As part of these experiments, we also analyzed replication activity of Pol IVR. As summarized in Fig. 3, βR was comparable to β+ with respect to supporting Pol III* and Pol IV replication. Likewise, Pol IVR displayed only modestly reduced replication activity compared to Pol IV+ in the presence of the wild-type β clamp. Based on these findings, we conclude that the Pol IV-clamp rim interaction is dispensable for Pol III* and Pol IV replication in vitro.
We next examined the ability of βR to support the Pol III*/Pol IV switch. The βR mutant clamp was able to retain a stalled Pol III* on the DNA template with a half life of ∼1 min (Fig. S1), which, although somewhat shorter than that observed with β+, was still in vast excess of the 15-s time frame required for the switching assay. As summarized in Fig. 2B, Pol IV was severely impaired for switching with a stalled Pol III* in the presence of βR. With the exception of the highest Pol IV level examined (50 nM), results were similar regardless of whether Pol IV-D103N was added before the missing dNTPs, or simultaneously. We next examined Pol IVR-D103N using a stalled Pol III* in complex with β+. As summarized in Fig. 2A, Pol IVR-D103N was severely impaired for switching. Taken together, results discussed above indicate that the ability of Pol IV to switch with Pol III* requires interaction of Pol IV with both the rim and cleft of the clamp.
Both the Cleft and Rim Contacts Contribute to Pol IV Function In Vivo.
Pol IV plays an important role in tolerating N2-dG adducts generated by nitrofurazone (NFZ) or 4-nitroquinilone 1-oxide (4-NQO) (24). As a result, E. coli strains deficient in Pol IV function (e.g., ΔdinBW2::cat) display increased sensitivity to NFZ and 4-NQO (24, 25) (see Fig. 4). In light of our results discussed above (Fig. 2), we hypothesized that strains expressing the Pol IVC or the Pol IVR mutants would be sensitive to both NFZ and 4-NQO, due to the impaired abilities of these mutants to switch with a stalled Pol III*. As a test of this hypothesis, we characterized NFZ and 4-NQO sensitivity of the Pol IV deficient strain JH100 bearing plasmids expressing physiological levels of different mutant Pol IV proteins containing mutations impairing the cleft or rim contacts. Western blot experiments confirmed that each of the mutant Pol IV proteins was expressed at a steady state level comparable to that of the Pol IV+ strain (Fig. 4A).
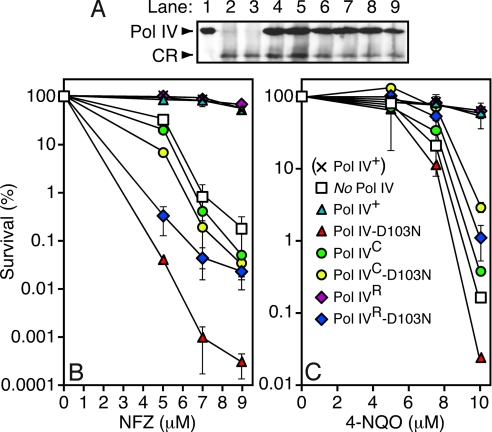
Fig. 4.
Ability of various mutant dinB alleles to complement NFZ and 4-NQO sensitivity of a ΔdinB E. coli strain. (A) Steady state levels of the indicated Pol IV proteins were measured by western blotting of whole cell extracts of strain JH100 (relevant genotype, ΔdinBW2::cat) bearing the indicated plasmid using a polyclonal anti-Pol IV antibody (25) and chemilumenescent detection (Pierce). Lane 1, purified Pol IV (5 ng); lane 2, no plasmid; lane 3, (pWSK29; empty vector control); lane 4, (pRM102; Pol IV+); lane 5, (pJH100; Pol IV-D103N); lane 6, (pJH101; Pol IVR); lane 7, (pJH102; Pol IVC); lane 8, (pJH103; Pol IVR-D103N); lane 9, (pJH104; Pol IVC-D103N). CR refers to a polypeptide which cross reacts with the anti-Pol IV antibody, and serves as a loading control. Sensitivity of strain AB1157 (× Pol IV+), or JH100 bearing the specified Pol IV-expressing plasmid (see legend in panel C), to the indicated concentration of NFZ (B) or 4-NQO (C) is shown. Results represent the average of 2 independent experiments, each performed with an independent transformant. Error bars represent the range.
