Abstract
Related studies showed that the protein PSF represses proto-oncogene transcription, and VL30–1 RNA, a mouse noncoding retroelement RNA, binds and releases PSF from a proto-oncogene, activating transcription. Here we show that this mechanism regulates tumorigenesis in human cells, with human RNAs replacing VL30–1 RNA. A library of human RNA fragments was used to isolate, by affinity chromatography, 5 noncoding RNA fragments that bind to human PSF (hPSF), releasing hPSF from a proto-oncogene and activating transcription. Each of the 5 RNA fragments maps to a different human gene. The tumorigenic function of the hPSF-binding RNAs was tested in a human melanoma line and mouse fibroblast line, by determining the effect of the RNAs on formation of colonies in agar and tumors in mice. (i) Expressing in human melanoma cells the RNA fragments individually promoted tumorigenicity. (ii) Expressing in human melanoma cells a shRNA, which causes degradation of the endogenous RNA from which an RNA fragment was derived, suppressed tumorigenicity. (iii) Expressing in mouse NIH/3T3 cells the RNA fragments individually resulted in transformation to tumorigenic cells. (iv) A screen of 9 human tumor lines showed that each line expresses high levels of several hPSF-binding RNAs, relative to the levels in human fibroblast cells. We conclude that human hPSF-binding RNAs drive transformation and tumorigenesis by reversing PSF-mediated repression of proto-oncogene transcription and that dysfunctional regulation of human hPSF-binding RNA expression has a central role in the etiology of human cancer.
The protein PSF (1) contains a DNA-binding domain (DBD) that binds to the regulatory region of a proto-oncogene and represses transcription, and 2 RNA-binding domains (RBDs) that bind VL30–1 RNA, releasing PSF from a repressed proto-oncogene and activating transcription (2–5). Mouse and human genomes encode homologous PSF proteins with ≈95% sequence identity, whereas the VL30–1 gene belongs to a family of mouse noncoding retroelement genes (6) that is not present in the human genome (7). To determine whether the PSF/RNA regulatory mechanism functions in human cells, a library of RNA fragments was constructed from the nuclear RNA repertoire of a human tumor cell, and the library was screened by affinity chromatography for RNAs that bind to human PSF (hPSF). The screen identified 5 hPSF-binding noncoding RNA fragments that release hPSF from a repressed proto-oncogene and activate transcription, similar to VL30–1 RNA. Each human RNA fragment maps to a matching sequence in a different human gene. The following experiments show that human hPSF-binding RNAs are involved in the control of tumorigenesis.
Results
Cloning and Mapping Human RNA Fragments That Bind to hPSF Protein.
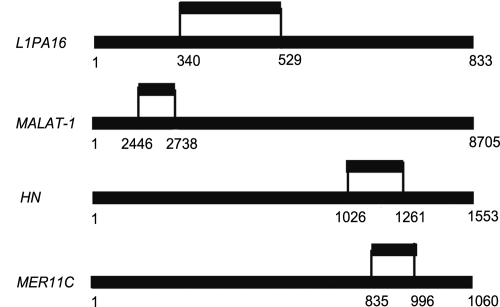
The finding that VL30–1 RNA, a mouse retroelement RNA that is not encoded in the human genome, binds selectively to hPSF protein and reverses repression of proto-oncogene transcription (2–5), prompted a search for human RNAs that have a similar function as VL30–1 RNA. The procedure involved synthesizing a library of RNA fragments from the nuclear RNA repertoire of a human melanoma line and selecting by affinity chromatography RNA fragments that bind to hPSF. The procedure yielded 5 such RNA fragments, 4 of which were mapped, by sequence identity, within 1 of the following genes: L1PA16, a non-LTR retroelement gene (8); MER11C, a LTR retroelement gene (9); MALAT-1, a noncoding gene (10, 11); or HN, a mitochondrial gene coding for the peptide humanin (12); a fifth fragment, not shown in the figure, maps to a region that has not been characterized (Fig. 1 and SI Text).
Fig. 1.
Locations of human hPSF-binding RNA fragments in the human genome, based on sequence identity within a gene. The upper line shows the RNA fragment, and the lower line shows the gene. The L1PA16 gene is on chromosome 3 at 177842604–177843436; the MALAT-1 gene is on chromosome 11 at 65021809–65030513; the HN gene is on chromosome M (mitochondrial) at 1685–3237; and the MER11C gene is on chromosome 11 at 50410308–50411367. A fifth fragment maps to chromosome 6 at 165453538–165453778 within an unidentified gene. Further details are in SI Text.
The sequence of the HN RNA fragment is 100% identical to a sequence in the mitochondrial 16S ribosomal RNA gene and is 85% identical to positions 21947595–21947823 on nuclear chromosome 17. Further testing showed that the HN RNA fragment is derived from the mitochondrial HN RNA and not from the nuclear RNA (SI Text). The mitochondrial HN RNA might be translocated to the nucleus or derived from a mitochondrial contamination in the nuclear preparation.
