Abstract
Helitrons are recently discovered eukaryotic transposons that are predicted to amplify by a rolling-circle mechanism. They are present in most plant and animal species investigated, but were previously overlooked partly because they lack terminal repeats and do not create target site duplications. Helitrons are particularly abundant in flowering plants, where they frequently acquire, and sometimes express, 1 or more gene fragments. A structure-based search protocol was developed to find Helitrons and was used to analyze several plant and animal genomes, leading to the discovery of hundreds of new Helitrons. Analysis of these Helitrons has uncovered mechanisms of element evolution, including end creation and sequence acquisition. Preferential accumulation in gene-poor regions and target site specificities were also identified. Overall, these studies provide insights into the transposition and evolution of Helitrons and their contributions to evolved gene content and genome structure.
Keywords: gene fragment acquisition, Helitron, insertion specificity, transposition
Helitrons are a new class of transposable elements (TEs) that were initially discovered by repeat-based computational analysis of the genome of the model plant Arabidopsis thaliana (1). Their structural homologies to genes encoding Rep/helicase-like and replication protein A (RPA)-like proteins suggest that they transpose by a rolling-circle mechanism, although this conclusion is not yet supported by experimental evidence. Because gemini viruses, some plasmids, and some bacterial transposons are known to replicate by a rolling-circle mechanism (2, 3), it is possible that Helitrons share a very ancient common ancestor with these other mobile sequences (4). Several Arabidopsis insertion elements that had been detected before the discovery and description of Helitrons were eventually found to be Helitrons, including Aie (5), AthE1 (6), Basho (7), and ATREP (8).
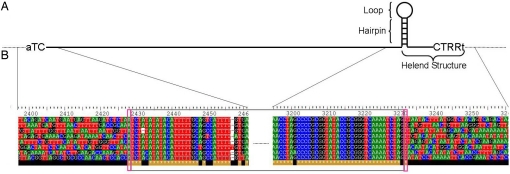
Helitrons are characterized by a 5′ TC terminus and a 3′ CTRR terminus that includes a predicted small hairpin structure near the 3′ CTRR end (Fig 1A). They are found to be preferentially inserted into the dinucleotide AT. Some elements encode Rep/helicase-like and RPA-like proteins that may be involved in the transposition process. The elements that encode Rep/helicase are considered putative autonomous elements.
Fig. 1.
(A) Helitron structure: 5′ TC and 3′ CTRR termini (shown in uppercase), a predicted small hairpin structure near the 3′ end, and insertion into the target dinucleotide AT (shown in lowercase). (B) The end sequences of 8 Helitrons (gray rectangle) from A. thaliana. 5′ start with TC, 3′ end with CTRR, insert between AT dinucluetide (red rectangles), and flanking sequences are shown. Sequences within the insertion site align very well and sequences outside of the insertion cannot be aligned because they are insertions at different genomic locations.
Since their original discovery in Arabidopsis, Helitrons have been found in 6 additional flowering plant species, and also in moss (9), fungi (10, 11), the worm Caenorhabditis elegans (1), sea urchin (11), fish (11, 12), and bats (13). They have been observed to often capture gene fragments, sometimes fragments from multiple genes that normally reside in unlinked chromosomal locations (14–18). Although the mechanism of gene fragment acquisition has not been determined, it apparently occurs at the DNA level because both introns and exons are found within the acquired DNA. Some of these collections of Helitron-acquired gene fragments can be found as chimeric transcripts (14, 19). In a few detected cases, acquired introns are spliced, sometimes alternatively, and the junctions between fragments can be processed as crude de novo introns (14, 20). This process is quite comparable to the model proposed by Gilbert (21) to explain the origin of introns. However, Gilbert's “exon shuffling,” the fusion of the first short templates for single peptide domains to create the potential to synthesize complex proteins out of dissimilar subunits, was proposed as a mechanism that existed primarily in the early days of life on earth, more than a billion years ago. In maize, Morgante et al. (19) calculated that there are more than 4,000 gene fragment acquisitions within Helitrons in a single maize inbred, suggesting that exon shuffling is a very active process right now in at least some flowering plant genomes.
In plants, gene fragment acquisition by TEs is not a process unique to Helitrons. Bs1 of maize was the first reported retrotransposon that contained sequences similar to a portion of a normal host gene, a plasma membrane proton ATPase (22–24), and many additional cases of similar phenomena have now been found (25). Novel sequence acquisition is also observed for DNA elements, especially in the Mutator system (26, 27). In the rice genome, over 3,000 Mutator elements were reported to contain fragments derived from more than 1,000 cellular genes and at least 5% appear to be expressed (27). Hence, some plant species appear to have a manic rate of genic sequence rearrangement, with the potential to create a vast array of novel genes and genetic functions (28).
