Abstract
α-Synuclein is a key protein in Parkinson's disease (PD) because it accumulates as fibrillar aggregates in pathologic hallmark features in affected brain regions, most notably in nigral dopaminergic neurons. Intraneuronal levels of this protein appear critical in mediating its toxicity, because multiplication of its gene locus leads to autosomal dominant PD, and transgenic animal models overexpressing human α-synuclein manifest impaired function or decreased survival of dopaminergic neurons. Here, we show that microRNA-7 (miR-7), which is expressed mainly in neurons, represses α-synuclein protein levels through the 3′-untranslated region (UTR) of α-synuclein mRNA. Importantly, miR-7-induced down-regulation of α-synuclein protects cells against oxidative stress. Further, in the MPTP-induced neurotoxin model of PD in cultured cells and in mice, miR-7 expression decreases, possibly contributing to increased α-synuclein expression. These findings provide a mechanism by which α-synuclein levels are regulated in neurons, have implications for the pathogenesis of PD, and suggest miR-7 as a therapeutic target for PD and other α-synucleinopathies.
Keywords: Parkinson's disease, neuroprotection, MPTP model, microRNA
Parkinson's disease (PD) is a common neurodegenerative disorder that affects 1% of the population over 65. It is characterized by disabling motor abnormalities including tremor, slow movements, rigidity, and poor balance. These impairments stem from the progressive loss of dopaminergic neurons in the substantia nigra pars compacta. Eventually, large percentages of patients develop dementia and hallucinations when the pathology involves other brain regions as well. Although the majority of Parkinson cases appear to be sporadic, the disorder runs in families in ≈15–20% of the cases. To date, 5 distinct genes have been identified to cause PD including α-synuclein, parkin, dj-1, pink1, and lrrk2 (1). Understanding how mutations in these genes cause neurodegeneration is crucial in the development of treatments that might slow or stop the disease progression.
α-Synuclein (α-Syn) is a key player in the pathogenesis of PD based on genetic, neuropathologic, and cellular/molecular lines of evidence (2–4). In addition to point mutations linked to dominantly inherited PD (5–7), mounting evidence suggests that elevated levels of α-Syn are deleterious to dopaminergic neurons. Individuals with multiplication of this gene locus develop PD with an earlier onset age and increasing severity associated with dementia in a gene dosage-dependent manner (8, 9), transgenic mice, Drosophila, and Caenorhabditis elegans overexpressing α-Syn manifest phenotypic changes reminiscent of the disease (10–12), and engineered cultured cells are made vulnerable by this protein (13, 14). In addition, a large-scale analysis in patients with PD and controls showed that variability in the α-Syn promoter region, which results in its up-regulation, is associated with an increased risk of PD (15). Besides these compelling data, postmortem investigations of PD and other α-synucleinopathies have demonstrated fibrillar α-Syn aggregates in Lewy bodies and Lewy neurites (16, 17). An immunization approach to clear the brain of the α-Syn burden has been shown to reduce the neurodegeneration in transgenic mice (18). Based on the aforementioned evidence, α-Syn overexpression appears to be a common mechanism for the pathogenesis of PD and other α-synucleinopathies.
microRNAs (miRNAs) are a class of endogenous 17–24 base-long single-stranded, noncoding RNAs that regulate gene expression in a sequence-specific manner in plants and animals. miRNAs are derived from long precursor transcripts by the action of nucleases Drosha and Dicer, and the resulting mature functional miRNAs bind to their target sequence in the 3′-untranslated region (UTR) of mRNA with imperfect complementarity and lead to repression of translation (19). Recently, miRNAs have been suggested to play important roles in diverse biological phenomena including development, oncogenesis, and brain functions (20, 21). Some miRNAs are specifically expressed and enriched in the brain and have been associated with memory, neuronal differentiation, and synaptic plasticity (22–24). The role of miRNAs in neurodegeneration has been suggested in several reports (25–28). Interestingly, miR-133b, which is specifically expressed in midbrain dopaminergic neurons and controls their maturation and function through its effect on the homeodomain transcription factor Pitx3, is deficient in PD brains (29), suggesting that miR-133b is essential for the maintenance of these neurons and could therefore play a role in PD pathogenesis. Here, we show that miR-7 represses α-Syn expression and inhibits α-Syn-mediated cell death.
Results
Identification of miR-7 as a Regulator of α-Syn Expression.