As summarized in Fig. 4B, NFZ sensitivity of the ΔdinB strain bearing the Pol IV+-expressing plasmid was indistinguishable from that of the AB1157 dinB+ parent strain, while the same strain bearing the empty control plasmid (ΔdinB control) was as much as ∼350-fold more sensitive. These findings confirm that our plasmid constructs express physiological levels of Pol IV. The strain expressing Pol IVC resembled the ΔdinB control, consistent with the model that Pol IV must contact the cleft of the clamp to catalyze bypass of N2-dG adducts in vivo. In contrast, the strain expressing Pol IVR was indistinguishable from the Pol IV+ strain (Fig. 4B), suggesting that the rim contact may not be required for tolerance of all NFZ-induced adducts in vivo. Comparable results were observed using 4-NQO (Fig. 4C).
Strains expressing Pol IV-D103N (dinB-D103N) display a hypersensitivity to NFZ and 4-NQO that can be as much as ∼1,000-fold more severe than that observed for a strain lacking Pol IV (ΔdinB) (24) (see Fig. 4 B and C). We hypothesized that this increased sensitivity was due to the fact that Pol IV-D103N was gaining access to the lesion via a Pol III*/Pol IV switch, but, due to its active site mutation, was unable to catalyze bypass and therefore caused cell death by blocking access of another cellular process capable of tolerating N2-dG lesions. We therefore constructed double mutants of Pol IV bearing both the D103N mutation, together with the Pol IVC or Pol IVR mutations, and measured their sensitivities to NFZ and 4-NQO. As summarized in Fig. 4B, both the Pol IVC and the Pol IVR mutations alleviated NFZ hypersensitivity of the Pol IV-D103N strain. In fact, the strains expressing Pol IVC-D103N or Pol IVR-D103N resembled the strain lacking Pol IV. Thus, both the rim and cleft contacts are required for hypersensitivity to NFZ. Comparable results were obtained using 4-NQO (Fig. 4C), but the respective levels of sensitivity differed slightly among the different Pol IV mutants, consistent with NFZ and 4-NQO inducing slightly different classes of DNA lesions. As discussed below, these findings suggest that Pol IV must contact both the rim and the cleft of the clamp to gain access to at least a subset of DNA lesions.
Discussion
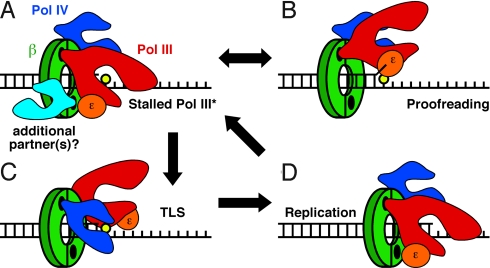
Results discussed in this report indicate that Pol IV switches with a stalled Pol III*, but not an actively replicating Pol III*. Based on the structure of the Pol IVLF-β clamp complex reported by Bunting et al. (13) (see Fig. 1), Pol IV contacts both the cleft and the rim of the clamp. Using mutant forms of Pol IV and β that bear substitutions impairing the cleft or rim contacts (Table 1 and Fig. S3), we demonstrated that the cleft contact was required for both Pol IV replication (Fig. 3) as well as the Pol III*/Pol IV switch (Fig. 2), consistent with previous reports (5, 6, 23). In contrast, the rim contact was not required for processive replication by either Pol III* or Pol IV (Fig. 3), but was crucial for the Pol III*/Pol IV switch (Fig. 2). Results of genetic experiments discussed below are consistent with this conclusion. Finally, a heterodimeric form of the β clamp (β+/βC) bearing 1 wild-type protomer in complex with a mutant (βC) lacking a functional cleft supported the Pol III*/Pol IV switch in a manner that was indistinguishable from the wild-type clamp, indicating that this switch required only 1 cleft on the clamp (Fig. 2B). These findings challenge the toolbelt model and suggest that Pol IV switches with a stalled Pol III* by first contacting the rim of the clamp adjacent to the cleft that is bound by Pol III*, before gaining control of the cleft that was bound by the α catalytic subunit of Pol III* (Fig. 5). Since a single cleft is sufficient for managing the actions of Pol III* and Pol IV, the second cleft, and/or other clamp surfaces may be available to simultaneously manage the actions of 1 or more additional partners (Fig. 5A). Further work is required to test this hypothesis.
