Abstract
Primary open-angle glaucoma (POAG) is the major type of glaucoma. To discover genetic markers associated with POAG, we examined a total of 1,575 Japanese subjects in a genome-wide association study (stage 1) and a subsequent study (stage 2). Both studies were carried out at a single institution. In the stage 1 association study, we compared SNPs between 418 POAG patients and 300 control subjects. First, low-quality data were eliminated by a stringent filter, and 331,838 autosomal SNPs were selected for analysis. Poorly clustered SNPs were eliminated by a visual assessment, leaving 255 that showed a significant deviation (P < 0.001) in the allele frequency comparison. In the stage 2 analysis, we tested these 255 SNPs for association in DNA samples from a separate group of 409 POAG and 448 control subjects. High-quality genotype data were selected and used to calculate the combined P values of stages 1 and 2 by the Mantel–Haenszel test. These analyses yielded 6 SNPs with P < 0.0001. All 6 SNPs showed a significant association (P < 0.05) in stage 2, demonstrating a confirmed association with POAG. Although we could not link the SNPs to the annotated gene(s), it turned out that we have identified 3 genetic loci probably associated with POAG. These findings would provide the foundation for future studies to build on, such as for the metaanalysis, to reveal the molecular mechanism of the POAG pathogenesis.
Keywords: diagnosis, SNP, GWAS, meta-analysis, glaucoma genetics
Glaucoma is a neurodegenerative disease of the eye, and it is one of the leading causes of blindness worldwide (1). It is characterized by a specific pattern of optic nerve degeneration and visual field defects. The diagnosis of glaucoma preceding the development of visual field defects is commonly made by observing optic nerve degeneration, which manifests, on fundus examination, as an enlarged optic disc cup and a damaged retinal nerve-fiber layer. Because early drug treatment, just after the onset of visual field damage, is quite effective in slowing the irreversible progression of glaucoma (2–4), routine fundus examinations of the optic nerve and visual field tests are desirable. However, because of the restriction of the medical costs and infrastructure to set a routine examination, especially in the preclinical state of glaucoma, it is necessary to create an alternative method for the early diagnosis of glaucoma.
Glaucoma shows familial aggregation, and its prevalence varies among different ethnic groups (5, 6). This epidemiological evidence strongly suggests that genetic factors play a significant role in the pathogenesis of glaucoma (5, 6). Indeed, previous linkage analyses implicated the genes for myocilin (MYOC) (7), optineurin (OPTN) (8), and WD-repeat domain 36 (WDR36) (9) in the pathogenesis of primary open-angle glaucoma (POAG), the major form of glaucoma. However, further analyses revealed that the frequencies of mutations in these genes were moderate and were associated with POAG in only a small fraction of patients (5). Therefore, the tasks remain to discover authentic and widely associated genetic factors for POAG and to use them for practical diagnostic or medical applications.
To analyze hundreds of thousands of SNPs in a genome-wide association study (GWAS) for POAG, we used DNA chips. This method permits the identification of genetic loci and genes associated with complex human traits, without a priori knowledge of the function or presumptive involvement of any gene in the disease pathway. To date, SNPs associated with over 40 kinds of diseases have been identified (for reviews, see refs. 10, 11). In ocular diseases, an association of lysyl oxidase-like 1 (LOXL1) gene polymorphisms with a minor type of secondary glaucoma, exfoliation glaucoma, was recently shown (12). Moreover, SNPs on complement factor H (CFH), C2-CFB, and a hypothetical gene, LOC387715, are associated with the onset of age-related macular degeneration (AMD) (13–15). Although it is still unclear how genes of the systemic immune system are involved in the pathogenesis of a local tissue disease, an additive effect was seen when CFH SNPs were combined with SNPs on the other susceptibility genes in predicting the risk for developing AMD (14, 15).
In this study, to identify genetic markers of POAG, we conducted a GWAS in 2 stages at the Kyoto Prefectural University of Medicine using data from a total of 1,575 Japanese POAG patients and control subjects without glaucoma. We obtained a few modestly associated SNPs with POAG belonging to 3 different loci of the genome. The results suggested that the SNPs and the loci identified in this study would be promising genetic markers for the further studies to reveal the molecular mechanism of POAG pathogenesis.
Results
GWAS Stage 1 Analysis.