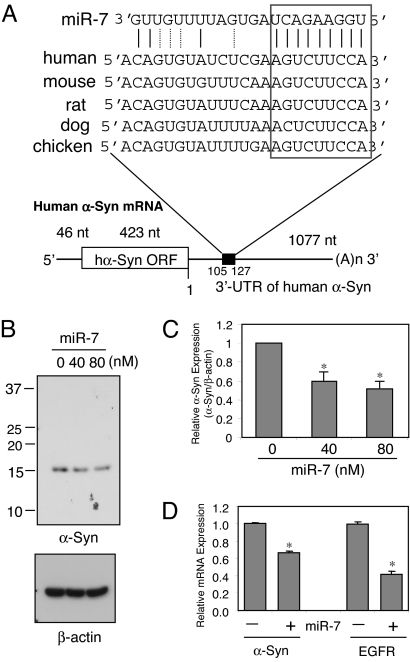
The human α-Syn gene has a 3′-UTR that is more than twice as long as its ORF, raising the possibility that it might contain posttranscriptional regulatory elements that would control its protein level. To search for miRNA(s) that might regulate human α-Syn expression, we used public prediction algorithms found in several web sites, such as miRbase (http://microrna.sanger.ac.uk/sequences/), Targetscan (http://www.targetscan.org/), Pictar (http://pictar.mdc-berlin.de/), and miRanda (http://www.microrna.org/microrna/home.do). A number of miRNA target sequences were predicted, but only one, miRNA-7 (miR-7), was common to all predictions. Expression of miR-7 was previously verified in human, mouse, and zebrafish and found to be especially enriched in the brain (30, 31). The seed match between miR-7 and α-Syn 3′-UTR is between bases 119 and 127, and this target region is conserved in human, mouse, rat, dog, and chicken (Fig. 1A). The free energy required for the interaction between miR-7 and its cognate α-Syn 3′-UTR binding site is −22.7 kcal/mol based on the Pictar prediction.
Fig. 1.
miR-7 regulates endogenous α-Syn levels. (A) Schematic diagram of human α-Syn mRNA containing the predicted conserved target site of miR-7. The seed match is indicated with the box. (B) miR-7 reduces the level of endogenous α-Syn in HEK293T cells. Cells were transfected with miR-7 precursor at the indicated concentrations and harvested at 48 h after transfection. A representative Western blot using SYN-1 antibody is shown. (C) Mean levels of α-Syn protein in B calculated from 3 independent experiments. (D) Quantitative RT-PCR analysis of α-Syn and EGFR mRNA expression in HEK293T cells. Cells were transfected with 40 nM miR-7 precursor or scrambled sequence and harvested 24 h later. Both mRNA expressions are normalized to GAPDH mRNA and are shown as a ratio to control (scrambled miR-7 sequence)-transfected cells. Experiments were done in triplicates. Data are shown as means ± SD. *, P < 0.01 for difference between control and miR-7-treated samples.
To test the role of miR-7 in regulating α-Syn protein expression, we transfected HEK293T cells, which express significant amounts of endogenous α-Syn, with premiR-7 at 2 concentrations. miR-7 reduced α-Syn expression dose-dependently: 40 nM miR-7 resulted in 40% reduction of α-Syn expression, and 80 nM miR-7 reduced it by 50% (Fig. 1 B and C). Endogenous levels of α-Syn in human dopaminergic neuroblastoma SH-SY5Y and mouse neuroblastoma NS20Y cells are too low to detect the effect of miR-7.
Next, we investigated the effect of miR-7 on α-Syn mRNA levels by quantitative RT-PCR. Transfection of HEK293T cells with 40 nM miR-7 significantly reduced α-Syn mRNA expression by 34% (Fig. 1D), indicating that miR-7 promotes the degradation of this target mRNA. As positive control in this experiment, miR-7 also reduced epidermal growth factor receptor (EGFR) mRNA expression by 57%, as reported previously (32).
Repression of α-Syn Expression by miR-7 Requires α-Syn 3′-UTR.
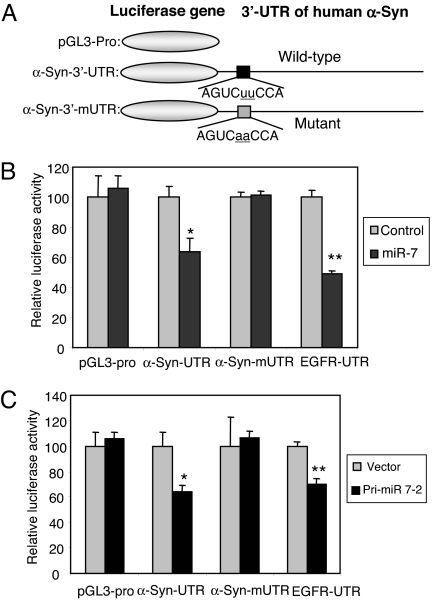
To test if miR-7 directly targets the 3′-UTR of α-Syn, this full-length 3′-UTR was inserted downstream of the firefly luciferase reporter gene (Fig. 2A) and cotransfected with premiR-7 into HEK293T cells. miR-7 significantly decreased luciferase activity from α-Syn-3′-UTR construct in a dose-dependent manner ( Fig. S1), but had no effect on pGL3-pro construct, which lacks α-Syn 3′-UTR (Fig. 2B). As positive control, miR-7 significantly inhibited luciferase activity from EGFR-3′-UTR-containing construct by 50% (Fig. 2B), consistent with previous reports (32, 33). The effect of miR-7 on the α-Syn 3′-UTR in a luciferase construct was also replicated in SH-SY5Y cells (Fig. S2). To confirm that the predicted target sequence of miR-7 in the α-Syn 3′-UTR is functional, this site was mutagenized as shown in Fig. 2A. Notably, miR-7 could not inhibit luciferase activity from the mutagenized α-Syn-3′-UTR construct (Fig. 2B), indicating that the predicted sequence is indeed a genuine binding site for miR-7. Further, we confirmed the effect of miR-7 on α-Syn expression using a pri-miR-7-2 vector expressing miR-7. Transfection of HEK293T cells with pri-miR-7-2 consistently inhibited luciferase activity from constructs having α-Syn-3′-UTR as well as EGFR-3′-UTR but not from α-Syn-3′-mUTR mutated at the miR-7 seed sequence (Fig. 2C). This experiment also shows that HEK293T cells can efficiently process pri-miR-7 to mature functional miR-7.