Results
A Structure-Based Approach for Helitron Identification.
The identification of Helitrons is difficult because of their few and tiny structural features. The approaches used up to now can be divided into 5 categories. One approach is to search for Rep/helicase or RPA-like protein homology (1, 11, 13, 29). The second approach is to identify Helitrons as de novo insertions (14, 15, 30). Insertions that mutated the sh2 gene and the ba1 gene in maize both turned out to be Helitron insertions. Another spontaneous mutation in morning glory was also caused by Helitron insertion. The third approach is to search for similarity to known Helitrons or known Helitron ends (1, 13, 30–33). The fourth approach involves characterizing identified repeats (1). There are a few programs that can find repetitive sequences in a given genome, such as RECON (34) and Spectral Repeat Finder (35). However, high levels of sequence diversity make it difficult to precisely define boundaries of Helitron elements found by such programs. This approach can only detect Helitron families that have abundances above a certain arbitrarily chosen copy number. The fifth approach is to search for violations of microcolinearity between genomes (16, 17, 19, 20). This method requires well-studied colinear regions from different haplotypes to define Helitron boundaries precisely and is very time and labor consuming.
To circumvent the limitations of previous approaches, a computer program called HelSearch was developed using the tiny structural features of Helitrons, and a requirement for at least 2 identical 3′ ends between separate elements, to find Helitrons in any genomic sequence (see Methods). A. thaliana, Medicago truncatula, Oryza sativa, Sorghum bicolor, Caenorhabditis elegans, and Drosophila melanogaster genomes were screened for Helitrons using this program. In addition, as a negative control, the genomic sequence of A. thaliana was pooled and randomly reconstructed and then searched with the same procedure. No Helitron was identified by HelSearch in the random sequence (Table 1).
Table 1.
Summary of Helitron discovery and description in six multicellular eukaryotes
| Organism | Common name | Genome screened, Mb | No. of intact elements | No. of families | No. of new families | Putative autonomous elements | No. of families with acquired gene fragment(s) | Total no. of elements | Genome abundance, % |
|---|---|---|---|---|---|---|---|---|---|
| Random sequence (ATH) | Negative control | 115 | 0 | 0 | 0 | − | 0 | − | 0 |
| Arabidopsis thaliana | Mustard weed | 115 | 281 | 10 | 2 | + | 2 | 1,242 | 1.30 |
| Medicago truncatula | Barrel medic | 243 | 230 | 10 | 6 | + | 3 | 1,386 | 1.29 |
| Oryza sativa spp. japonica | Rice | 389 | 651 | 23 | 18 | + | 7 | 6,947 | 2.09 |
| Sorgum bicolor | Sorghum | 748 | 608 | 11 | 11 | + | 2 | 4,875 | 3.00 |
| Caenorhabditis elegans | Nematode worm | 100 | 281 | 4 | 0 | + | 0 | 600 | 2.30 |
| Drosophila melanogaster | Fruit fly | 154 | 0 | 0 | 0 | − | 0 | – | 0 |
In 2 small genomes, those of A. thaliana and C. elegans, Helitrons have been well characterized (1). A total of 281 intact elements from 10 Helitron families were identified in Arabidopsis by HelSearch, including 2 new families, and 281 intact elements from 4 families were found in the C. elegans genome (no new family) (see Tables S1–S5 for details). Families were defined as containing elements that shared the same intact 3′ end, and subfamilies as those with the same intact 5′ ends and the same intact 3′ end. The Helitrons in A. thaliana and C. elegans make up 1.3 and 2.3% of their nuclear genomes, respectively, a number consistent with previous calculations (1). Two Helitron families in Arabidopsis were found to have acquired gene fragments, compared to 1 family with gene fragments reported in previous studies (36).
Few Helitrons have been reported in Medicago, rice, and sorghum. HelSearch identified 230, 651, and 608 intact elements from 10, 23, and 11 families, respectively, in these 3 species. Only 4 families in Medicago and 3 in rice were previously known. Helitrons compose about 1.3, 2.1, and 3.0% of the Medicago, rice, and sorghum genomes, respectively. Three families in Medicago, 7 families in rice, and 2 families in sorghum were found to have captured gene fragments. Helitrons seem to acquire gene fragments of all types, as assessed by GO category analysis (see Table S6 for details). Putative autonomous elements were found in all of the above-mentioned 5 genomes. No intact Helitron was found in the D. melanogaster genome. Table 1 provides a summary of these results.