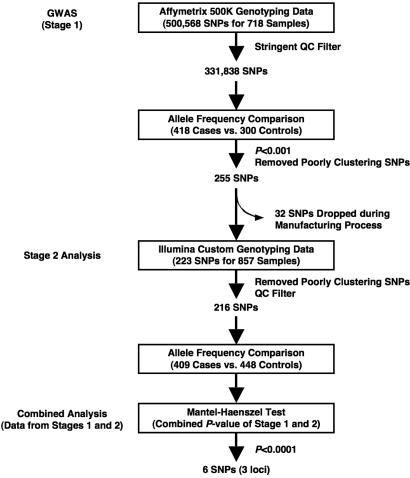
We performed the GWAS for stage 1 by screening 500,568 SNPs to discover genetic markers associated with POAG. We then attempted to reduce the false-positive associations from the results of stage 1 using an independent population in stage 2. Finally, we combined the results of stages 1 and 2 by the Mantel–Haenszel test to evaluate the SNPs identified in this study (Fig. 1). In both stages, we performed allele frequency comparison to analyze combined P values of stages 1 and 2 by the Mantel–Haenszel test.
Fig. 1.
Project overview for the discovery of genetic markers of POAG. Several hundred SNPs of P < 0.001 selected in the GWAS (stage 1) were screened with another study population (stage 2). We identified 6 SNPs of P < 0.0001, evaluated by the combined P values of stages 1 and 2 and by the Mantel–Haenszel test. See text for details.
Based on our power calculation (see SI Results and Fig. S1), we analyzed 718 samples, from 418 POAG patients (case subjects) and 300 control subjects without glaucoma (controls), in stage 1. The clinical characteristics of the study subjects are shown in Table 1. Between the case subjects and controls, no significant difference was observed in gender ratio (female/male: 1.0 vs. 1.3), but a significant difference was observed in their age at the time of blood sampling [64.6 ± 13.5 (n = 418) vs. 51.1 ± 13.9 (n = 300)], which was also seen at the age at diagnosis [58.3 ± 13.4 (n = 324) vs. 51.1 ± 13.9 (n = 300)].
Table 1.
Clinical characteristics of case and control samples
| Stage 1 |
Stage 2 |
|||||
|---|---|---|---|---|---|---|
| Case | Control | P value | Case | Control | P value | |
| No. subjects participating in case-control analyses | 418 | 300 | 409 | 448 | ||
| Female/male ratio | 1.0 (418) | 1.3 (300) | 0.17* | 1.0 (409) | 1.8 (448) | <0.05* |
| Age (years) at: | ||||||
| Blood sampling | 64.6 ± 13.5 (418) | 51.1 ± 13.9† (300) | <0.05‡ | 61.9 ± 13.9 (409) | 55.2 ± 14.7† (448) | <0.05‡ |
| Diagnosis | 58.3 ± 13.4 (324) | 51.1 ± 13.9† (300) | <0.05‡ | 55.8 ± 13.9 (301) | 55.2 ± 14.7† (448) | 0.57‡ |
| Family history of glaucoma, % | 26.5 (392) | 0 (282) | 21.2 (353) | 0 (400) | ||
Numbers in parentheses are the total numbers of subjects whose samples were used for the analysis. Data are indicated as mean ± SD.
*P value of χ 2 test for case and control comparisons.
†Age at blood sampling and diagnosis was the same for the control subjects.
‡P value of Student's t test for case and control comparisons.
After genotyping the 718 samples, we selected 331,838 SNPs for further analysis, using the stringent criteria chosen for our quality-control (QC) filter (see SI Results and Fig. 1). To identify SNPs associated with POAG, we compared the allele frequency of each SNP between case and control samples. In a quantile-quantile plot, the observed P value deviated from the expected P value between P = 10−3 and P = 10−4. Therefore, we set the threshold at P = 10−3 for further analysis (Fig. S2). In total, 431 SNPs showed P < 0.001 in the allele frequency comparison (Fig. S2). We then visually checked the 2D cluster plots of these 431 SNPs and selected 255 SNPs as candidates (Fig. 1 and Fig. S3A); 176 SNPs were not clearly associated with clusters (Fig. S3B). There were no SNPs with significant associations after Bonferroni's correction. Although 1 (rs11056970) of the 255 SNPs in the 10−4 < P < 10−3 group showed a significant deviation (P < 10−10) from the Hardy–Weinberg equilibrium (HWE) in the control population, no significant HWE deviation (P > 10−2) was observed among the 21 SNPs in the P < 10−4 group. We evaluated the SNPs neighboring these 21 SNPs on the 500K array set and found a similar P value (P = 10−3-10−4) throughout the linkage disequilibrium (LD) block. These results supported a high confidence in the genotyping results for these SNPs.
Stage 2 Analysis.
In stage 2, we analyzed the 255 SNPs of P < 10−3 identified in stage 1 that formed good clusters (Fig. 1). To reduce the false-positive associations in stage 1, we used another population of 857 samples, from 409 case subjects and 448 controls, for the stage 2 analysis (see SI Results). Because 32 SNPs were dropped during the manufacturing of the custom array, we evaluated the remaining 223 SNPs (Fig. 1).