Fig. 2.
miR-7 acts on the 3′-UTR of α-Syn mRNA. (A) Schematic diagrams of human α-Syn 3′-UTR luciferase constructs showing wild-type and mutant (α-Syn-3′-mUTR) miR-7 target sequences. (B) HEK293T cells were transfected with luciferase constructs shown in A and 40 nM miR-7 and harvested 24 h later. Luciferase activities normalized to β-galactosidase activity are shown as a percentage of control (scrambled miR-7 sequence). (C) miR-7 expressed from plasmid pri-miR-7-2 inhibits luciferase activity in HEK293T cells. Transfected cells were harvested 24 h later. *, P < 0.05; **, P < 0.01 for difference between control and miR-7-treated samples.
miR-7 Inhibitor Up-Regulates α-Syn Expression.
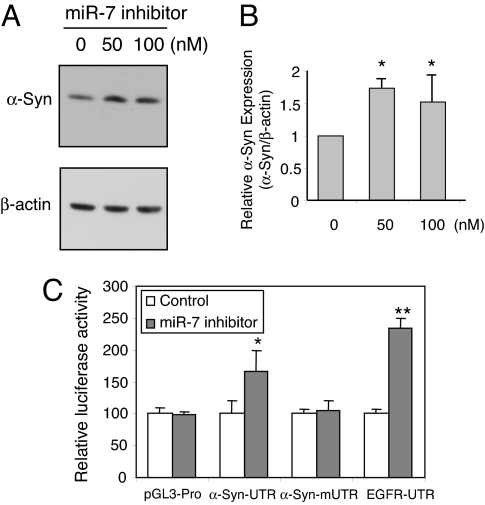
Anti-miR miRNA inhibitors are chemically modified (2′-O-methyl), single-stranded nucleic acids designed to specifically bind to and inhibit endogenous miRNAs. By using this specific inhibitor against miR-7, we found that endogenous level of α-Syn protein was significantly increased in SH-SY5Y cells (Fig. 3 A and B). In addition, luciferase activity from the plasmid containing α-Syn-3′-UTR also increased significantly in the presence of miR-7 inhibitor, whereas those from constructs having α-Syn mutant 3′-UTR (α-Syn-3′-mUTR) or no 3′-UTR (pGL3-pro) were not affected (Fig. 3C). Once again, as positive control, EGFR-3′-UTR responded by increased expression in the presence of miR-7 inhibitor. These results confirm that miR-7 inhibits α-Syn expression through the 3′-UTR, and further suggest that endogenous miR-7 actively suppresses α-Syn expression in SH-SY5Y cells, which could be blocked by miR-7 inhibitor.
Fig. 3.
miR-7 inhibitor increases α-Syn protein level. (A) SH-SY5Y cells were transfected with the indicated concentrations of anti-miR-7 inhibitor (2′-O-methyl) and harvested 48 h later. A representative Western blot using SYN-1 antibody is shown. (B) Mean amount of α-Syn measured from 3 independent experiments was normalized to β-actin. (C) SH-SY5Y cells transfected with luciferase constructs and 50 nM 2′-O-methyl inhibitor of miR-7 and harvested 24 h later. Luciferase values normalized to β-galactosidase activity are shown as a percentage of control (scrambled miR-7 inhibitor). *, P < 0.05; **, P < 0.01 for difference between control and miR7 inhibitor-treated samples.
MirR-7 Protects Against α-Syn-Induced Proteasome Inhibition and Cytotoxicity.