Fig. 5.
Model for the Pol III*/Pol IV switch. Pol IV is shown in dark blue, Pol III* is shown in red, its ε subunit is in orange, and a putative third Pol (or partner) is in light blue. For simplicity, our model does not depict the other Pol III* subunits, and represents a single DNA primer/template junction. See text for additional details regarding the model.
Of relevance to our model (Fig. 5), both Pol IIIα and Pol IV use a CBM in their finger domain to contact the clamp (13, 26). As a result, Pol III likely associates with the β clamp in a manner that is reminiscent of the Pol IVLF-clamp complex (Fig. 5A). Thus, if Pol IV is bound to the rim of the clamp adjacent to the cleft that is occupied by Pol III, then Pol IIIα could potentially reside in between Pol IV and the ε proofreading subunit of Pol III (Fig. 5A). This structural arrangement may favor shuttling of the DNA template between the α catalytic and ε proofreading subunits of Pol III for repeated cycles of polymerization and exonuclease proofreading, effectively limiting access of Pol IV to the template until such time as TLS is required (see Fig. 5). Conformational change(s) in either Pol III* and/or the β clamp induced by the lesion may serve to signal the Pol III*/Pol IV switch. Alternatively, the Pol III*/Pol IV switch may also be influenced by the affinity of Pol IV for the lesion present in the template strand, as suggested previously for eukaryotic Pol η (27). Our finding that the Pol IVLF was unable to switch with a stalled Pol III* (Fig. 2A) is consistent with the need for Pol IV to bind the DNA template to switch with Pol III*.
The strain expressing Pol IVR was indistinguishable from the Pol IV+ control strain with respect to NFZ and 4-NQO sensitivity (Fig. 4), suggesting that Pol IV may not require contact with the rim of the clamp to catalyze bypass of all N2-dG adducts in vivo. Strains expressing Pol IV-D103N display a hypersensitivity to NFZ and 4-NQO, which was suggested to result from an ability of the mutant Pol IV to interfere with 1 or more cellular processes that act to tolerate NFZ- and 4-NQO-induced lesions, such as Pol switching (24). Importantly, hypersensitivity was not observed in strains expressing double mutant Pol IV proteins containing both D103N and rim (Pol IVR-D103N) or cleft (Pol IVC-D103N) substitutions (Fig. 4). One possible explanation for these apparently conflicting results is that Pol IV can gain access to lesions by at least 2 distinct mechanisms, 1 of which involves switching with Pol III*, and a second mechanism that is independent of the rim contact, and may not involve switching with Pol III*. Based on the phenotype of the Pol IVC strain, the cleft contact is required for both proposed mechanisms. The rim contact is likely crucial for Pol IV to switch with Pol III* stalled at a lesion. In the absence of this switch (e.g., mutations affecting the rim contact), Pol III replication is presumably reprimed downstream of the lesion, leaving a ssDNA gap (1). Our finding that the rim contact was dispensable for Pol IV replication in vitro (Fig. 3 A and B) suggests that Pol IV could gain access to the lesion in the ssDNA gap independent of the rim contact, particularly if the lesion was adjacent to the 3′-primer terminus. Thus, the lack of NFZ and 4-NQO sensitivity of the Pol IVR strain may be due to the ability of the Pol IVR mutant to tolerate NFZ- and 4-NQO-induced lesions in ssDNA gaps without having to switch with Pol III*. In contrast, hypersensitivity of the Pol IV-D103N strain to NFZ and 4-NQO suggests that it gains access to lesions, but due to the active site mutation is unable to catalyze bypass, and as a result acts to block another cellular process capable of tolerating the damage. Our finding that hypersensitivity is alleviated by either the rim or cleft mutations suggests that Pol IV-D103N gains access via the switching mechanism discussed above (Fig. 5). Alternatively, Pol IV catalysis may contribute to Pol switching following TLS in vivo, in which case, the Pol IV-D103N mutant may become “locked” on the lesion. In this case, disruption of the rim or cleft contacts may prevent Pol IV-D013N from gaining access via a switch with Pol III*. Further work is required to test these hypotheses. Regardless, our finding that both the rim and cleft contacts were required for hypersensitivity suggests that Pol IV requires these contacts to access at least a subset of the NFZ- and 4-NQO-induced lesions, possibly by switching with a stalled Pol III*.