The clinical characteristics of the 409 POAG patients and 448 control subjects in the stage 2 study are shown in Table 1. In this population, there was a significant difference in the gender ratio in case vs. control subjects (female/male: 1.0:1.8). Although a significant difference was also observed in age at the time of blood sampling [61.9 ± 13.9 (n = 409) vs. 55.2 ± 14.7 (n = 448)], there was no age difference at the time of diagnosis [55.8 ± 13.9 (n = 301) vs. 55.2 ± 14.7 (n = 448)].
Using samples from these subjects, we selected 216 SNPs with high-quality genotyping data from among the 223 SNPs analyzed and used them in the subsequent analysis (Fig. 1).
Combined Analysis of Study Stages 1 and 2.
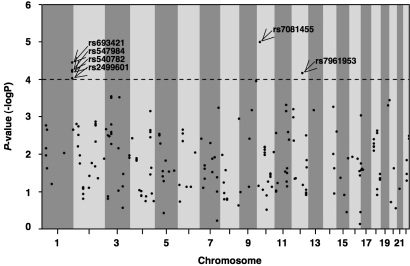
To evaluate the SNPs identified in this study, we compared the allelic frequency of each SNP between case and control samples in stage 2 and calculated their combined P values from stages 1 and 2 by the Mantel–Haenszel test with Yates' correction (Figs. 1 and 2). From the Mantel–Haenszel test, we obtained 6 SNPs with P < 10−4 (Fig. 2). Because all these SNPs showed P < 0.05 in stage 2 (Table 2), we considered their association to be confirmed (Fig. 1). Detailed information about these SNPs is summarized in Table 2 and Table S1. The combined P values ranged from 1.0 × 10−5 to 9.0 × 10−5 with an odds ratio (OR) between 1.33 and 1.49 (Table 2). One SNP was intronic, and the others were intergenic (Table 2). Four SNPs (rs547984, rs540782, rs693421, and rs2499601) were located on the same LD block. Although the rs7961953 SNP had a relatively low HWE P value (P = 0.004) in the stage 2 control group (Table S1), the genotyping data fit the 2D cluster plot (Fig. S3C). We evaluated the possible joint contributions of 6 candidate SNPs in a preliminary analysis (Tables S2 and S3 and Fig. S4). We observed that the ORs of the candidate SNPs increased when they were combined (Fig. S4).
Fig. 2.
Distribution of the combined P values of stages 1 and 2 calculated by the Mantel–Haenszel test. Comparison of the combined P values for allele frequency for 216 SNPs of P < 0.001 in stage 1 plotted against chromosomes in numerical order. Horizontal line, P = 0.0001 in the Mantel–Haenszel test. Arrows indicate SNPs with P < 0.0001.
Table 2.
List of candidate genetic markers from the Mantel-Haenszel test of SNPs from stages 1 and 2
| dbSNP ID | Chr | SNP type | Nearest gene | Stage 1 |
Stage 2 |
Mantel-Haenszel test (stages 1 and 2) |
|||
|---|---|---|---|---|---|---|---|---|---|
| P value | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | ||||
| rs547984 | 1 | Intergenic | ZP4 | 0.00033 | 1.47 (1.19–1.81) | 0.02536 | 1.24 (1.03–1.50) | 0.00006 | 1.34 (1.16–1.54) |
| rs540782 | 1 | Intergenic | ZP4 | 0.00037 | 1.47 (1.19–1.81) | 0.02536 | 1.24 (1.03–1.50) | 0.00006 | 1.34 (1.16–1.54) |
| rs693421 | 1 | Intergenic | ZP4 | 0.00029 | 1.48 (1.20–1.83) | 0.01839 | 1.26 (1.04–1.52) | 0.00004 | 1.35 (1.17–1.56) |
| rs2499601 | 1 | Intergenic | ZP4 | 0.00058 | 1.45 (1.17–1.79) | 0.02679 | 1.24 (1.02–1.50) | 0.00009 | 1.33 (1.15–1.53) |
| rs7081455 | 10 | Intergenic | PLXDC2 | 0.00005 | 1.70 (1.31–2.19) | 0.02005 | 1.33 (1.05–1.69) | 0.00001 | 1.49 (1.25–1.77) |
| rs7961953 | 12 | Intronic | DKFZp762A217 | 0.00096 | 1.48 (1.17–1.86) | 0.01482 | 1.30 (1.05–1.60) | 0.00007 | 1.37 (1.18–1.61) |
P values are for allele frequency comparison between case and control. Chr, chromosome; CI, confidence interval.