Release of hPSF from the GAGE6 Regulatory Region by the hPSF-Binding RNA Fragments.
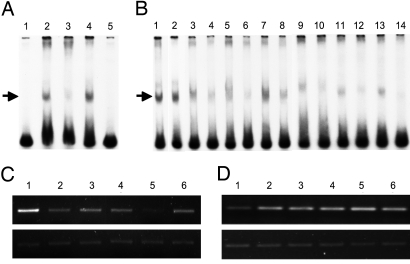
Earlier studies showed that hPSF binds selectively to a 61-bp sequence at the 5′-end of the regulatory region of the human proto-oncogene GAGE6 (4, 13), which is used here as the target DNA for hPSF in all experiments. The 5 human hPSF-binding RNA fragments release hPSF from the GAGE6 regulatory DNA in vitro and in vivo, and activate GAGE6 transcription (Fig. 2).
Fig. 2.
Release of hPSF from the GAGE6 regulatory DNA by hPSF-binding RNAs. (A) Binding of hPSF to GAGE6 regulatory DNA. 32P-labeled GAGE6 regulatory DNA was mixed with a nuclear extract of YU-SIT1 melanoma cells, and 20 min later an anti-PSF monoclonal, or an anti-ATCB monoclonal antibody that does not bind hPSF, was added. The samples were incubated for 20 min at room temperature and fractionated by PAGE, and the gel was autoradiographed. Lane 1: GAGE6 regulatory DNA; Lane 2: GAGE6 regulatory DNA + nuclear extract; Lane 3: GAGE6 regulatory DNA + nuclear extract + anti-PSF antibody; Lane 4: GAGE6 regulatory DNA + nuclear extract + anti-ATCB antibody; Lane 5: GAGE6 regulatory DNA + E. coli protein. The high mobility bands at the bottom of the gel indicate free GAGE6 regulatory DNA, and the low mobility bands marked by an arrow indicate the GAGE6 regulatory DNA/hPSF complex. (B) Release of hPSF from GAGE6 regulatory DNA in vitro by hPSF-binding RNAs. 32P-labeled GAGE6 regulatory DNA was incubated with a nuclear extract of YU-SIT1 cells, and 20 min later a hPSF-binding RNA or control RNA was added. Lane 1: GAGE6 regulatory DNA + nuclear extract without added RNA; Lanes 2–14: GAGE6 regulatory DNA + nuclear extract with added RNA; Lane 2: control RNA encoded by pcDNA-3.1; Lanes 3 and 4: mouse VL30–1 RNA; Lanes 5 and 6: L1PA16 RNA fragment; Lanes 7 and 8: MALAT-1 RNA fragment; Lanes 9 and 10: HN RNA fragment; Lanes 11 and 12: unidentified RNA fragment; Lanes 13 and 14: MER11C RNA fragment. The high mobility bands at the bottom of the gel indicate the free GAGE6 regulatory DNA, and the low mobility bands marked by an arrow indicate the GAGE6 regulatory DNA/hPSF complex. The molar ratio of RNA to GAGE6 regulatory DNA was 30 in Lanes 3, 5, 7, 9, 11, and 13, and 100 in Lanes 2, 4, 6, 8, 10, 12, and 14. (C) Release of hPSF from GAGE6 regulatory DNA in vivo by human hPSF-binding RNA fragments. The parental yusac line was stably transfected with plasmid pcDNA-3.1 and plasmid pcDNA-3.1 encoding 1 of the human hPSF-binding RNA fragments, and yusac-pcDNA3.1 and yusac-RNA lines were cloned. An anti-PSF antibody was used to immunoprecipitate hPSF from the cell lines, and the amount of GAGE6 regulatory DNA co-precipitated with hPSF was assayed by semiquantitative PCR. Upper shows the immunoprecipitated GAGE6 regulatory DNA, and Lower shows the amount of total GAGE6 regulatory DNA used to normalize the amount of total DNA in the samples. Lane 1: yusac-pcDNA3.1 control; Lane 2: yusac-L1PA16; Lane 3: yusac-MALAT-1; Lane 4: yusac-HN; Lane 5: yusac-unidentified RNA; Lane 6: yusac-MER11C. (D) Activation of GAGE6 transcription in vivo by human hPSF-binding RNA fragments. The amount of GAGE6 mRNA was determined by semiquantitative RT-PCR. Upper shows the amount of GAGE6 cDNA, and Lower shows the amount of ATCB cDNA used to normalize the total RNA in the samples. The cell lines for Lanes 1–6 are the same as for C.
Tumorigenicity of the Human hPSF-Binding RNA Fragments.