A minimum total number of elements was estimated by searching for conserved 3′ ends. In Arabidopsis, C. elegans, Medicago, rice, and sorghum genomes, there are at least 1,200, 600, 1,300, 7,000, and 5,000 Helitrons, respectively.
Although some elements from different genomes belong to the same family, it does not necessarily mean they are the most related elements, because families are defined by a shared 3′ end sequence. For instance, Medicago Helitrons exhibit only 2 independent gene acquisition events, both quite ancient. Because of changes in the 3′ end, they have evolved into what we now call different families.
Because our approach can only identify elements that have at least 2 copies in the genome, a linear regression analysis was performed to estimate the number of intact single copy elements that are likely to have been missed (Fig. S1). By this approach, single copy families of Helitrons in A. thaliana, O. sativa, M. truncatula, and S. bicolor were estimated to number ≈19, 162, 9, and 271, respectively. Hence, HelSearch is predicted to find the great majority of elements in each of these genomes, but to identify less than half of the different element families. It should be noted, however, that these low-copy-number elements have been missed by all previous searches.
Helitron Distribution Along Plant and Animal Chromosomes.
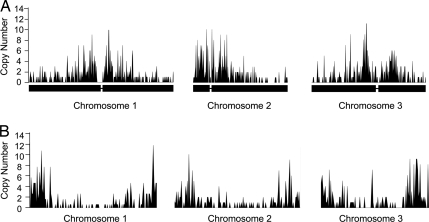
The identified Helitrons in the Arabidopsis, rice, and C. elegans genomes were mapped along the sequenced genomes in these species. The results indicate that Helitrons in Arabidopsis are enriched in gene-poor pericentromeric regions (Fig. 2A, Fig. S2A), showing a similar pattern to that previously seen for LTR retrotransposons and opposite to the DNA transposons that have been found to be preferentially associated with gene-rich regions in those genomes (37, 38). Similarly, Helitrons in the C. elegans genome were found to be most abundant in the gene-poor terminal regions of each chromosome (Fig. 2B, Fig. S2B). In C. elegans, DNA transposons are relatively more abundant in the gene-poor chromosome termini, as now observed for Helitrons (39–41). Rice, on the other hand (Fig. S2C), exhibited a less ordered pattern of Helitron distribution, with some pericentromeric regions rich in these elements and others less so.
Fig. 2.
Distribution of Helitrons on 3 chromosomes of A. thaliana (A) and 3 chromosomes of C. elegans (B). x axis indicates chromosome locations while the y axis shows copy numbers of Helitrons (both intact and fragmented elements) per 100 kb. Narrow lines in A on the x axis indicate the positions of centromeres.
Helitron Structures and Specificities.
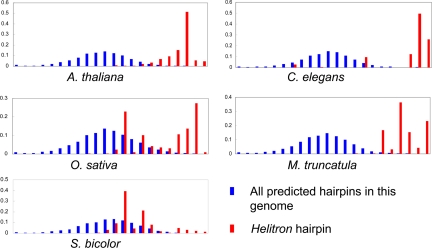
The hairpins of Helitrons have higher predicted melting temperatures than are found in similar hairpins that are not associated with Helitrons (Fig. 3). For reasons unknown, the 2 eudicot plants (Arabidopsis and Medicago) and C. elegans demonstrated a much higher and uniform range of predicted Helitron melting temperatures (Tm) than did the 2 monocots (rice and sorghum).
Fig. 3.
Melting temperature distributions of predicted hairpins in the 5 studied genomes. Blue denotes predicted hairpins across the entire genome, and red denotes predicted hairpins in identified Helitrons. The x axis indicates predicted melting temperature (0–100 °C) while the y axis shows the frequency of predicted hairpins with that predicted Tm.
Fifty base pairs upstream and downstream of all intact Helitron insertions were used to screen for insertion specificities beyond the flanking AT dinucleotide. The results (Fig. S3) indicate that insertion regions are relatively A/T rich. The region from the insertion sites to about 12 bp downstream shows an 82% A plus T abundance. As a control, AT dinucleotide sites were chosen randomly from each genome, and 50 bp both upstream and downstream were scored for A/T composition. A χ2 test comparing A/T content to Helitron insertion sites at each position was performed and the results (Fig. S3) demonstrated that most of the positions from the insertion site to 12 bp downstream and 3 bp upstream were significantly more A/T rich than would have been expected by chance.