The analysis of potential confounding effects by clinical factors such as age, sex, and medical histories showed no significant differences with the genotypes of these 6 candidate SNPs (see SI Results), suggesting that the P values obtained here were specific results from the case-control comparison.
Population Stratification Analysis.
To assess the population heterozygosity, we analyzed the stratification of the populations used in stages 1 and 2. There was no significant difference in population stratification between the case and control subjects of the 2 stages (Fig. S5).
Discussion
In this study, we successfully obtained 6 candidate genetic markers modestly associated with POAG by conducting a GWAS in 2 stages that used independent study populations totaling 1,575 Japanese subjects.
We divided samples from the 1,575 subjects into an initial screening for the GWAS (stage 1) and a stage 2 study (Fig. 1). Because our aim in this study was to identify steady genetic markers of common variants that were significantly associated with the pathogenesis of POAG, we focused on ensuring that the power of our 2-stage association study would be sufficient to detect SNPs possessing reasonably high disease allele frequency and genotype-associated relative risk (Fig. S1). Our design thus resulted in a statistical power sufficient (≥80%) to detect an association with a genotypic relative risk of 2.0 at P = 1 × 10−7 if the disease allele frequency was in the range of 10 to 40%, as long as the number of the samples genotyped in stage 1 exceeded 50% (Fig. S1A). For a genotypic relative risk of greater than 1.8, the power was sufficient to detect an association with a disease allele frequency of 0.25 (Fig. S1B). In our study, the combined P values of the candidate SNPs ranged from 1.0 × 10−5 to 9.0 × 10−5 with the minor allele frequency (MAF) and the respective ORs were between 0.20 and 0.49 or between 1.33 and 1.49 (Table 2). Because the simulated powers were maintained even when we reduced the significance level to P = 10−4 (Fig. S1 C and D), if we regarded a disease allele as a minor allele and assumed that the genotype relative risk was the value of an OR, our sample setting for the 2-stage association study based on the power calculation was ample for our purposes.
In addition to the sample size, statistical power is affected by the accuracy of the clinical data and genotyping results. In our study, 3 ophthalmologists belonging to the same institution selected the POAG patients and control subjects who met our strict criteria so as to reduce any diagnostic variations among the observers. To make certain that our control samples were from volunteers without glaucoma or suspected glaucoma, we performed multiple ophthalmic tests, including a visual field test and fundus examination, for more than 1,100 control volunteers as well as for the POAG patients. To be sure that we excluded samples from volunteers who might be at risk for glaucoma, we preferentially chose volunteers with no evidence of glaucomatous anomalies (category I in case and control selection). Because the prevalence rate of POAG in the Japanese is 3.9% among people older than 40 years of age, as determined by a recent robust epidemiology study called the Tajimi Study (16), volunteers who were 40 years of age or older having a normal diagnosis with a slightly larger cup-to-disc ratio (category II) were considered to have little risk for developing glaucoma, and their samples were included. Finally, we excluded more than 400 volunteers with small optic abnormalities (all category III and some category II) from our study because they did not fit our criteria.
Genotyping errors tend to lead to a false-positive association with a significantly lower P value. In our preliminary GWAS analysis, which was done using a standard QC filter (≥85% of call rate per SNP in case and control samples and ≥5% MAF in case and control samples), we observed a large number of SNPs that showed very low P values throughout the genome (see SI Results and Fig. S6A). When we analyzed the data precisely, we found that hundreds of high-ranked SNPs showed both a significantly low call rate (Fig. S6B) and a large difference in call rate between the case and control samples (Fig. S6C). Most of these SNPs showed obvious genotyping errors and poor clustering (Figs. S6 D and E). Therefore, we adopted our stringent QC filter to remove such low-quality data and carefully checked the 2D cluster plots in both stages 1 and 2, even after applying the QC filter (Fig. 1). Using our stringent filter, the genotype concordance after extracting high-quality data from the different genotyping systems used in stages 1 and 2 was 99.8%. We verified this by genotyping 216 SNPs using 104 samples (52 control and 52 case samples) by both genotyping methods (see SI Results). This result supports a high confidence value for the genotyping data that we obtained after QC filtering and visual checking of the 2D cluster plots.
More recently, Yamaguchi-Kabata et al. (17) reported that Japanese population stratification could mainly be divided into 2 clusters: the main islands of Hondo and Ryukyu from Okinawa. They suggested that the false-positive rates in GWASs would be acceptable when the samples were collected from Hondo in Japan, indicating that the population stratification within the region was relatively small. In this study, we collected all the samples at a single institution in the middle part of Hondo. As expected, we observed no obvious population stratification between case and control samples in stages 1 and 2 (Fig. S5). These data indicated that P values obtained here were specific results from the case-control comparison.