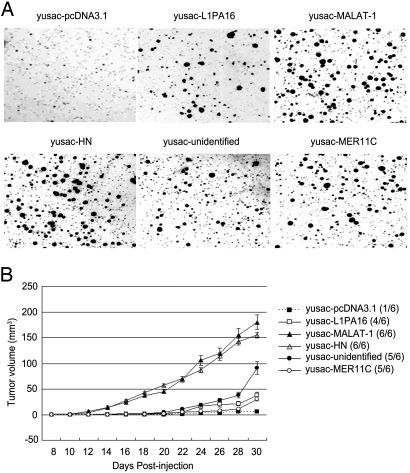
The human melanoma line yusac was transfected with a plasmid pcDNA-3.1 encoding a human hPSF-binding RNA fragment, or with plasmid pcDNA-3.1 as a control, and the transfected cells were tested for colony formation in agar and tumor formation in nude mice. The yusac-pcDNA3.1 control cells formed minute colonies in agar and did not form tumors, or formed small tumors in the mice, in contrast to the yusac-RNA cells that formed large colonies in agar and large tumors in the mice (Fig. 3).
Fig. 3.
Tumorigenic effect of increasing expression of human hPSF-binding RNA fragments in the human melanoma line yusac. The yusac cells were stably transfected with plasmid pcDNA-3.1 as a control or pcDNA-3.1 encoding the indicated human RNA fragment. (A) Colony formation in agar. The agar was seeded with 5 × 103 cells and solidified in petri dishes, and the colonies were stained and photographed after 2 weeks. (B) Tumor formation in nude mice. For each cell line, 3 mice were injected on opposite sides of the neck, using 5 × 105 cells on each side. The number of injection sites that formed tumors is shown to the right (n/6), and the average size of the tumors on each day is shown in the curves on the left. The error bars show the mean value ± SE (n = 6). The procedures for A and B are described in Materials and Methods.
Tumorigenicity of an Intact hPSF-Binding RNA.
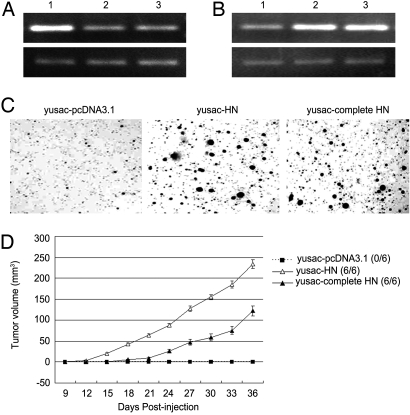
To determine whether an intact hPSF-binding RNA, as well as a fragment of the RNA, is tumorigenic, yusac melanoma cells were transfected with plasmid pcDNA-3.1 encoding the HN RNA fragment or intact HN RNA, or with plasmid pcDNA-3.1 as a control. The transfected cells were tested for binding of hPSF to GAGE6 and transcription of GAGE6, and formation of colonies in agar and tumors in mice. The HN RNA fragment and complete HN RNA showed similar tumorigenicity, except that tumor formation in mice was stronger with the HN RNA fragment than the complete HN RNA, probably because the RNA fragment is transcribed more efficiently (Fig. 4). Another experiment showed that the tumorigenicity of yusac cells decreased when the cells were transfected with a shRNA construct that initiates degradation of HN RNA by an RNAi (Fig. 5). These results indicate that an intact hPSF-binding RNA, as well as a hPSF-binding fragment of the RNA, can drive tumorigenesis.
Fig. 4.
Tumorigenic effect of increasing expression of the hPSF-binding HN RNA fragment or intact HN RNA in the human melanoma line. The yusac cells were stably transfected with plasmid pcDNA-3.1 as a control or pcDNA-3.1 encoding the hPSF-binding HN RNA fragment or the intact HN RNA. (A) Release of hPSF from GAGE6 regulatory DNA. hPSF was immunoprecipitated from the transfected yusac cells with an anti-PSF antibody, and the amount of GAGE6 regulatory DNA co-precipitated with hPSF was determined by semiquantitative PCR. Upper shows the immunoprecipitated GAGE6 regulatory DNA, and Lower shows the amount of total GAGE6 regulatory DNA used to normalize the total DNA in the samples. Lane 1: yusac-pcDNA3.1 control; Lane 2: yusac-HN cells; Lane 3: yusac-complete HN cells. (B) Activation of GAGE6 transcription. The amount of GAGE6 mRNA was determined by semiquantitative RT-PCR. Upper shows the amount of GAGE6 cDNA, and Lower shows the amount of ATCB cDNA used to normalize the total RNA in the samples. The cells in Lanes 1–3 are the same as in A. (C) Colony formation in agar. The agar was seeded with 5 × 103 cells and solidified in petri dishes, and the colonies were stained and photographed after 2 weeks. (D) Tumor formation in nude mice. For each cell line, 3 mice were injected on opposite sides of the neck, using 5 × 105 cells on each side. The number of injection sites that formed tumors is shown to the right (n/6), and the average size of the tumors on each day is shown in the curves on the left. The error bars show the mean value ± SE (n = 6). The procedures for C and D are described in Materials and Methods.