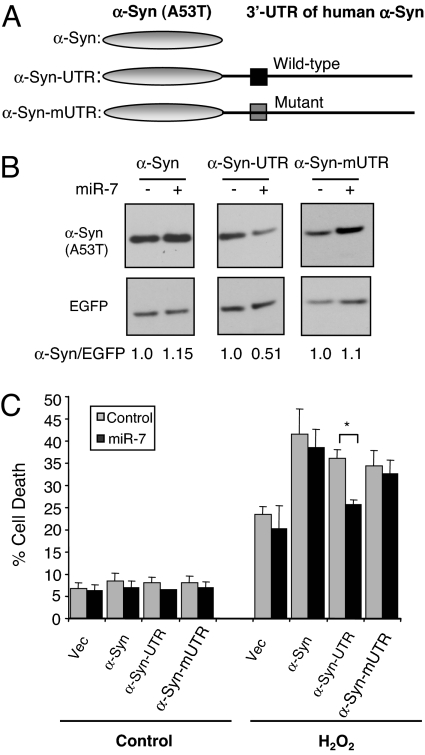
α-Syn overexpression and particularly its PD causing A53T mutant increases the susceptibility of cells to oxidative stress (14, 34), which is a consistent abnormality in the PD brain (5). Further, the A53T mutant form of α-Syn reportedly impairs proteasome activity (34–36). To investigate if miR-7 regulates α-Syn-mediated proteasome inhibition and cytotoxicity, we generated constructs expressing the α-Syn ORF harboring the A53T mutation with either its wild-type 3′-UTR, miR-7 target site mutant 3′-UTR, or no 3′-UTR (Fig. 4A). Cotransfecting miR-7 along with these constructs significantly reduced α-Syn (A53T) expression from the wild-type 3′-UTR containing plasmid, but not from the construct lacking the 3′-UTR or from the construct with miR-7 target site mutation (Fig. 4B). As shown in Fig. S3, A53T mutant α-Syn expression resulted in inhibition of chymotryptic proteasome activity regardless of the presence of the 3′-UTR, whereas coexpression of miR-7 significantly recovered this proteasome activity only in the presence of α-Syn wild-type 3′-UTR.
Fig. 4.
miR-7-mediated reduction of α-Syn level protects against cell death. (A) Schematic diagram of A53T mutant α-Syn construct without its 3′-UTR, with wild-type 3′-UTR, or miR-7 target site mutant 3′-UTR. (B) Mouse neuroblastoma NS20Y cells were transfected with constructs shown in A along with 50 nM miR-7 or control (scrambled miR sequence). pEGFP-C1 was used as internal control for normalization of transfection efficiency. A representative Western blot using LB509 antibody is shown. The relative levels of α-Syn normalized to EGFP expression were calculated based on band density using NIH Image J software with each scrambled miR transfected sample set to 1.0. (C) NS20Y cells were transfected with α-Syn (A53T) constructs in the presence of scrambled miR (control) or presence of miR-7. pSV-β-Gal plasmid was used as internal control for normalization of transfection efficiency. After transfection, cells were treated with 200 μM H2O2 for 16 h. Experiments were performed in triplicates and repeated independently 3 times. *, P < 0.05 for difference between control and miR-7-treated samples.
Next, the cytotoxicity of A53T mutant α-Syn was tested. Because NS20Y cells do not manifest toxicity to the mere expression of α-Syn (A53T), they were also challenged with hydrogen peroxide. As expected, A53T α-Syn transfected cells demonstrated increased sensitivity to H2O2 compared with empty vector transfected cells (Fig. 4C). Notably, in the presence of miR-7 and α-Syn with its wild-type 3′-UTR, H2O2-induced cell death was significantly reduced down to the toxicity seen with empty vector transfected cells (Fig. 4C). However, miR-7 could not suppress H2O2-induced cell death in cells that expressed α-Syn with miR-7 target site mutant 3′-UTR, α-Syn without 3′-UTR, or in empty vector transfected cells. Thus, miR-7 completely protects against A53T mutant α-Syn mediated susceptibility to H2O2 in an α-Syn 3′-UTR dependent manner. Taken together, these results indicate that miR-7 inhibits α-Syn expression and thereby protects against α-Syn-mediated proteasome impairment and susceptibility to oxidative stress.
miR-7 Expression in the Mouse Brain and in Brain Cell Types.
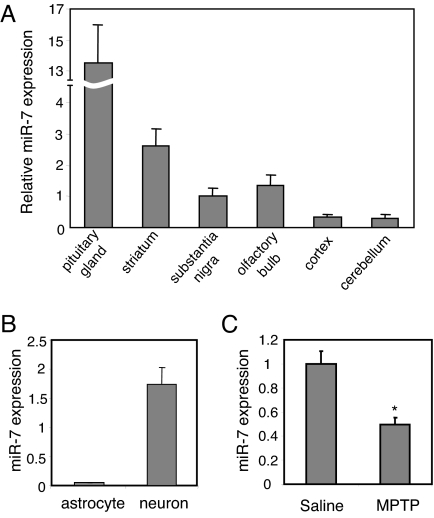
Nigral dopaminergic neurons are vulnerable in PD where accumulated α-Syn might cause toxicity. To study the possibility that α-Syn expression in the substantia nigra could be regulated by miR-7, we investigated the levels of mature miR-7 in this region and several other regions of the mouse brain by quantitative real-time RT-PCR (qRT-PCR) analysis (Fig. 5A). As reported (30, 37), miR-7 expression is highest in the pituitary gland, validating the quality and specificity of this experiment. Interestingly, the substantia nigra, striatum where nigral dopaminergic nerve terminals project, and olfactory bulb, which are neural systems affected in PD, have relatively higher miR-7 levels compared with the cerebral cortex and cerebellum, suggesting perhaps that dysregulation of miR-7 in these regions might relate to PD pathogenesis.