Thus, beginning with stringent diagnostic criteria, we successfully used our polished genotype data to obtain 6 candidate genetic markers that were modestly associated with POAG. Because 4 SNPs (rs547984, rs540782, rs693421, and rs2499601) showed a strong LD with each other, we ultimately obtained 3 genetic loci associated with a potential functional determinant of POAG pathogenesis. Interestingly, of these 3 loci, none was associated with the previously reported associated genes MYOC (7), OPTN (8), and WDR36 (9). Under our conditions, all the SNPs associated with these genes were dropped in stage 1 because they did not pass the P < 10−3 filter. P values of 6 candidate SNPs in stage 2 were not so low when compared with those in stage 1 (Table 2). Slight difference, such as the ratio of classic POAG and normal tension glaucoma (NTG) subtypes, between the population of stages 1 and 2 might affect the differences of these P values.
To evaluate the 3 genetic loci that we determined to be associated with POAG, we performed high-density genotyping around the SNP (rs7081455) that showed the highest association in our study. Although we obtained a POAG-associated SNP (allele frequency comparison, P < 0.05) on the same LD block, we were still not able to link the SNP to the annotated gene(s).
Recently, Thorleifsson et al. (12) performed a GWAS and demonstrated that 3 SNPs on LOXL1 are strongly associated with exfoliation glaucoma, which is one of the fewer types of open-angle glaucoma compared to the POAG, in 2 populations of subjects from Iceland and Sweden. Although the risk haplotype differs with different populations, a strong association between the SNPs on LOXL1 and exfoliation glaucoma has been replicated in many studies (18–26), including ours of Japanese subjects (27), even when the sample size was relatively small. However, in our GWAS, in which the power was sufficient to detect SNPs with a high genotype risk ratio (Fig. S1), none of the SNPs was adjacent to LOXL1. Our data thus indicate that POAG may be more complicated than exfoliation glaucoma, because multiple SNPs with a moderate OR appear to be involved in its pathogenesis.
POAG manifests as 2 subtypes: POAG with high intraocular pressure (IOP; classic POAG subtype) and POAG with normal IOP (NTG subtype). Because the clinical states of both subtypes overlap almost completely, they are usually categorized as a single disease. However, most Japanese POAG patients have the NTG subtype, which is a unique epidemiological distribution, compared with other populations (16). Previous reports showed that toll-like receptor 4 (28) was associated with the NTG subtype and that NCK adaptor protein 2 (29) was the nearest gene from the locus associated with the NTG subtype revealed by the association study using an SNP or microsatellite marker, respectively. These 2 studies were attempting to reveal the molecular mechanism of the NTG-specific pathogenesis by focusing on a gene or locus based on previous knowledge, whereas the current study design aims to identify causative gene(s) for the common mechanism of both classic POAG and NTG subtypes by whole-genome screening. Because we combined the patients from both subtypes as a single case group to obtain a larger sample size, the genetic loci identified in this study are most likely to be components of the molecular mechanism underlying a particular neurodegenerative pathway. If we divided the samples into the classic POAG and NTG subtypes and carried out separate GWASs with enough power to detect associations, we might be able to identify genetic loci or molecule(s) that are associated with mechanisms of pathogenesis and/or progression specific to the NTG subtype, such as the genes described in the previous reports (28, 29), at least some of which would probably be related to the control of IOP.
In this study, we obtained 6 candidate SNPs located on the 3 different loci that are modestly associated with the pathogenesis of POAG. This conclusion was achievable only because we used (i) an adequate distribution of the limited subjects available in the 2-stage association study, based on the power simulation; (ii) stringent diagnostic criteria to distinguish clearly between POAG patients and control subjects without glaucoma; and (iii) a stringent data filter along with a careful visual check of the 2D cluster plots so as to restrict the genotyping data to a meaningful subset. However, we could not link these SNPs and their surroundings within the 3 loci to specific gene(s), which might have helped us to elucidate the molecular mechanism of POAG pathogenesis. It is also worth noting that other loci associated with POAG may have been dropped from this study, owing to the impaired SNP density caused by the stringent QC filter and/or because some are latent rare variants that could not be detected at all by the current study design. Along with the 3 loci discovered in this study, the identification of loci we missed, by performing large-scale association studies, replication study using an independent cohort, and subsequent in-depth sequencing, could provide a complete set of genetic markers useful for diagnosing POAG as well as for revealing the molecular mechanism of its pathogenesis.
Materials and Methods
Case and Control Subjects.
Enrollment of participants and blood sampling.
