BIOPHYSICS AND COMPUTATIONAL BIOLOGY Correction for “Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs,” by Shunichi Kosugi, Masako Hasebe, Masaru Tomita, and Hiroshi Yanagawa, which appeared in issue 25, June 23, 2009, of Proc Natl Acad Sci USA (106:10171–10176; first published June 11, 2009; 10.1073/pnas.0900604106).
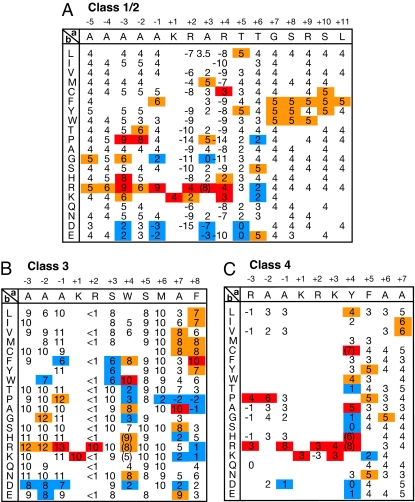
The authors note that on page 10172, the legend for Fig. 1 appeared incorrectly in part. The fourth sentence, “Activity scores were determined as in Fig. S1 -GUS-GFP reporters fused to NLSs with scores (1 and 2), (3–5), (6 and 7), and (8–10) exhibit nuclear, partially nuclear, nuclear plus cytoplasmic, and cytoplasmic phenotypes, respectively” should instead read “Activity scores were determined as in Fig. S1 -GUS-GFP reporters fused to NLSs with scores (1 and 2), (3–5), (6 and 7), and (8–10) exhibit cytoplasmic, nuclear plus cytoplasmic, partially nuclear, and nuclear phenotypes, respectively.” The figureand its corrected legend appear below.
Fig. 1.
Activity-based profiles of classical and noncanonical classes of NLSs optimized for yeast. (A) A profile of a classical class 2 monopartite NLS. A single amino acid residue within the template sequence indicated at the top in the matrix was replaced with various other residues indicated in the left column, and the nuclear import activity was assayed in yeast. Activity scores were determined as in Fig. S1 -GUS-GFP reporters fused to NLSs with scores (1 and 2), (3–5), (6 and 7), and (8–10) exhibit cytoplasmic, nuclear plus cytoplasmic, partially nuclear, and nuclear phenotypes, respectively. This template NLS has an activity score of 4, a middle level of NLS activity. Scores with higher, slightly higher, and lower activities than an average value for each position are shown in red, orange, and blue, respectively. At several mutational positions, modified templates with a different level of basal activity were used to obtain more dispersed scores. Blanks represent undetermined scores. A value that may overlap the scores of class 4 NLSs is given in parentheses. (B) A profile of noncanonical class 3 monopartite NLS. Values that may overlap the scores of class 2 NLS are given in parentheses. (C) A profile of a noncanonical class 4 monopartite NLS.