Fig. 5.
miR-7 expression in the mouse brain and in brain cell types. (A) Total RNA samples isolated from brain regions were used for qRT-PCR analysis of miR-7. Amounts are calculated relative to the level in substantia nigra. (B) Expression levels of miR-7 measured in primary neurons and astrocytes by qRT-PCR. (C) Mice were challenged with MPTP for 5 days, and 2 weeks later ventral midbrain tissue was subjected to qRT-PCR analysis of miR-7 normalized to U6 level. *, P < 0.05 for difference between saline and MPTP-treated mice.
Because of the heterogeneous cell population of the brain, which includes neurons and astroglia, we investigated which cell type expresses miR-7. qRT-PCR analysis showed that the expression of miR-7 in neurons is 40-fold higher than in astrocytes (Fig. 5B). This result was confirmed by in situ hybridization of primary neurons and astrocytes. The signal intensity of the specific miR-7 probe was significantly stronger in neurons compared with the scrambled sequence probe used as control. On the other hand, the intensity of miR-7 in situ signal in astrocytes was not much higher than that of the scrambled probe (Fig. S4). These data indicate that miR-7 is expressed primarily in neurons. Consistent with the notion that a certain miRNA should co-localize with its target mRNAs to fine tune expression of the target genes rather than have a mutually exclusive expression pattern (38), we detected α-Syn mRNA in neurons but not in astrocytes. These results suggest that miR-7 negatively regulates α-Syn expression in neurons.
Next, we investigated if the level of miR-7 is reduced in PD models, which might result in enhanced α-Syn expression. As reported previously (39), MPP+ treatment of SH-SY5Y cells increased α-Syn mRNA levels as quantified by qRT-PCR (Fig. S5A). Further, this treatment resulted in a significant reduction in miR-7 levels (Fig. S5B), suggesting that enhanced α-Syn expression in MPP+-challenged cells could be due to reduced miR-7 levels. In mice, Vila et al. reported that α-Syn is up-regulated following MPTP-intoxication (40). To determine if the latter observation is mediated through changes in miR-7, we measured miR-7 levels in the ventral midbrain of MPTP-intoxicated mice by qRT-PCR. As shown in Fig. 5C, subchronic MPTP administration resulted in a 50% reduction of miR-7 expression. This finding raises the possibility that the reduction in miR-7 expression is involved in degeneration of the nigrostriatal system in the MPTP model, likely through up-regulation of α-Syn expression.
Discussion
Increased α-Syn gene (SNCA) dosage due to locus multiplication leads to autosomal dominantly inherited PD, suggesting that higher concentration of α-Syn protein in neurons is involved in the pathogenesis of PD. In sporadic PD, it is conceivable that various genetic or environmental factors that up-regulate α-Syn expression can be potential culprits as well. Recent evidence suggests that miRNAs regulate a plethora of genes and are involved in many disease states ranging from cancer to neurodegeneration (20). In this report, we provide experimental evidence for a specific miRNA species that directly represses α-Syn protein levels and protects against α-Syn-mediated cytotoxicity. We found that miR-7, which is a brain-enriched miRNA, binds to the 3′-UTR of α-Syn mRNA in a sequence-dependent manner and significantly inhibits its translation. GenBank BLAST search revealed that miR-7 is found in human, mouse, rat, zebrafish, and fly, suggesting that it regulates biological functions conserved between vertebrates and invertebrates. Antisense inhibition of miR-7 has been found to down-regulate cell growth and increase apoptosis (41), suggesting that miR-7 has a protective role. The latter finding is consistent with our observation that miR-7 suppresses α-Syn-mediated cell death. In contrast, miR-7 can also have tumor suppressor-like characteristics in glioblastomas. It potently down-regulates the EGF receptor (EGFR) as well as upstream players of the Akt pathway. Additionally, miR-7 is down-regulated in human glioblastoma tissue relative to surrounding normal brain (33). The apparent discrepancy between the anti- and pro- cell death activity of miR-7 might reflect the complex regulatory role of this microRNA, requiring additional investigations into its biology in different cellular contexts.
miR-7 is transcribed from 3 loci in the human genome and 1 locus of the mouse genome. miR-7-1 is located within an intron of the HNRNPK gene on chromosome 9, which encodes a ribonucleoprotein. Profiling microRNA expression in various tissues has found miR-7 highly expressed in the pituitary gland, presumably because miR-7-3 is transcribed from an intron of pituitary gland-specific factor 1a (PGSF1) gene (37). In the present study, we showed that miR-7 is expressed in the nigra and striatal tissue of mice, and it is decreased following MPTP exposure, suggesting that its relative under-expression in those brain regions may relate to the pathogenesis of PD possibly through α-Syn accumulation. Further analysis of miR-7 in PD brains can shed light on its regulation and in turn provide insight about its control of α-Syn levels and PD pathogenesis.