New End Creation and Sequence Acquisition.
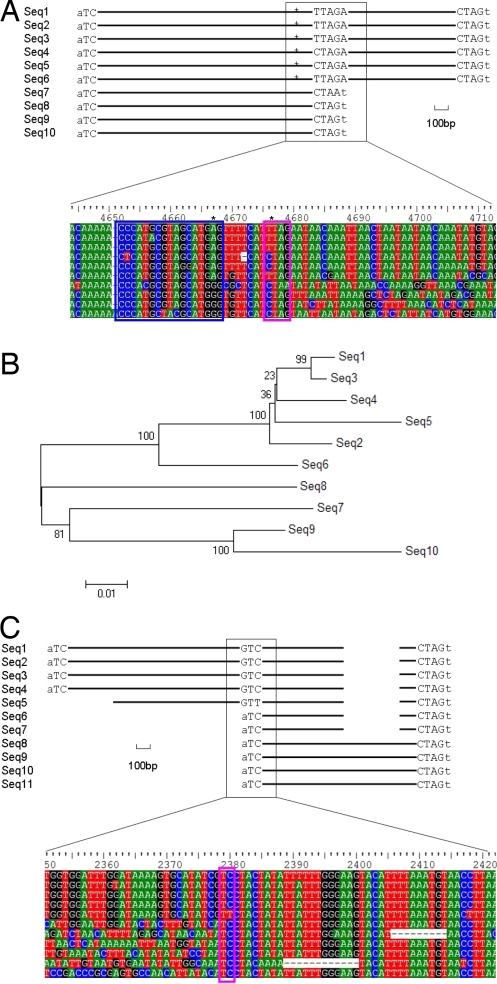
Analysis of the complete set of Helitrons in Arabidopsis indicated that these elements can acquire new sequences by recognizing either a new 3′ termination site or a new 5′ start site. Fig. 4A shows an example of a Helitron family with a new 3′ end. Because of their lesser homology to each other [84–90% pairwise identity over all the elements (Fig. 4A and data not shown)], Helitrons in sequences 7 to 10 are proposed to have the ancestral structure. Helitrons represented by sequences 1 to 6 are expected to be recently derived, as they exhibit 90–98% similarity. This proposition is supported by neighbor joining tree analysis with 1,000 bootstrap replicates (Fig. 4B). If one element had a mutation in the hairpin, noted as the first star from the left, that damaged the rolling-circle stop signal, then a transpositional replication event might not stop at the original termination site. However, by chance, another potential stop site will sometimes be present somewhere downstream. A Helitron thus acquires another stop and a few hundred base pairs of sequence to complete the transposition event, giving rise to the Helitrons in sequences 1 to 6.
Fig. 4.
A mechanism for Helitron sequence acquisition and new end creation. (A) Helitrons in Arabidopsis pick up a new 3′ end. Stars indicate sequence divergence in the helend. A pink rectangle encloses the 3′ CTRR of Helitrons, while the blue rectangle shows the predicted Helitron hairpins. (B) Neighbor joining tree for sequences in A, with 1,000 bootstrap replicates. (C) Helitrons in Arabidopsis acquire new 5′ termini. The pink rectangle shows the 5′ TC of these Helitrons.
In Fig. 4C, sequences 1 through 5 appear to represent Helitrons that are now recognizing a new 5′ start site without any mutation to the normal 5′ start sites. That 1–5 are recently derived is supported by the fact that Helitrons in sequences 1–5 have 96–98% similarity, while the Helitrons represented by sequences 6–11 exhibit 88–96% similarity.
Hence, it appears that Helitrons can acquire flanking sequences by recognizing a new 5′ start site or a new 3′ termination site. We observed similar end sequence acquisitions for Helitron families in other species, including Hup in rice and Hip in sorghum (data not shown).
Discussion
Efficient Discovery of Helitron Transposable Elements.
HelSearch proved superior to any of the 5 previous methods of identifying Helitrons. In the A. thaliana genome, where Helitrons are fairly well studied, new Helitron families were still identified. For genomes where Helitrons have been identified but are not very well characterized such as Medicago and rice, the new approach uncovered a large number of families that were not previously detected.