All procedures were conducted in accordance with the Helsinki Declaration. This study was approved by the Institutional Review Board of Kyoto Prefectural University of Medicine. All participants provided written informed consent after an explanation of the nature and possible consequences of the study, and they were interviewed to determine their familial history of glaucoma and other ocular or general diseases. A total of 1,591 Japanese participants were recruited to give peripheral blood samples for this study between March 2005 and December 2007. Because 16 of the 1,591 sets of genotyping data were dropped during the genotyping process (see SI Text), the data derived from 1,575 participants were used. Blood samples were assigned an anonymous code by a third person who was blinded to both the blood sampling and genotyping. Genomic DNA was isolated from the blood, and Epstein-Barr virus–transformed lymphocytes were prepared to serve as a future resource of genomic DNA (see SI Text).
Selection of case subjects and control subjects.
We recruited the case patients with POAG and control subjects without glaucoma for this study at the University Hospital of Kyoto Prefectural University of Medicine (Kyoto, Japan). Both the case and control groups in stages 1 and 2 received the following serial ophthalmic examinations for diagnosis: silt-lamp and fundus examination, gonioscopy, visual field tests, and IOP measurements. The anterior chamber angle was examined by means of a slit-lamp, based on the method of van Herick et al. (30). The ocular fundus was examined using a confocal scanning laser ophthalmoscope (HRT-II; Heidelberg Engineering GmbH), scanning laser polarimeter (GDx-VCC; Carl Zeiss Meditec), and fundus photography (TRC-NW200; Topcon). The visual field was tested by frequency-doubling technology perimetry (Matrix; Carl Zeiss Meditec) using program N-30/Humphrey automated perimetry with program 30–2 SITA Fast and/or Standard (Carl Zeiss Meditec) and/or Goldmann perimetry (Haag–Streit) for both study groups. The IOP was measured by a noncontact tonometer (RKT-7700; Nidek) for the selection of control subjects and by a Goldmann applanation tonometer (Haag–Streit) for both study groups. Three ophthalmologists (Y.I., S.K., and K.M.) diagnosed glaucoma in the patients, based on the diagnosis standard (31). The baseline IOPs of case groups in stages 1 and 2 were 15.5 ± 3.9 mmHg (mean ± SD, n = 157) and 15.5 ± 3.1 mmHg (mean ± SD, n = 154), respectively, whereas the mean deviations of Humphrey perimetry were −10.7 ± 8.6 dB (n = 319) and −10.1 ± 8.1 dB (n = 261), respectively. We could not obtain all the cases' baseline IOP or mean deviation of visual field data because of the severity of the disease. Further information for selecting case and control subjects is detailed in SI Text.
GWAS (Stage 1).
SNP genotyping.
We first genotyped the whole-genome SNPs of 425 case and 301 control samples on an Affymetrix GeneChip Mapping 500K Array Set, following the manufacturer's instructions. The protocols for obtaining SNP data are described in SI Text.
Criteria for SNP selection.
From 500,568 SNPs (262,264 and 238,304 SNPs in the Nsp I and StyI arrays, respectively), a total of 331,838 autosomal SNPs were selected for association analysis based on our stringent QC filter, which had the following criteria: (i) ≥90% call rate per SNP in case and control samples, (ii) ≤5% call rate difference between case and control samples for each SNP, and (iii) ≥5% MAF in case and control samples. After the association analysis, we visually checked the 2D cluster plots of the genotypes for each P < 10−3-ranked SNP (431 SNPs) to remove SNPs that clustered poorly. The scoring system for assessing the 2D cluster plots is detailed in SI Text. We finally selected 255 SNPs as the stage 1 candidates.
Stage 2 Analysis.
SNP genotyping.
We next attempted to replicate the genotyping of the 255 candidate SNPs identified in stage 1 using samples from another population of 410 case subjects and 455 control subjects by means of the iSelect Custom Infinium Genotyping system (Illumina). The protocols for obtaining SNP data are described in SI Text.
Criteria for SNP selection.
From the 223 SNPs, a total of 216 SNPs were selected for the association analysis based on our QC filter: (i) ≥90% of call rate per SNP for both case and control, respectively, and (ii) ≥5% of MAF for both case and control. To validate the genotyping accuracy in stages 1 and 2, we genotyped the 216 SNPs in 104 (52 case and 52 control) samples using both an Affymetrix 500K Array Set and the iSelect system and analyzed the genotype concordance between the 2 systems (see SI Text).
Population Stratification.
To analyze the stratification of our stage 1 and 2 populations, we used STRUCTURE version 2.2 software (http://pritch.bsd.uchicago.edu/software.html). Detailed protocols are described in SI Text.
Statistical Analysis.