To better understand the mechanisms regulating α-Syn expression, it will be necessary to expand the list of miRNAs that target α-Syn 3′-UTR. Based on the prediction algorithms of Target scan, Pictar, and miRanda, another miRNA species, miR-153, is predicted to bind to the α-Syn 3′-UTR with its seed sequence complementary to nucleotides 462–468. This sequence is conserved in human, mouse, rat, dog, and chicken. Considering that miR-153 is also highly expressed in brain (37), it is conceivable that it can regulate α-Syn expression even though its duplex formation is less stable than that of miR-7. As the expression of many genes is regulated by the combination of several miRNAs (27), investigating the role of miR-153 in α-Syn regulation could be informative.
The present finding pointing to the importance of the 3′-UTR and miRNA target sites in controlling α-Syn expression also raises the possibility of polymorphisms at these sites contributing to PD susceptibility. Several polymorphisms in the 3′-UTR of the human α-Syn genes are reported in GenBank, but not in the target sites of miR-7 or miR-153. Instead polymorphic variation (rs10024743) lies in the potential target site of miR-34b, even though its association with PD is not known. It remains to be determined if this polymorphism correlates with resistance to miR-34b action leading to higher α-Syn protein levels. Interestingly, a polymorphic variation in the miR-433 binding site of the fibroblast growth factor 20 (FGF20) gene was recently reported to confer risk of PD, which was attributed to increased FGF20 levels and indirectly enhanced α-Syn expression (42).
Several studies have implicated miRNAs in brain diseases. For example, a mutation in the target site of miR-189 in the human SLITRK1 gene has been shown to be associated with Tourette's syndrome (43). In addition, conditional deletion of Dicer in murine postmitotic Purkinje cells resulted in progressive loss of miRNAs, cerebellar degeneration, and ataxia (44). miRNAs regulate the expression of ataxin1, amyloid precursor protein (APP), and BACE1 and have been suggested to contribute to neurodegenerative disorders (26, 27, 45). These observations and our present findings raise the possibility that mutations in miR-binding sites or in miR genes themselves can trigger neurodegenerative disease.
Inhibitors of α-Syn expression are attractive therapeutic targets for PD and other α-synucleinopathies. The results shown here provide a target that can accomplish this objective and potentially slow or halt PD progression.
Materials and Methods
Cell Culture, Transfection, and Luciferase Assay.
Human embryonic kidney cell line HEK293T and human neuroblastoma cell line SH-SY5Y were obtained from American Type Culture Collection (ATCC), and mouse neuroblastoma cell line NS20Y was a kind gift from Dr. Marshall Nirenberg [National Heart, Lung and Blood Institute (NHLBI), National Institutes of Health (NIH), Bethesda, MD]. All cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS. PremiRNA-7 and miRNA-7 inhibitor were purchased from Ambion. Transfections were performed using Lipofectamine 2000 reagent (Invitrogen), according to the supplier's instructions. For luciferase assay, cells were cotransfected with luciferase reporter constructs and internal control construct pSV-β-galactosidase in the absence or presence of miR-7 or premiR-7 at the indicated concentrations. After cell lysis, luciferase activity was measured with Steady-Glo Luciferase Assay System (Promega) using Luminometer (Victor2; PerkinElmer). β-Galactosidase activity was measured with the β-Galactosidase Enzyme Assay System (Promega) and used to normalize luciferase activity. Experiments were performed in triplicates.
Plasmids.
3′-UTR containing reporter plasmids were constructed by inserting the human α-Syn 3′-UTR into the XbaI site located downstream of the luciferase gene in the pGL3 promoter plasmid (Promega) and named α-Syn-3′-UTR. In addition, this 3′-UTR was inserted downstream of human wild-type or A53T mutant α-Syn coding sequence. α-Syn 3′-UTR was amplified from human adult brain cDNA library (Invitrogen) with primers 5′-CTCTAGAGAAATATCTTTGCTCCCAGTT-3′ and 5′-CTCTAGACATGGTCGAATATTATTTATTGTC-3′, and cloned into pCR2.1-TOPO plasmid (Invitrogen) before subcloning into pGL3 promoter plasmid and α-Syn-expressing plasmids. The plasmid α-Syn-mUTR containing mutation of the miR-7 target site in the α-Syn 3′-UTR was created using the QuikChange site-directed mutagenesis kit (Stratagene) with primers 5′-TCTCGAAGTCAACCATCAGCAG-3′ and 5′-CTGCTGATGGTTGACTTCGAGA-3′, and DNA sequences were confirmed. Control plasmids EGFR containing 3′-UTR of EGF receptor, and pri-miR-7-2 expressing premiR-7-2 with 600-bp flanking sequences, were gifts from Dr. Benjamin Purow (University of Virginia, Charlottesville, VA) (33).