The HelSearch approach yielded 3 false positives in the rice genome, all LTR retrotransposons having the same terminal structures as Helitrons. Ten previously identified Helitrons were missed in the Arabidopsis genome, 20 in the C. elegans genome, and 2 in the rice genome. All of the false negatives from the Arabidopsis and rice genomes are putative autonomous elements, and thus are easily detected by a homology-based search for Helitron-encoded proteins. Those missed from the C. elegans genome have an unusual Helitron 3′ end structure (namely, no gap between the predicted hairpin and the conserved 3′ end sequences). In most of the previous Helitron discovery procedures, many intact elements were missed because they have low-copy numbers, have accumulated many mutations including big insertions/deletions, or are unusually large in size (8–20 kb). It can be difficult to build multiple sequence alignments because of these Helitron properties. The sensitivity of the structure-based approach (correctly identified/(correctly identified + false negatives)) is 93%, and the specificity (correctly identified/(correctly identified + false positives)) is 99%.
There are several reasons why all other techniques for Helitron discovery have been less sensitive and/or more error prone. A Rep/helicase protein-based search yields a large number of false negatives, because the majority of Helitrons are nonautonomous elements. A similarity-based search will not identify any new families and will thus work poorly in newly studied genomes. Such programs (31, 33) only capture variations within the known families and are likely to provide incorrect annotation on nested Helitrons. A repeat-based search requires extensive manual curation to identify Helitron families, an overwhelming task in large genomes with substantial DNA repetition such as sorghum or maize, and misses the great majority of families because they have a low-copy number.
On the basis of the overall sensitivity and specificity, the structure-based approach to identify Helitron elements is quite successful and especially useful to identify Helitron elements in a newly characterized genome. However, because at least 2 copies are needed to make an alignment, single copy Helitrons will be missed. Finally, the HelSearch program does not identify Helitron fragments for families that contain no intact copies. As with most informatic approaches, a full set of tools are best used to provide a comprehensive discovery process. HelSearch, accompanied by homology searches to Rep/Helicase and known Helitrons, would be an especially comprehensive strategy.
Distributions of Helitrons Within and Between Genomes.
In all 3 genomes where these elements could be comprehensively mapped, those with both near-complete sequence descriptions and a large number of Helitrons, it was observed that Helitrons preferentially accumulate in gene-poor regions. It is not clear whether this is caused by insertion specificities, possibly slower rates of DNA removal in gene-poor heterochromatic regions, and/or selection against Helitron insertions in gene-rich regions. These questions can best be answered in a genome with highly active Helitrons, so that de novo insertions could be analyzed before the action of natural selection or sequence degradation and removal.
It is apparent that different eukaryotic genomes can accumulate very different numbers of Helitrons. In this study, the genomes of sorghum (≈750 Mb), Medicago (≈250 Mb sequenced out of ∼460 Mb), rice (≈400 Mb), Arabidopsis (≈120 Mb), Drosophila (≈150 Mb), and C. elegans (≈100 Mb) were predicted to contain a respective minimum of 22, 2, 8, 2, 0, and 2 Mb of Helitrons. None of these elements appear to be major contributors to genome size in any species studied. Moreover, their absolute quantities do not correlate with genome size, indicating that the host characteristics that allow different levels and rates of LTR retrotransposon amplification and removal (42) do not act on Helitrons in an absolutely parallel manner.
Helitron Properties.
Although all of the eukaryotic genomes investigated had a wealth of possible Helitron ends, as indicated by the presence of the short terminal consensus sequences and a nearby 3′ hairpin, it was found that Helitrons tended to have hairpins with a high predicted Tm. In the rolling-circle transposition model (1), helicase unwinds the double-stranded DNA and the hairpin serves as a stop signal to terminate the transposition event. A high melting temperature may allow a Helitron hairpin to serve as a particularly powerful stop signal for transcription and/or rolling-circle replication. Roles for the non-Helitron short hairpins deserve to be investigated, although it should be noted that the randomly reconstructed Arabidopsis sequence also yielded many such short hairpins (about one-third as many as seen in the real Arabidopsis genome). Thus, some may have no function but may be an unavoidable outcome of other issues of sequence arrangement. It should be noted, however, that HelSearch's requirement for identification of 2 of the same 3′ ends to prove an element was a Helitron will mean that some single copy intact Helitrons are missed. Hence, it is possible that all high Tm hairpins of the approximate size found in Helitrons are actually associated with Helitrons. It is also true, from the results with rice and sorghum, that some fairly low Tm hairpins can function in Helitrons, but these may often be newly created elements ends that have not yet been fully selected for a high Tm.
All transposable elements exhibit a degree of insertion specificity, some for specific sequences but more commonly (at least in eukaryotes) for a specific set of chromatin-associated proteins, like silencers or RNA polymerase subunits. Because of its mechanism of rolling-circle transposition, it is possible that the A/T-rich insertion specificity observed for Helitrons would facilitate helicase unwinding associated with the next rounds of transposition. Interestingly, the AT-richness bias is mostly for the region 3′ to the insertion site. This should give Helitrons a bias not only for the region of their insertion but also for their orientation.