Indiani et al. (28) recently described the ability of Pol II and Pol IV to exchange with Pol III* present within the replisome in vitro. This finding is reminiscent of the dynamic processivity described for the T4 and T7 systems (29, 30). Dynamic processivity refers to the rapid exchange between extra or auxiliary copies of the replicative Pol and the DNA template. Taken together, these studies indicate that Pols access the replication fork more dynamically than previously thought. Related to this topic, E. coli strains bearing certain mutations in the β clamp that affect its interactions with DnaX, Pol IIIα, and/or the DNA template display conditional lethal phenotypes that are largely alleviated by inactivation of 1 or more specialized Pol, particularly Pol IV (15, 31–35). Taken together, findings discussed above suggest that Pol IV may gain frequent access to the replication fork during normal growth, providing the replisome with the capacity to tolerate certain classes of lesions via TLS. The finding that Pol IV is capable of catalyzing relatively accurate TLS past certain lesions makes this model more palatable (24, 36). The steady state level of Pol IV is ∼350 nM in the SOS-repressed state, which is close to the apparent KD of Pol IV for the β clamp (∼460 nM), and rises to ∼3 μM following SOS induction (25), and so it is present at sufficient levels to associate with clamp independent of SOS induction. Viewed in this way, the Pol IV sensitivity of mutant strains impaired for Pol III* function may be due to impaired replication resulting from excessive Pol III*/Pol IV switching. This may occur as a result of reduced Pol III HE processivity caused by the mutations, resulting in more frequent stalling of the replisome. Uchida et al. (37) recently suggested that SOS-induced levels of Pol IV act to displace Pol III from the clamp as part of a DNA damage checkpoint system. It is unclear whether the modest levels of Pol IV used in this report effect displacement of Pol III*. In light of the fact that Pol IIIα and Pol IV possess similar affinities for clamp in solution (Table 1), displacement of Pol III may result in part from direct competition between Pol IIIα and SOS-induced levels of Pol IV for binding to the clamp. However, 1 or more mechanisms must exist to regulate the ability of Pol IV to displace Pol III from the clamp as SOS induction on its own fails to inhibit replication. Further work is required to understand the relationship between the proposed checkpoint function of Pol IV, and our model for the Pol III*/Pol IV switch. Analysis of additional site-specific heterodimeric clamp proteins will help to further dissect the mechanism(s) by which the clamp helps to coordinate the actions of Pol III* and Pol IV, as well as additional partners acting on DNA.
Materials and Methods
Proteins and Reagents.
Pol IIIα (16), Pol III* ([τ2γδδ'ψχ][αεθ]2) (15), γ3δδ' (14), SSB (38), Pol IV+ (15), β+ (16), N-terminally his6- and heart muscle kinase (HMK)-tagged β+ (16), his6- and HMK-tagged βC (14, 39), and the his6- and HMK-tagged β+/N-terminally myc-tagged βC (β+/βC) and his6- and HMK-tagged β+/myc-tagged β+ (β+/β+) heterodimers (14) were purified as described in the indicated reference. Other mutant proteins were cloned using the Quickchange strategy (Stratagene). Primers used for Quickchange mutagenesis were from Sigma-Genosys, are their sequences are shown in Table S1. His6- and HMK-tagged βR bears E93K-L98K substitutions and was purified as described previously (39). Recombinant forms of Pol IV-D103N, Pol IVC-D103N, Pol IVR, and Pol IVR-D103N lack tags. These proteins were purified using the same protocol as that used for Pol IV+ (15). Pol IVLF contains an N-terminal his10-tag, and was purified as described (13). M13mp18 ssDNA was purified from phage as described previously (40), PAGE-purified SP20 (5′-ACG CCT GTA GCA TTC CAC AG-3′) was from Sigma-Genosys, and ultra pure dNTPs were from GE Healthcare.
Pol III*/Pol IV Switch Assay.