Western Blot Analysis.
Cell lysates were analyzed by western blotting as described previously (46) using mouse monoclonal anti-α-Syn antibodies SYN-1 (BD Transduction Lab) and LB509 (Zymed), mouse monoclonal β-actin antibody (Sigma), and rabbit polyclonal anti-GFP antibody (Santa Cruz Biotechnology). Band densities were quantified using National Institutes of Health (NIH) Image J.
Quantitative Real-time RT-PCR.
For qRT-PCR of α-Syn, total RNAs were prepared from cells with TRIzol reagent (Gibco BRL) according to the manufacturer's instructions. Reverse transcription reaction was performed with 1 μg total RNA per 20 μL reaction. Real-time PCR was performed in triplicates with Taqman PCR Mix (Applied Biosystems) in the ABI's 7900HT Fast Real-Time PCR System (Applied Biosystems). All primers were purchased from Applied Biosystems. The level of α-Syn mRNA expression was normalized to GAPDH mRNA. For qRT-PCR of miR-7, brains from 12-week-old C57/B6 mice were used to prepare total RNA from various regions with miRCURY RNA isolation kit (Exiqon) according to the manufacturer's protocol. Reverse transcription was done with miRCURY LNA first-strand cDNA kit (Exiqon), and qRT-PCR was performed with miRCURY LNA miR-7 and U6 primer set (Exiqon) using miRCURY LNA SYBR Green master mix (Exiqon) according to the manufacturer's instruction.
Cell Death Assay.
NS20Y cells transfected with α-Syn-expressing constructs in the absence or presence of miR-7 were exposed to H2O2, and cell death was assessed by lactate dehydrogenase (LDH) Cytotoxicity Detection kit (Roche Molecular Biochemicals) as described in ref. 47.
MPTP Administration.
Male C57/B6 mice, 12 weeks of age, received IP injections of MPTP-HCl (30 mg/kg/day of free base; Sigma) (n = 3) or saline (n = 2) for 5 consecutive days and killed 14 days after the last injection (40). Animals were perfused with saline, and brains were harvested. The ventral midbrain area was dissected, total RNA extracted and used for qRT-PCR. Three separate qRT-PCRs were performed on each brain sample. Animal experiments were approved by the Robert Wood Johnson Medical School Institutional Animal Care and Use Committee.
Statistical Analysis.
Statistical significance between control and experimental values was determined using Student's t test (paired, 2-tailed). All data are expressed as means ± SD.
For additional details of materials and methods used, see SI Text.
Supplementary Material
Acknowledgments.
E.J. is supported by the American Parkinson Disease Association and the Foundation of University of Medicine and Dentistry of New Jersey. M.M.M. is the William Dow Lovett Professor of Neurology.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906277106/DCSupplemental.
References
- 1.Thomas B, Beal MF. Parkinson's disease. Hum Mol Gene. 2007;2:R183–R194. doi: 10.1093/hmg/ddm159. [DOI] [PubMed] [Google Scholar]
- 2.Mouradian MM. Recent advances in the genetics and pathogenesis of Parkinson disease. Neurology. 2002;58:179–185. doi: 10.1212/wnl.58.2.179. [DOI] [PubMed] [Google Scholar]
- 3.Eriksen JL, Dawson TM, Dickson DW, Petrucelli L. Caught in the act: Alpha-synuclein is the culprit in Parkinson's disease. Neuron. 2003;40:453–456. doi: 10.1016/s0896-6273(03)00684-6. [DOI] [PubMed] [Google Scholar]
- 4.Lim KL, Dawson VL, Dawson TM. The cast of molecular characters in Parkinson's disease: Felons, conspirators, and suspects. Ann N Y Acad Sci. 2003;991:80–92. doi: 10.1111/j.1749-6632.2003.tb07465.x. [DOI] [PubMed] [Google Scholar]
- 5.Polymeropoulos MH, et al. Mutation in the α-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 6.Kruger R, et al. Ala30Pro mutation in the gene encoding α-synuclein in Parkinson's disease. Nat Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 7.Zarranz JJ, et al. The new mutation, E46K, of α-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 8.Singleton AB, et al. α-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 9.Farrer M, et al. Comparison of kindreds with parkinsonism and α-synuclein genomic multiplications. Ann Neurol. 2004;55:174–179. doi: 10.1002/ana.10846. [DOI] [PubMed] [Google Scholar]
- 10.Masliah E, et al. Dopaminergic loss and inclusion body formation in α-synuclein mice: Implications for neurodegenerative disorders. Science. 2000;287:1265–1269. doi: 10.1126/science.287.5456.1265. [DOI] [PubMed] [Google Scholar]
- 11.Feany MB, Bender WW. A Drosophila model of Parkinson's disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 12.Lakso M, et al. Dopaminergic neuronal loss and motor deficits in Caenorhabditis elegans overexpressing human α-synuclein. J Neurochem. 2003;86:165–172. doi: 10.1046/j.1471-4159.2003.01809.x. [DOI] [PubMed] [Google Scholar]