Element Evolution and the Acquisition of New Element Sequences.
One of the great mysteries of Helitron function is how they acquire new internal sequences. When these sequences are parts of genes, the acquisition has an increased potential for the creation of a new gene. Studies in maize suggested that gene fragments are sequentially acquired, perhaps during transposition, and can occur at both ends (20). The process observed for both 3′ and 5′ end sequence capture in this study suggested the possibility that Helitrons may skip the original end to thereby acquire new sequences. Of course, this may not be the only way for Helitrons to acquire new sequences. For instance, an integrase like that seen in integrons might initiate a site-specific recombination event that would lead to some gene capture events (43), although an integrase of this type has not yet been reported to be encoded within any Helitrons.
The rolling-circle transposition model proposes that the hairpin serves as a stop signal during Helitron transposition. With the presence of other Helitrons in a genome, and other Helitron-end like sequences, it is likely that an end-like sequence could be acquired from many genomic locations, including from nearby Helitrons to create chimeric elements.
Future Prospects.
For Helitrons, the issues of transposition mechanism, gene acquisition processes, and the fates of acquire gene fragments remain unresolved. Future research will need to approach these issues and can be done best in species that have a high likelihood of containing active Helitrons. At this time, maize is a particularly strong candidate for an optimal species for these studies because of the presence of recently created mutations (14, 15) and the abundance of haplotype variation associated with Helitron presence/absence (19). The use of HelSearch on the maize genome is underway (L.Y. and J.L.B., unpublished results) and is yielding a great wealth of new Helitrons for functional, structural, and evolutionary analysis.
Methods
Structure-Based Helitron Identification.
A. thaliana genomic sequence build 6 (TAIR6), M. truncatula sequence version 1.0, O. sativa ssp. japonica cultivar Nipponbare sequence version 4.0, S. bicolor sequence Sbi1 assembly, C. elegans sequence build WS144, and D. melanogaster sequence build 4.1 were downloaded to screen for Helitrons. A random sequence assembly was also generated with the same genome size and GC content as the A. thaliana genome by randomly rearranging each nucleotide into a full genome pseudomolecule.
The program “HelSearch” was designed to search for CTRRT in genomic sequence first. To narrow the results, insertion site T was included in this search. The proposed Helitron end (helend) structure is composed of a minimum of 6 hairpin pairs (2 mismatches allowed) upstream of the CTRR, a 2- to 4-bp loop, and 5–8 bp between the hairpin and CTRR. The program was developed in PERL to search for these features in a given genome. Identified candidate helends were grouped together by their hairpin structure. Flanking sequences were obtained for each helend, and multiple alignments by CLUSTALW were performed for those sequences within each group. These alignments were inspected manually to define boundaries of Helitron elements. Two or more sequences with a clear 5′ end at the TC dinucleotide and clear 3′ boundary were defined as Helitron elements (Fig. 1B). BLASTX screening of the NCBI nonredundant protein database was used to find gene fragments acquired by Helitrons, with a required expect value of e-10, or e-5 if a homology was found in a different species (self hits, hypothetical proteins, and tranposase proteins were ignored).
The HelSearch program can be downloaded at http://lxyang.myweb.uga.edu/helsearch1.0.tar.gz and http://sourceforge.net/projects/helsearch/. All Helitron sequences described in this article can be downloaded at http://lxyang.myweb.uga.edu/All_Helitrons_AT_CE_MT_OS_SB.tar.gz.
Helitron Family and Subfamily Assignment.
There is no current knowledge involving Helitron cis or trans activation, so it is not possible to categorize Helitrons as nonautonomous or autonomous members that respond to the same transposition function. So, a family classification was instead assigned as a description of general ancestry, wherein sequences with the most similar 3′ ends (30 bp with at least 80% identity) were classified as members of the same family and sequences with the most similar 5′ ends (30 bp with at least 80% identity) were classified as members of the same subfamily. A short word starting with “H” was assigned as the name for each Helitron family. The same family name, when used for elements from different species, indicates that the shared name describes elements with the same 3′ end (80% identity over 30 bp).
Helitron Properties.
Predicted melting temperatures of hairpins were calculated by the melt program in the UNAFold 3.3 software package (44). The program was run on Windows. Parameters were set as follows: DNA molecule, sodium concentration 1, magnesium concentration 0. Flanking sequences (50 bp both upstream and downstream) of all intact Helitron insertion sites were used to calculate base composition, with PICTOGRAM.