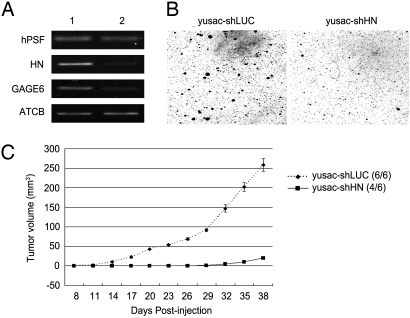
Fig. 5.
Tumor suppression of yusac melanoma cells by an RNAi that degrades a hPSF-binding RNA. The yusac cells were stably transfected with a plasmid encoding a luciferase shRNA as a control (yusac-shLUC) or a HN-specific shRNA that initiates degradation of HN RNA by an RNAi (yusac-shHN). (A) Transcription of hPSF, HN, GAGE6, and ATCB, which is a constitutive RNA standard. Lane 1, yusac-shLUC cells; Lane 2, yusac-shHN cells. (B) Colony formation in agar. The agar was seeded with 1 × 104 cells and solidified in petri dishes, and the colonies were stained and photographed after 4 weeks. (C) Tumor formation in nude mice. For each cell line, 3 mice were injected on opposite sides of the neck, using 2 × 106 cells on each side. The number of injection sites that formed tumors is shown to the right (n/6), and the average size of the tumors on each day is shown in the curves on the left. The error bars show the mean value ± SE (n = 6). The procedures for B and C are described in Materials and Methods.
Transformation of Mouse NIH/3T3 Cells by Human hPSF-Binding RNA Fragments.
The strong conservation of the PSF molecule between humans and mice (95% sequence identity) suggests that human hPSF-binding RNAs could function similarly in mice and human cells. Accordingly, the tumorigenic function of the human RNA fragments was tested in mouse NIH/3T3 fibroblast cells, which can be transfected more efficiently than human fibroblast cells. The NIH/3T3 cells were transfected separately with plasmid pcDNA-3.1 as a control or a plasmid pcDNA-3.1 encoding a hPSF-binding RNA fragment, and the transfected cells were tested for the formation of colonies in agar and tumors in mice. The results show that the NIH/3T3-RNA lines, in contrast to the NIH/3T3-pcDNA3.1 control line, form colonies in agar and tumor-like growths in mice (Fig. 6); histological examination of the growths confirmed the presence of tumor cells. These results indicate that the human hPSF-binding RNA fragments can transform NIH/3T3 cells.
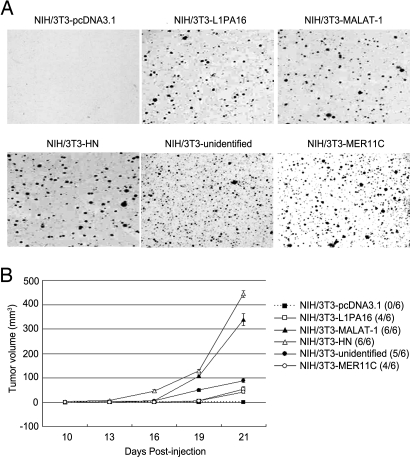
Fig. 6.
Transformation of NIH/3T3 cells by expression of a human hPSF-binding RNA fragment. The NIH/3T3 cells were stably transfected with plasmid pcDNA-3.1 as a control or pcDNA-3.1 encoding the indicated human RNA fragment. (A) Colony formation in agar. The agar was seeded with 1 × 104 cells and solidified in petri dishes, and the colonies were stained and photographed after 3 weeks. (B) Tumor formation in nude mice. For each cell line, 3 mice were injected on opposite sides of the neck, using 2.5 × 105 cells on each side. The number of injection sites that formed tumors is shown to the right (n/6), and the average size of the tumors on each day is shown in the curves on the left. The error bars show the mean value ± SE (n = 6). The procedures for A and B are described in Materials and Methods.
Expression of Human hPSF-Binding RNAs and hPSF in Human Fibroblast Cells and Tumor Cells.
In the next experiment, the relative amounts of the hPSF-binding RNAs were measured by real-time RT-PCR in 2 human fibroblast lines and 9 human tumor lines (Fig. 7), Each tumor line expresses high levels of several hPSF-binding RNAs, as compared to the levels in the fibroblast lines, and the pattern of expression differs among the tumor lines, suggesting that different groups of hPSF-binding RNAs contribute to the tumorigenicity of each tumor line.
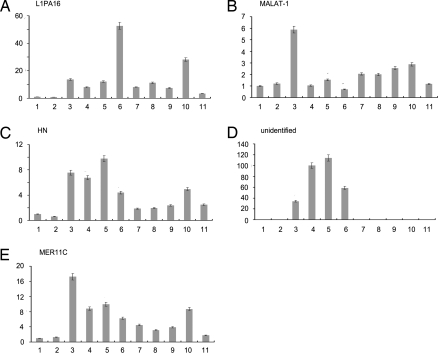
Fig. 7.