- 13.Kanda S, Bishop JF, Eglitis MA, Yang Y, Mouradian MM. Enhanced vulnerability to oxidative stress by α-synuclein mutations and C-terminal truncation. Neuroscience. 2000;97:279–284. doi: 10.1016/s0306-4522(00)00077-4. [DOI] [PubMed] [Google Scholar]
- 14.Junn E, Mouradian MM. Human α-synuclein over-expression increases intracellular reactive oxygen species levels and susceptibility to dopamine. Neurosci Lett. 2002;320:146–150. doi: 10.1016/s0304-3940(02)00016-2. [DOI] [PubMed] [Google Scholar]
- 15.Maraganore DM, et al. Collaborative analysis of α-synuclein gene promoter variability and Parkinson disease. J Am Med Assoc. 2006;296:661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 16.Spillantini MG, et al. α-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 17.Spillantini MG, Goedert M. The α-synucleinopathies: Parkinson's disease, dementia with Lewy bodies, and multiple system atrophy. Ann NY Acad Sci. 2000;920:16–27. doi: 10.1111/j.1749-6632.2000.tb06900.x. [DOI] [PubMed] [Google Scholar]
- 18.Masliah E, et al. Effects of α-synuclein immunization in a mouse model of Parkinson's disease. Neuron. 2005;46:857–868. doi: 10.1016/j.neuron.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 19.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 20.Gartel AL, Kandel ES. miRNAs: Little known mediators of oncogenesis. Semin Cancer Biol. 2008;18:103–110. doi: 10.1016/j.semcancer.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 22.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 23.Miska EA, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bilen J, Liu N, Burnett BG, Pittman RN, Bonini NM. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol Cell. 2006;24:157–163. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 26.Hebert SS, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/β-secretase expression. Proc Natl Acad Sci USA. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y, et al. miR-19, miR-101 and miR-130 co-regulate ATXN1 levels to potentially modulate SCA1 pathogenesis. Nat Neurosci. 2008;11:1137–1139. doi: 10.1038/nn.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, et al. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bak M, et al. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 32.Webster RJ, et al. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 33.Kefas B, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 34.Jiang H, et al. Parkinson's disease genetic mutations increase cell susceptibility to stress: Mutant α-synuclein enhances H2O2- and Sin-1-induced cell death. Neurobiol Aging. 2007;28:1709–1717. doi: 10.1016/j.neurobiolaging.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 35.Petrucelli L, et al. Parkin protects against the toxicity associated with mutant α-synuclein: Proteasome dysfunction selectively affects catecholaminergic neurons. Neuron. 2002;36:1007–1019. doi: 10.1016/s0896-6273(02)01125-x. [DOI] [PubMed] [Google Scholar]
- 36.Stefanis L, Larsen KE, Rideout HJ, Sulzer D, Greene LA. Expression of A53T mutant but not wild-type α-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. J Neurosci. 2001;21:9549–9560. doi: 10.1523/JNEUROSCI.21-24-09549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farh KK, et al. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 38.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalivendi SV, et al. α-synuclein up-regulation and aggregation during MPP+-induced apoptosis in neuroblastoma cells: Intermediacy of transferrin receptor iron and hydrogen peroxide. J Biol Chem. 2004;279:15240–15247. doi: 10.1074/jbc.M312497200. [DOI] [PubMed] [Google Scholar]
- 40.Vila M, et al. α-synuclein up-regulation in substantia nigra dopaminergic neurons following administration of the parkinsonian toxin MPTP. J Neurochem. 2000;74:721–729. doi: 10.1046/j.1471-4159.2000.740721.x. [DOI] [PubMed] [Google Scholar]
- 41.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G, et al. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of α-synuclein. Am J Hum Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abelson JF, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 44.Schaefer A, et al. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hebert SS, et al. MicroRNA regulation of Alzheimer's amyloid precursor protein expression. Neurobiol Dis. 2009;33:422–428. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 46.Junn E, Ronchetti RD, Quezado MM, Kim SY, Mouradian MM. Tissue transglutaminase-induced aggregation of α-synuclein: Implications for Lewy body formation in Parkinson's disease and dementia with Lewy bodies. Proc Natl Acad Sci USA. 2003;100:2047–2052. doi: 10.1073/pnas.0438021100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Junn E, Jang WH, Zhao X, Jeong BS, Mouradian MM. Mitochondrial localization of DJ-1 leads to enhanced neuroprotection. J Neurosci Res. 2009;87:123–129. doi: 10.1002/jnr.21831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.