To manage all the genotyped data, we used LaboServer (World Fusion Co., Ltd.) as a laboratory information management system. We used LaboServer, Microsoft Office Excel 2003 (Microsoft), and R for the statistical analysis. The power calculation was performed using CaTS software (www.sph.umich.edu/csg/abecasis/CaTS/index.html) (see SI Text).
The frequency of alleles in case and control samples was compared using the basic allele test. The OR and the upper and lower limits of the 95% confidence interval of each SNP were calculated for the allele possessing a higher frequency in the case samples than in the control samples. The HWE was evaluated by the χ2 test. Quantile–quantile plots were generated by ranking the observed values from minimum to maximum and plotting them against their expected values (32). To examine the possible confounding effects of several factors, such as age, gender, history of systemic disease, and reported risk of glaucoma, we assessed the correlations between the clinical profile values and the genotype data from the case and control samples by one-way ANOVA or χ2 test (33, 34). The Mantel–Haenszel test with Yates' correction was performed as described previously (35). All the numerical data were expressed as the mean ± SD.
Supplementary Material
Acknowledgments.
We thank all the patients and volunteers who enrolled in our study. We also thank Ms. Sayaka Ohashi, Ms. Naoko Saito, and Ms. Yuko Konoshima for processing blood samples and performing genotyping; Mrs. Hiromi Yamada, Ms. Aiko Hashimoto, Ms. Keiko Nirasawa, and Mrs. Akemi Tanaka for their assistance in clinical information analysis; Mr. Ryuichi Sato and Ms. Fumiko Sato (World Fusion, Tokyo, Japan) for management of the genotype data; Mr. Hiroshi Inoue for the logistic regression analysis and excellent advice regarding statistical analysis; and Ms. Tomoko Tsuda for excellent secretarial assistance. This work was supported by grants from the Collaborative Development of Innovative Seeds of the Japan Science and Technology Agency (M.K. and K.T.); Ministry of Health, Labor, and Welfare of Japan (K.M., S.K. and K.T.), and Santen Pharmaceutical Co., Ltd (S.K. and K.T).
Footnotes
Conflict of interest statement: This study has been completed under the Collaborative Research Agreement executed by Kyoto Prefectural University of Medicine and Santen Pharmaceutical Co., Ltd. All materials and information produced through this study are the parts of the co-owned intellectual properties. T.T., M.F., and M.K. are employees of Santen Pharmaceutical Co., Ltd.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906397106/DCSupplemental.
References
- 1.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heijl A, et al. Reduction of intraocular pressure and glaucoma progression: Results from the Early Manifest Glaucoma Trial. Arch Ophthalmol. 2002;120:1268–1279. doi: 10.1001/archopht.120.10.1268. [DOI] [PubMed] [Google Scholar]
- 3.Leske MC, et al. Factors for glaucoma progression and the effect of treatment: The Early Manifest Glaucoma Trial. Arch Ophthalmol. 2003;121:48–56. doi: 10.1001/archopht.121.1.48. [DOI] [PubMed] [Google Scholar]
- 4.Leske MC, Heijl A, Hyman L, Bengtsson B. Early Manifest Glaucoma Trial: Design and baseline data. Ophthalmology. 1999;106:2144–2153. doi: 10.1016/s0161-6420(99)90497-9. [DOI] [PubMed] [Google Scholar]
- 5.Hewitt AW, Craig JE, Mackey DA. Complex genetics of complex traits: The case of primary open-angle glaucoma. Clin Exp Ophthalmol. 2006;34:472–484. doi: 10.1111/j.1442-9071.2006.01268.x. [DOI] [PubMed] [Google Scholar]
- 6.Fan BJ, Wang DY, Lam DS, Pang CP. Gene mapping for primary open angle glaucoma. Clin Biochem. 2006;39:249–258. doi: 10.1016/j.clinbiochem.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Stone EM, et al. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 8.Rezaie T, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- 9.Monemi S, et al. Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Hum Mol Genet. 2005;14:725–733. doi: 10.1093/hmg/ddi068. [DOI] [PubMed] [Google Scholar]
- 10.Topol EJ, Murray SS, Frazer KA. The genomics gold rush. J Am Med Assoc. 2007;298:218–221. doi: 10.1001/jama.298.2.218. [DOI] [PubMed] [Google Scholar]
- 11.Kingsmore SF, Lindquist IE, Mudge J, Gessler DD, Beavis WD. Genome-wide association studies: Progress and potential for drug discovery and development. Nat Rev Drug Discovery. 2008;7:221–230. doi: 10.1038/nrd2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorleifsson G, et al. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science. 2007;317:1397–1400. doi: 10.1126/science.1146554. [DOI] [PubMed] [Google Scholar]