Helitron Abundance.
The HelSearch program identifies only intact elements (i.e., those with both a conserved 3′ end and a conserved 5′ end). To find the genome contribution of all Helitron elements, both intact and fragmented, in a given genome, a BLAST search against an entire genome was performed using all intact elements. Hits with at least 100 bp of 80% identity were counted to calculate genome contribution. The total number of 3′ ends (30 bp with at least 80% identity to 3′ ends of all intact elements) was used to estimate the minimum total number of elements in a given genome.
Supplementary Material
Acknowledgments.
We thank Dr. Renyi Liu and Dr. Clementine Vitte for advice and training on issues regarding the discovery of transposable elements, gene fragments, and genes in plant genome sequence data. This research was supported by National Science Foundation Grant DBI 0607123.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905563106/DCSupplemental.
References
- 1.Kapitonov V, Jurka J. Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci USA. 2001;98:8714–8719. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mendiola M, Bernales I, Cruz F. Differential roles of the transposon termini in IS91 transposition. Proc Natl Acad Sci USA. 1994;91:1922–1926. doi: 10.1073/pnas.91.5.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stenger D, Revington G, Stevenson M, Bisaro D. Replicational release of geminivirus genomes from tandemly repeated copies: Evidence for rolling-circle replication of a plant viral DNA. Proc Natl Acad Sci USA. 1991;88:8029–8033. doi: 10.1073/pnas.88.18.8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murad L, et al. The origin and evolution of geminivirus-related DNA sequences in Nicotiana. Heredity. 2004;92:352–358. doi: 10.1038/sj.hdy.6800431. [DOI] [PubMed] [Google Scholar]
- 5.Doutriaux M, Couteau F, Bergounioux C, White C. Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol Gen Genet. 1998;257:283–291. doi: 10.1007/s004380050649. [DOI] [PubMed] [Google Scholar]
- 6.Surzycki S. Characterization of repetitive DNA elements in Arabidopsis. J Mol Evol. 1999;48:684–691. doi: 10.1007/pl00006512. [DOI] [PubMed] [Google Scholar]
- 7.Le Q, Wright S, Yu Z, Bureau T. Transposon diversity in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:7376–7381. doi: 10.1073/pnas.97.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapitonov V, Jurka J. Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica. 1999;107:27–37. [PubMed] [Google Scholar]
- 9.Rensing S, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 10.Hood M. Repetitive DNA in the automictic fungus Microbotryum violaceum. Genetica. 2005;124:1–10. doi: 10.1007/s10709-004-6615-y. [DOI] [PubMed] [Google Scholar]
- 11.Poulter R, Goodwin T, Butler M. Vertebrate helentrons and other novel Helitrons. Gene. 2003;313:201–212. doi: 10.1016/s0378-1119(03)00679-6. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Q, et al. Helitron transposons on the sex chromosomes of the Platyfish Xiphophorus maculatus and their evolution in animal genomes. Zebrafish. 2006;3:39–52. doi: 10.1089/zeb.2006.3.39. [DOI] [PubMed] [Google Scholar]
- 13.Pritham E, Feschotte C. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc Natl Acad Sci USA. 2007;104:1895–1900. doi: 10.1073/pnas.0609601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lal S, Giroux M, Brendel V, Vallejos C, Hannah L. The maize genome contains a Helitron insertion. Plant Cell. 2003;15:381–391. doi: 10.1105/tpc.008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta S, Gallavotti A, Stryker G, Schmidt R, Lal S. A novel class of Helitron-related transposable elements in maize contain portions of multiple pseudogenes. Plant Mol Biol. 2005;57:115–127. doi: 10.1007/s11103-004-6636-z. [DOI] [PubMed] [Google Scholar]
- 16.Lai J, Li Y, Messing J, Dooner H. Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc Natl Acad Sci USA. 2005;102:9068–9073. doi: 10.1073/pnas.0502923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Messing J. Maize haplotype with a helitron-amplified cytidine deaminase gene copy. BMC Genet. 2006;7:52–64. doi: 10.1186/1471-2156-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Dooner H. Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc Natl Acad Sci USA. 2006;103:17644–17649. doi: 10.1073/pnas.0603080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgante M, et al. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 20.Brunner S, Pea G, Rafalski A. Origins, genetic organization and transcription of a family of nonautonomous helitron elements in maize. Plant J. 2005;43:799–810. doi: 10.1111/j.1365-313X.2005.02497.x. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert W. The exon theory of genes. Cold Spring Harb Symp Quant Biol. 1987;52:901–905. doi: 10.1101/sqb.1987.052.01.098. [DOI] [PubMed] [Google Scholar]