Switching assays were modeled loosely after the method described by Indiani and colleagues (6). Briefly, reactions (20 μL) containing replication assay buffer (20 mM Tris-HCl, pH 7.5, 8.0 mM MgCl2, 0.1 mM EDTA, 5 mM DTT, 1 mM ATP, 5% glycerol, 0.8% BSA) were supplemented with 133 μM dCTP/dGTP, 4 μM SSB, 40 nM β clamp, and 5.0 nM SP20/M13mp18 template. Reactions were initiated by addition of 5 nM Pol III*, followed by incubation for 2 min at 37 °C. Wild-type or mutant Pol IV was then added either 15 s before or simultaneously with [3H-dTTP]-dNTPs (133 μM, 68.5 or 111.7 cpm/pmol). After addition of dNTPs, reactions were incubated for 15 s at 37 °C. Reactions were quenched by spotting onto DE81 paper (VWR), followed by washing 4 times in 0.5 M sodium phosphate, and replication activity was quantitated by liquid scintillation spectroscopy, as described (40).
Interactions of β Clamp with Pol IIIα and Pol IV.
Interactions involving the his6-tagged β clamp and untagged Pol IIIα, or untagged Pol IV were measured by SPR using a BIAcore X instrument (GE Healthcare) as described previously (14). Kinetic constants were derived from SPR traces using the Langmuir binding model included in the BIAevaluation software (version 4.1; GE Healthcare). Interactions of Pol IVLF, as well as Pol IVLF derivatives bearing either the 303VWP305→303AGA305 (rim) or the Δ346–351 (cleft) mutations with the β clamp were measured using Superose-12 gel filtration, as described previously (15).
NFZ and 4-NQO Sensitivity.
E. coli strain AB1157 (E. coli genetic stock center), or JH100 [AB1157 with ΔdinBW2::cat (24)], bearing different dinB-expressing plasmids (see legend to Fig. 4), were cultured in LB medium supplemented with 150 μg/mL ampicillin (Amp) for ∼16 h at 37 °C. Appropriate dilutions were spread onto LB agar plates containing Amp and increasing concentrations of NFZ or 4-NQO (Sigma), followed by incubation overnight at 37 °C. Colony forming units were counted, and survival at each concentrations of NFZ or 4-NQO was determined relative to the LB-Amp control, which was set equal to 100%.
Supplementary Material
Acknowledgments.
We thank Graham Walker (Massachusetts Institute of Technology, Cambridge, MA) for his generous gift of the ΔdinBW2::cat allele; Brian Lang for expert advice regarding analysis of SPR data; and the members of our laboratory for numerous helpful discussions. This work was supported by National Institutes of Health Grants GM066094 (to M.D.S.) and F31GM073586 (to S.K.S.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903460106/DCSupplemental.
References
- 1.Friedberg EC, et al. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. [Google Scholar]
- 2.Friedberg EC, Feaver WJ, Gerlach VL. The many faces of DNA polymerases: Strategies for mutagenesis and for mutational avoidance. Proc Natl Acad Sci USA. 2000;97:5681–5683. doi: 10.1073/pnas.120152397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: How do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 4.Lehmann AR, et al. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Furukohri A, Goodman MF, Maki H. A dynamic polymerase exchange with Escherichia coli DNA polymerase IV replacing DNA polymerase III on the sliding clamp. J Biol Chem. 2008;283:11260–11269. doi: 10.1074/jbc.M709689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Indiani C, et al. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell. 2005;19:805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Burgers PM, Kornberg A, Sakakibara Y. The dnaN gene codes for the beta subunit of DNA polymerase III holoenzyme of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:5391–5395. doi: 10.1073/pnas.78.9.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson A, O'Donnell M. Cellular DNA replicases: Components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 9.Lopez de Saro FJ, O'Donnell M. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc Natl Acad Sci USA. 2001;98:8376–8380. doi: 10.1073/pnas.121009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Dalrymple BP, et al. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc Natl Acad Sci USA. 2001;98:11627–11632. doi: 10.1073/pnas.191384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeruzalmi D, et al. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell. 2001;106:417–428. [PubMed] [Google Scholar]
- 13.Bunting KA, Roe SM, Pearl LH. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. EMBO J. 2003;22:5883–5892. doi: 10.1093/emboj/cdg568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scouten Ponticelli SK, Duzen JM, Sutton MD. Contributions of the individual hydrophobic clefts of the Escherichia coli beta sliding clamp to clamp loading, DNA replication, and clamp recycling. Nucleic Acids Res. 2009;37:2796–2809. doi: 10.1093/nar/gkp128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maul RW, Ponticelli SK, Duzen JM, Sutton MD. Differential binding of Escherichia coli DNA polymerases to the beta-sliding clamp. Mol Microbiol. 2007;65:811–827. doi: 10.1111/j.1365-2958.2007.05828.x. [DOI] [PubMed] [Google Scholar]