Expression of human hPSF-binding RNAs in human cells. (A) L1PA16. (B) MALAT-1. (C) HN. (D) Unidentified. (E) MER11C. Bar 1: BJ: fibroblast. Bar 2: HPF: primary foreskin fibroblast. Bars 3–6: A2058, SKmel28, YU-SIT1, and yusac melanoma lines. Bars 7–9: SMMC-7402, SMMC-7721, and HepG2: hepatocellular carcinoma lines. Bar 10: MCF7: breast tumor line. Bar 11: A549 non-small cell lung tumor line. Bars 2–11 show the amounts of hPSF-binding RNAs relative to the amount in Bar 1. The procedure is described in Materials and Methods.
Because a decrease in hPSF protein, as well as an increase in hPSF-binding RNAs, could contribute to the tumorigenicity of human tumor cells, the relative amounts of hPSF mRNA were measured in the fibroblast and tumor lines (Fig. 8). The amount of hPSF mRNA did not decrease in 8 of the 9 tumor lines tested, compared to the fibroblast lines, and decreased only partially in the remaining tumor line, suggesting that a decrease of hPSF protein does not contribute to the tumorigenicity of human tumor cells.
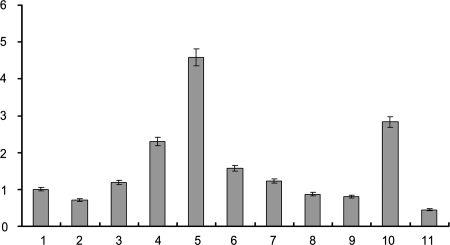
Fig. 8.
Expression of hPSF in human cells. The cells for each bar are the same as in Fig. 7. Bars 2–11 show the amounts of hPSF mRNA relative to the amount in Bar 1.
Discussion
Related studies described a mechanism for reversible regulation of proto-oncogene transcription, involving the protein hPSF that binds to the regulatory region of a proto-oncogene and represses transcription, and VL30–1 RNA, a mouse noncoding retroelement RNA, that binds to hPSF, forming a hPSF/RNA complex that dissociates from a proto-oncogene, activating transcription (2–5). Because VL30–1 RNA is not encoded in the human genome (7), we screened by affinity chromatography a library of RNA fragments, derived from the RNA of a human tumor cell nucleus, for RNAs that bind to hPSF. The screen isolated 5 such human RNAs that release hPSF from a proto-oncogene and activate transcription, similar to VL30–1 RNA (Figs. 1 and 2, and SI Text). Each hPSF-binding RNA fragment maps to a matching sequence in a human gene, of which 4 have been characterized: L1PA16 and MER11c are noncoding retroelement genes, MALAT-1 is a noncoding gene, and HN encodes humanin, a small peptide (8–12); the HN RNA fragment is derived from the noncoding 3′UTR of the HN gene.
The VL30–1 RNA is a 4.9-kb molecule containing 2 short homologous sequences, 1 with 22 bases and another with 30 bases, that are responsible for the binding of VL30–1 RNA to hPSF (3). Accordingly, the human hPSF-binding RNA fragments could contain short sequences that bind to hPSF. A scan of the human RNA fragments failed to identify a conserved sequence that could serve as a common motif for binding to hPSF, which suggests that the hPSF-binding sequences differ among the RNAs, or the binding to hPSF involves another property of the RNAs.
A role for the human hPSF-binding RNAs in tumorigenesis was tested in a human melanoma line. One procedure involved transfecting a plasmid encoding a hPSF-binding RNA fragment or the endogenous RNA from which a fragment was derived; another procedure involved transfecting a plasmid encoding a shRNA that causes degradation of an endogenous hPSF-binding RNA. The tumorigenicity of the transfected melanoma cells was assayed by colony formation in agar and tumor formation in nude mice. The results showed that the first procedure, which increased expression of the encoded RNA fragments or endogenous RNA, increased tumorigenicity (Figs. 3 and 4), and the second procedure, which decreased expression of the targeted endogenous RNA, decreased tumorigenicity (Fig. 5), indicating that hPSF-binding RNAs drive tumorigenesis in human tumor cells.
Another test for a tumorigenic role of human hPSF-binding RNAs was done with the mouse fibroblast line NIH/3T3, which expresses a mouse PSF protein that has ≈95% sequence identity with hPSF. The fibroblast cells were transfected separately with plasmids encoding the human hPSF-binding RNA fragments (NIH/3T3-RNA cells) or a control plasmid (NIH/3T3-pcDNA3.1 cells), each controlled by a CMV promoter, and the transfected cells were assayed for tumorigenicity. The results showed that the NIH/3T3-RNA cells, but not the control cells, were transformed to tumorigenic cells (Fig. 6), providing further support for a tumorigenic role of the human hPSF-binding RNAs. The HN RNA and MALAT-1 RNA fragments showed the strongest tumorigenic effects both in NIH/3T3 cells and a human tumor line (Fig. 3).