- 22.Bureau T, White S, Wessler S. Transduction of a cellular gene by a plant retroelement. Cell. 1994;77:479–480. doi: 10.1016/0092-8674(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 23.Jin Y, Bennetzen JL. Integration and nonrandom mutation of a plasma membrane proton ATPase gene fragment within the Bs1 retroelement of maize. Plant Cell. 1994;6:1177–1186. doi: 10.1105/tpc.6.8.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmgren M. Capturing of host DNA by a plant retroelement: Bs1 encodes plasma membrane H+-ATPase domains. Plant Mol Biol. 1994;25:137–140. doi: 10.1007/BF00023232. [DOI] [PubMed] [Google Scholar]
- 25.Wang W, et al. High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell. 2006;18:1791–1802. doi: 10.1105/tpc.106.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talbert L, Chandler VL. Characterization of a highly conserved sequence related to mutator transposable elements in maize. Mol Biol Evol. 1988;5:519–529. doi: 10.1093/oxfordjournals.molbev.a040510. [DOI] [PubMed] [Google Scholar]
- 27.Jiang N, Bao Z, Zhang X, Eddy S. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 28.Bennetzen JL. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr Opin Genet Dev. 2005;15:621–627. doi: 10.1016/j.gde.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 29.Arkhipova I, Meselson M. Diverse DNA transposons in rotifers of the class Bdelloidea. Proc Natl Acad Sci USA. 2005;102:11781–11786. doi: 10.1073/pnas.0505333102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi J, Hoshino A, Park K, Park I, Iida S. Spontaneous mutations caused by a Helitron transposon, Hel-It1, in morning glory, Ipomoea tricolor. Plant J. 2007;49:924–934. doi: 10.1111/j.1365-313X.2006.03007.x. [DOI] [PubMed] [Google Scholar]
- 31.Du C, Caronna J, He L, Dooner H. Computational prediction and molecular confirmation of Helitron transposons in the maize genome. BMC Genomics. 2008;9:51–60. doi: 10.1186/1471-2164-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tempel S, Nicolas J, El Amrani A, Couee I. Model-based identification of Helitrons results in a new classification of their families in Arabidopsis thaliana. Gene. 2007;403:18–28. doi: 10.1016/j.gene.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 33.Sweredoski M, DeRose-Wilson L, Gaut B. A comparative computational analysis of nonautonomous Helitron elements between maize and rice. BMC Genomics. 2008;9:467–479. doi: 10.1186/1471-2164-9-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bao Z, Eddy S. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma D, Issac B, Raghava G, Ramaswamy R. Spectral repeat finder (SRF): Identification of repetitive sequences using Fourier transformation. Bioinformatics. 2004;20:1405–1412. doi: 10.1093/bioinformatics/bth103. [DOI] [PubMed] [Google Scholar]
- 36.Hollister J, Gaut B. Population and evolutionary dynamics of Helitron transposable elements in Arabidopsis thaliana. Mol Biol Evol. 2007;24:2515–2524. doi: 10.1093/molbev/msm197. [DOI] [PubMed] [Google Scholar]
- 37.Wright S, Agrawal N, Bureau T. Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res. 2003;13:1897–1903. doi: 10.1101/gr.1281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng Q, et al. Sequence and analysis of rice chromosome 4. Nature. 2002;420:316–320. doi: 10.1038/nature01183. [DOI] [PubMed] [Google Scholar]
- 39.Waterston R. Genome sequence of the nematode C. elegans: A platform for investigating biology. The C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 40.Surzycki S, Belknap W. Repetitive-DNA elements are similarly distributed on Caenorhabditis elegans autosomes. Proc Natl Acad Sci USA. 2000;97:245–249. doi: 10.1073/pnas.97.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duret L, Marais G, Biemont C. Transposons but not retrotransposons are located preferentially in regions of high recombination rate in Caenorhabditis elegans. Genetics. 2000;156:1661–1669. doi: 10.1093/genetics/156.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vitte C, Bennetzen JL. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc Natl Acad Sci USA. 2006;103:17638–17643. doi: 10.1073/pnas.0605618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall R, Collis C. Mobile gene cassettes and integrons: Capture and spread of genes by site-specific recombination. Mol Micro. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 44.Markham N, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;(33):577–581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.