- 13.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maller J, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38:1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 15.Ross RJ, et al. Genetic markers and biomarkers for age-related macular degeneration. Expert Rev Ophthalmol. 2007;2:443–457. doi: 10.1586/17469899.2.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwase A, et al. The prevalence of primary open-angle glaucoma in Japanese: The Tajimi Study. Ophthalmology. 2004;111:1641–1648. doi: 10.1016/j.ophtha.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi-Kabata Y, et al. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: Effects on population-based association studies. Am J Hum Genet. 2008;83:445–456. doi: 10.1016/j.ajhg.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aragon-Martin JA, et al. Evaluation of LOXL1 gene polymorphisms in exfoliation syndrome and exfoliation glaucoma. Mol Vis. 2008;14:533–541. [PMC free article] [PubMed] [Google Scholar]
- 19.Challa P, et al. Analysis of LOXL1 polymorphisms in a United States population with pseudoexfoliation glaucoma. Mol Vis. 2008;14:146–149. [PMC free article] [PubMed] [Google Scholar]
- 20.Fan BJ, et al. DNA sequence variants in the LOXL1 gene are associated with pseudoexfoliation glaucoma in a U.S. clinic-based population with broad ethnic diversity. BMC Med Genet. 2008;9:5. doi: 10.1186/1471-2350-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fingert JH, et al. LOXL1 mutations are associated with exfoliation syndrome in patients from the midwestern United States. Am J Ophthalmol. 2007;144:974–975. doi: 10.1016/j.ajo.2007.09.034. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi H, Gotoh N, Ueda Y, Nakanishi H, Yoshimura N. Lysyl oxidase-like 1 polymorphisms and exfoliation syndrome in the Japanese population. Am J Ophthalmol. 2008;145:582–585. doi: 10.1016/j.ajo.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 23.Mossbock G, et al. Lysyl oxidase-like protein 1 (LOXL1) gene polymorphisms and exfoliation glaucoma in a Central European population. Mol Vis. 2008;14:857–861. [PMC free article] [PubMed] [Google Scholar]
- 24.Ozaki M, et al. Association of LOXL1 gene polymorphisms with pseudoexfoliation in the Japanese. Invest Ophthalmol Vis Sci. 2008;49:3976–3980. doi: 10.1167/iovs.08-1805. [DOI] [PubMed] [Google Scholar]
- 25.Pasutto F, et al. Association of LOXL1 common sequence variants in German and Italian patients with pseudoexfoliation syndrome and pseudoexfoliation glaucoma. Invest Ophthalmol Vis Sci. 2008;49:1459–1463. doi: 10.1167/iovs.07-1449. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, et al. Genetic association of LOXL1 gene variants and exfoliation glaucoma in a Utah cohort. Cell Cycle. 2008;7:521–524. doi: 10.4161/cc.7.4.5388. [DOI] [PubMed] [Google Scholar]
- 27.Mori K, et al. LOXL1 genetic polymorphisms are associated with exfoliation glaucoma in the Japanese population. Mol Vis. 2008;14:1037–1040. [PMC free article] [PubMed] [Google Scholar]
- 28.Shibuya E, et al. Association of toll-like receptor 4 gene polymorphisms with normal tension glaucoma. Invest Ophthalmol Vis Sci. 2008;49:4453–4457. doi: 10.1167/iovs.07-1575. [DOI] [PubMed] [Google Scholar]
- 29.Akiyama M, et al. Microsatellite analysis of the GLC1B locus on chromosome 2 points to NCK2 as a new candidate gene for normal tension glaucoma. Br J Ophthalmol. 2008;92:1293–1296. doi: 10.1136/bjo.2008.139980. [DOI] [PubMed] [Google Scholar]
- 30.Van Herick W, Schwartz A. Estimation of width of angle of anterior chamber incidence and significance of the narrow angle. Am J Ophthalmol. 1969;68:626–629. doi: 10.1016/0002-9394(69)91241-0. [DOI] [PubMed] [Google Scholar]
- 31.European Glaucoma Society. Terminology and Guidelines for Glaucoma. 2nd Ed. Savona, Italy: Dogma; 2003. pp. 1–152. [Google Scholar]
- 32.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 33.Miyamoto Y, et al. A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet. 2007;39:529–533. doi: 10.1038/2005. [DOI] [PubMed] [Google Scholar]
- 34.Ozaki K, et al. A functional SNP in PSMA6 confers risk of myocardial infarction in the Japanese population. Nat Genet. 2006;38:921–925. doi: 10.1038/ng1846. [DOI] [PubMed] [Google Scholar]
- 35.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.