A screen of 9 human tumor lines showed that all of the lines express elevated levels of several hPSF-binding RNAs, relative to the levels in human fibroblast lines, (Fig. 7), although each of the RNAs should activate transcription of the same set of proto-oncogenes repressed by hPSF. Transfecting the human melanoma line yusac, which expresses elevated levels of 4 of the hPSF-binding RNAs, with a plasmid encoding a shRNA that causes degradation of 1 of the RNAs, strongly suppressed tumorigenicity of the cells (Fig. 5), suggesting that the hPSF-binding RNAs expressed in a tumor cell function cooperatively.
Tumorigenesis in human tumor cells could be driven by the increased expression of hPSF-binding RNAs (Fig. 7), or by a decreased expression of hPSF, or both. Expression of hPSF mRNA was lower only in 1 tumor line (Fig. 8), suggesting that the hPSF-binding RNAs are principally responsible for driving tumorigenesis in human tumor cells. Earlier studies showed that the hPSF gene in the cervical tumor line HeLa has a deletion of the DNA-binding domain (4), which is the probable cause of tumorigenesis by preventing repression of proto-oncogenes. Further screens of human tumors for mutations that block the synthesis or function of hPSF are needed to determine the significance of hPSF mutations in human cancer.
The hPSF-binding RNA fragment derived from HN RNA was tested by EMSA gel electrophoresis for binding to proteins in a total nuclear extract from a human tumor cell. The results showed an intense radiolabeled HN RNA band at the position of hPSF, as expected, and numerous additional bands at other positions that were much weaker, indicating that HN RNA, and probably other hPSF-binding RNAs, can bind strongly to hPSF and weakly to other proteins. The significance of the weak binding of a human hPSF-binding RNA to proteins other than hPSF is not known.
In conclusion, the study reported here describes a mechanism that controls tumorigenesis in human cells, involving repression of proto-oncogene transcription by hPSF protein and reversal of proto-oncogene repression by human hPSF-binding RNAs. The key variable of the mechanism appears to be an increase in the level of hPSF-binding RNAs, which drives transformation and tumorigenesis. We propose that dysfunctional regulation of human hPSF-binding RNAs has a central role in the etiology of human cancer, and that elucidating the molecular biology of this regulatory mechanism is a major challenge for cancer research.
Materials and Methods
Purification of Human PSF (hPSF) Protein.
The hPSF protein labeled with a His tag was synthesized in E. coli BL21 cells transfected with a pET-28a expression vector (Novagen) encoding the protein, and was purified using TALON affinity resin (BD Clontech).
Cell Lines.
BJ: human fibroblast; HPF: human primary foreskin fibroblasts; A2058, SKmel28, YU-SIT1, and yusac: human melanoma; SMMC-7402, SMMC-7721, and HepG2: human hepatocellular carcinoma; MCF7: human breast cancer; A549: human non-small cell lung cancer; NIH/3T3: mouse fibroblast. The cells were cultured in DMEM or RPMI1640 supplemented with 10% FBS in a 5% CO2 incubator at 37°C.
Construction of a cDNA Library Encoding Human RNA Fragments Derived from the Nuclear RNA Repertoire of the Human Melanoma Line Yusac.
The yusac cells were suspended in hypotonic buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT) and disrupted with 20 strokes of a Dounce homogenizer. The nuclei were isolated by centrifuging for 5 min at 3,300 × g, and the nuclear RNA was extracted and purified by TRIzol reagent (Invitrogen). The cDNAs were synthesized from the RNAs using random hexamers and cloned into the PCR-blunt-TOPO vector (Invitrogen).
Isolation and Cloning of Human Nuclear RNA Fragments that Bind to hPSF.
The RNA fragments encoded by the cDNA library were synthesized using T7 RNA polymerase. The RNA fragments were incubated with purified His-tagged hPSF, which showed a single band at the position of ≈100 kDa in SDS/PAGE, in binding buffer (10 mM HEPES, pH 7.6, 5 mM MgCl2, 1 mM DTT, 100 mM KCl) and gently mixed at 4°C for 3 h. The RNA fragments that bound to His-tagged hPSF were collected on TALON affinity resin and extracted using TRIzol reagent. The cDNAs were synthesized by RT-PCR and then cloned into the PCR-blunt -TOPO vector. The procedure was repeated 5 times. One hundred clones from the final cDNAs pools were sequenced, and 5 individual RNA fragments were identified. cDNA encoding the hPSF-binding RNA fragments were cloned into pcDNA-3.1 vector.
Extraction of Nuclear Extract from YU-SIT1 Cells and Total Protein Extract from E. coli.