- 16.Duzen JM, Walker GC, Sutton MD. Identification of specific amino acid residues in the E. coli beta processivity clamp involved in interactions with DNA polymerase III, UmuD and UmuD′. DNA Repair (Amst) 2004;3:301–312. doi: 10.1016/j.dnarep.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Masutani C, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 18.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 19.Pages V, Fuchs RP. How DNA lesions are turned into mutations within cells? Oncogene. 2002;21:8957–8966. doi: 10.1038/sj.onc.1206006. [DOI] [PubMed] [Google Scholar]
- 20.Stukenberg PT, Studwell-Vaughan PS, O'Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- 21.Shwartz H, Livneh Z. Dynamics of termination during in vitro replication of ultraviolet-irradiated DNA with DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem. 1987;262:10518–10523. [PubMed] [Google Scholar]
- 22.Wagner J, et al. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- 23.Lenne-Samuel N, Wagner J, Etienne H, Fuchs RP. The processivity factor beta controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo. EMBO Rep. 2002;3:45–49. doi: 10.1093/embo-reports/kvf007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarosz DF, et al. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 25.Kim SR, et al. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol Genet Genomics. 2001;266:207–215. doi: 10.1007/s004380100541. [DOI] [PubMed] [Google Scholar]
- 26.Lamers MH, et al. Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III. Cell. 2006;126:881–892. doi: 10.1016/j.cell.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 27.McCulloch SD, et al. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 28.Indiani C, Langston LD, Yurieva O, Goodman MF, O'Donnell M. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc Natl Acad Sci USA. 2009;106:6031–6038. doi: 10.1073/pnas.0901403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, et al. The dynamic processivity of the T4 DNA polymerase during replication. Proc Natl Acad Sci USA. 2004;101:8289–8294. doi: 10.1073/pnas.0402625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamdan SM, et al. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27:539–549. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 31.Sutton MD. The Escherichia coli dnaN159 mutant displays altered DNA polymerase usage and chronic SOS induction. J Bacteriol. 2004;186:6738–6748. doi: 10.1128/JB.186.20.6738-6748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutton MD, Duzen JM. Specific amino acid residues in the beta sliding clamp establish a DNA polymerase usage hierarchy in Escherichia coli. DNA Repair (Amst) 2006;5:312–323. doi: 10.1016/j.dnarep.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Sutton MD, Duzen JM, Maul RW. Mutant forms of the Escherichia coli beta sliding clamp that distinguish between its roles in replication and DNA polymerase V-dependent translesion DNA synthesis. Mol Microbiol. 2005;55:1751–1766. doi: 10.1111/j.1365-2958.2005.04500.x. [DOI] [PubMed] [Google Scholar]
- 34.Maul RW, et al. Role of Escherichia coli DNA polymerase I in conferring viability upon the dnaN159 mutant strain. J Bacteriol. 2007;189:4688–4695. doi: 10.1128/JB.00476-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maul RW, Sutton MD. Roles of the Escherichia coli RecA protein and the global SOS response in effecting DNA polymerase selection in vivo. J Bacteriol. 2005;187:7607–7618. doi: 10.1128/JB.187.22.7607-7618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan B, et al. Efficient and accurate bypass of N2-(1-carboxyethyl)-2′-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc Natl Acad Sci USA. 2008;105:8679–8684. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchida K, et al. Overproduction of Escherichia coli DNA polymerase DinB (Pol IV) inhibits replication fork progression and is lethal. Mol Microbiol. 2008;70:608–622. doi: 10.1111/j.1365-2958.2008.06423.x. [DOI] [PubMed] [Google Scholar]
- 38.Lusetti SL, et al. The RecF protein antagonizes RecX function via direct interaction. Mol Cell. 2006;21:41–50. doi: 10.1016/j.molcel.2005.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutton MD, Narumi I, Walker GC. Posttranslational modification of the umuD-encoded subunit of Escherichia coli DNA polymerase V regulates its interactions with the beta processivity clamp. Proc Natl Acad Sci USA. 2002;99:5307–5312. doi: 10.1073/pnas.082322099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heltzel JMH, et al. Sliding clamp-DNA interactions are required for viability and contribute to DNA polymerase management in Escherichia coli. J Mol Biol. 2009;387:74–91. doi: 10.1016/j.jmb.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.