The nuclei of YU-SIT1 cells were isolated as described above, equilibrated 30 min in extraction buffer (20 mM HEPES, pH 7.9, 25% glycerol, 1.5 mM MgCl2, 0.6M KCl, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT) with continuous gentle mixing, and then centrifuged at 25,000 × g for 30 min. The supernatant was dialyzed against 50 volume of dialysis buffer (20 mM HEPES, pH 7.9, 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT). The dialysate was centrifuged for 20 min, and the supernatant was used as the nuclear extract. E. coli was lysed by sonication for 5 min followed by 30 min centrifugation at 13,000 × g. The supernatant was dialyzed against 50 volume of dialysis buffer. The dialysate was centrifuged for 20 min at 13,000 × g, and the supernatant was used as the total protein extract of E. coli.
Antibody Supershift Assay and Competitive EMSA.
The 61-bp hPSF-binding sequence at the 5′-end of the GAGE6 regulatory region (4) was labeled using 32P-dCTP. Ten femptomoles of 32P-labeled GAGE6 DNA fragment was incubated with 2 μg of a nuclear extract from YU-SIT1 cells or total extract of E. coli protein at 25°C for 20 min. For the antibody supershift assay, 1 μg of anti-PSF monoclonal antibody (Sigma-Aldrich) or an equal amount of anti-ATCB antibody was added, and the samples were incubated at 25°C for 20 min. For the competitive EMSA experiments, hPSF-binding RNAs were added to GAGE6 regulatory DNA at a RNA/DNA molar ratio of 30 and 100; the RNA transcribed from plasmid pcDNA-3.1 was used as the control. The samples were incubated at 25°C for 20 min and fractionated by 5% PAGE, and the gel was autoradiographed.
Cloning the Complete HN cDNA.
The HN cDNA was synthesized from yusac cells by RT-PCR, using the primers 5′-ataaagcttccaaacccactccaccttact-3′ and 5′-tatctcgagtcttaacaaaccctgttcttgg-3′, and the cDNA was cloned into the restriction sites HindIII and XhoI of plasmid pcDNA-3.1.
Plasmid-Mediated RNA Interference for HN mRNA.
The complementary sense and anti-sense oligonucleotides were synthesized, annealed, and cloned into BamH1/HindIII sites of pGenesil-1 plasmid (Wuhan Genesil Biotechnology Co.). The recombinant plasmid encodes shRNA with a 19-mer stem derived from the RNA target site, and initiates the RNAi in vivo. The sense and anti-sense oligonucleotides for HN shRNA construction were 5′-gatccgcgacctcgatgttggatcattcaagagatgatccaacatcgaggtcgtttttta-3′ and 5′-agcttaaaaaacgacctcgatgttggatcatctcttgaatgatccaacatcgaggtcgcg-3′, and the recombinant plasmid was named pGenesil-shHN. The sense and anti-sense oligonucleotides for luciferase shRNA construction were: 5′-gatccgtagcgcggtgtattatacttcaagagagtataatacaccgcgctactttttta-3′ and 5′-agcttaaaaaagtagcgcggtgtattatactctcttgaagtataatacaccgcgctacg-3′. The recombinant vector was named pGenesil-shLUC and used as the control plasmid. The target sequences are italic.
Stable Transfection of Yusac and NIH/3T3 Cell Lines.
The yusac and NIH/3T3 cells were transfected with pcDNA3.1 plasmids encoding the human hPSF-binding RNA fragments, and stable cell lines expressing higher levels of the RNA fragments (NIH/3T3-RNA and yusac-RNA lines) were established. The control cells were transfected with an empty pcDNA3.1 plasmid (yusac-pcDNA3.1 and NIH/3T3-pcDNA3.1 control lines). To construct the HN shRNA cell line yusac-shHN, the yusac cells were transfected with plasmid pGenesil-shHN, and a stable transfectant was isolated. The control cell line yusac-shLUC was generated by transfecting yusac cells with plasmid pGenesil-shLUC. The transfection reagent Lipofectamine 2000 (Invitrogen) was used for all transfections. The expression levels of the targeted RNAs were assayed by semiquantitative RT-PCR.
Chromatin Immunoprecipitation (ChIP) and Semiquantitative PCR Assay for the Release of hPSF from GAGE6 Regulatory DNA by hPSF-Binding RNAs.
The ChIP assay was done using yusac-pcDNA3.1 and yusac-RNA cells as described (4). A monoclonal anti-PSF antibody (Sigma-Aldrich) was used to immunoprecipitate hPSF, and the GAGE6 regulatory DNA in the precipitate was analyzed using semiquantitative PCR, using the primers 5′-gccttctgcaaagaagtcttgcgc-3′ and 5′-atgcgaattcgaggctgaggcagacaat-3′. The amount of total GAGE6 regulatory DNA was used to normalize the amount of total DNA in the samples.
Semiquantitative RT-PCR Assay for GAGE6 Transcription in Yusac Cells.
The yusac-RNA lines, yusac-shHN line, and control lines were tested for the effect of hPSF-binding RNAs on GAGE6 transcription using semiquantitative RT-PCR. The GAGE6 primers were 5′-ggtgtgagtgtgaagatggtcctga-3′ and 5′-ccaacataggagcagcctgcatcat-3′. The hPSF primers were 5′-gcggaggagcaatgaacatggga-3′ and 5′-cacttcccatcatggaaccactc-3′. The ATCB control primers were 5′-ctcaccgagcgcggctaca-3′ and 5′-ctcctgcttgctgatccacat-3′.
Colony Formation in Soft-agar.
The cells were suspended in 1 mL of 0.3% melted agar in DMEM + 10% FBS and plated in 60-mm dishes overlayed with 0.5% agar in the same medium. The resulting colonies were stained with nitro blue tetrazolium and scanned 1 day later with an Epson Perfection 3200 scanner.
Tumorigenesis Assays.
Nude mice 5 weeks old were maintained under pathogen-free conditions. The mice were anesthetized and injected s.c. into bilateral sites on the neck with cells in 0.1 mL PBS using a 25-gauge needle. Tumor size was monitored every second or third day, and tumor volume was calculated by the formula 4/3π(radius)3. The mice were killed at the end of an experiment, and the tumors were sectioned for histology.
Expression of hPSF-Binding RNAs and hPSF in Human Cells by Real-Time RT-PCR.
The human fibroblast cells and tumor cells listed above were tested for expression of the hPSF-binding RNAs and hPSF mRNA by real-time RT-PCR, and the results were recorded as threshold cycle numbers (Ct), which were normalized against an internal control (ATCB mRNA) and calculated as fold changes. The RT-PCR primers were as follows: (i) L1PA16: 5′-tattctttatccagtccaccac-3′ and 5′-gaaataaatcattctacccaaaag-3′; (ii) MALAT-1: 5′-cggaagtaattcaagatcaagag-3′ and 5′-actgaatccacttctgtgtagc-3′; (iii) HN: 5′-gcaagacgagaagaccctatg-3′ and 5′-agtagttcgctttgactggtga-3′; (iv) MER11C: 5′-aaacttgctgattttgtggctt-3′ and 5′-tgttggctggtctgtgaaata-3′; (v) unidentified: 5′-tttgacacgagtgactgtattttgaa-3′ and 5′-atttatgaattgtcacaggacctct-3′; (vi) ATCB: 5′-ctcctccctggagaagagcta-3′ and 5′-ccttctgcatcctgtcggcaa-3′; (vii) hPSF: 5′-gcggaggagcaatgaacatggga-3′ and 5′-cacttcccatcatggaaccactc-3′
Supplementary Material
Acknowledgments.
This work was supported by National Natural Science Foundation of China Grants 30700459 and 90919005, Ph.D. Programs Foundation of the Ministry of Education of China Grant 20070610170, and Scientific Research Foundation for Returned Scholars, Ministry of Education of China Grant 20071108-18-9.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906005106/DCSupplemental.
References
- 1.Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Develop. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 2.Song X, et al. Retroviral-mediated transmission of a mouse VL30 RNA to human melanoma cells promotes metastasis in an immunodeficient mouse model. Proc Natl Acad Sci USA. 2002;99:6269–6273. doi: 10.1073/pnas.092112199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song X, Sui A, Garen A. Binding of mouse VL30 retrotransposon RNA to PSF protein induces genes repressed by PSF: Effects on steroidogenesis and oncogenesis. Proc Natl Acad Sci USA. 2004;101:621–626. doi: 10.1073/pnas.0307794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song X, Sun Y, Garen A. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc Natl Acad Sci USA. 2005;102:12189–12193. doi: 10.1073/pnas.0505179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garen A, Song X. Regulatory roles of tumor-suppressor proteins and noncoding RNAs in cancer and normal cell functions. Int J Cancer. 2008;122:1687–1689. doi: 10.1002/ijc.23285. [DOI] [PubMed] [Google Scholar]
- 6.French NS, Norton JD. Structure and functional properties of mouse VL30 retrotransposons. Biochim Biophys Acta. 1997;1352:33–47. doi: 10.1016/s0167-4781(97)00009-2. [DOI] [PubMed] [Google Scholar]
- 7.Song X, Sun Y, Garen A Correction. Proc Natl Acad Sci USA. 2005;102:16905. [Google Scholar]
- 8.Smit AF, Toth G, Riggs AD, Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan DJ, Jurka J, Solus JF, Duncan CH. Medium reiteration frequency repetitive sequences in the human genome. Nucleic Acids Res. 1991;19:4731–4738. doi: 10.1093/nar/19.17.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji P, et al. MALAT-1, a novel noncoding RNA, and thymosin b4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 11.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2006;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 12.Tajima H, et al. Evidence for in vivo production of humanin peptide, a neuroprotective factor against Alzheimer's disease-related insults. Neuroscience Lett. 2002;324:227–231. doi: 10.1016/s0304-3940(02)00199-4. [DOI] [PubMed] [Google Scholar]
- 13.DeBacker O, et al. Characterization of the GAGE genes that are expressed in various human cancers and in normal testis. Cancer Res. 1999;59:3157–3165. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.