Abstract
Kinesin I can walk on a microtubule for distances as long as several micrometers. However, it is still unclear how this molecular motor can remain attached to the microtubule through the hundreds of mechanochemical cycles necessary to achieve this remarkable degree of processivity. We have addressed this issue by applying ensemble and single-molecule fluorescence methods to study the process of kinesin stepping, and our results lead to 4 conclusions. First, under physiologic conditions, ≈75% of processively moving kinesin molecules are attached to the microtubule via both heads, and in this conformation, they are resistant to dissociation. Second, the remaining 25% of kinesin molecules, which are in an “ATP waiting state” and are strongly attached to the microtubule via only one head, are intermittently in a conformation that cannot bind ATP and therefore are resistant to nucleotide-induced dissociation. Third, the forward step in the kinesin ATPase cycle is very fast, accounting for <5% of the total cycle time, which ensures that the lifetime of this ATP waiting state is relatively short. Finally, by combining nanometer-level single-molecule fluorescence localization with higher ATP concentrations than used previously, we have determined that in this ATP waiting state, the ADP-containing head of kinesin is located 8 nm behind the attached head, in a location where it can interact with the microtubule lattice. These 4 features reduce the likelihood that a kinesin I motor will dissociate and contribute to making this motor so highly processive.
Keywords: fluorescence, motility, gating, fluorescence imaging with 1-nm accuracy, processivity
Processive molecular motors, such as kinesin I, myosin V, and myosin VI, walk along their tracks in a hand-over-hand manner, with at least one head attached to the track at all times (1–4). Although this arrangement is necessary for processivity, it is not sufficient. What is also required is a mechanism that allows the two heads to communicate their enzymatic state to each other. This process, which is referred to as “gating,” ensures that both heads do not simultaneously assume a weak-binding conformation and hence dissociate prematurely.
How gating is accomplished by kinesin remains controversial. Several reports, using solution kinetics or single-molecule measurements, found evidence that nucleotide binding to the lead head of a doubly attached kinesin conformation is gated by intramolecular strain (4–8). For strain to work effectively as a gating mechanism, kinesin must attach to the microtubule with both heads for an appreciable fraction of its cycle time. Although this mechanism is consistent with prior models of the kinesin ATPase cycle (9–11), a more recent study has questioned whether strain can play any role in the gating process of kinesin (12). Alsonso et al. (12) found that, like microtubules, unpolymerized αβ tubulin could also activate the ATPase of kinesin by accelerating ADP release from only 1 of the 2 heads of kinesin. This finding was interpreted to mean that gating can occur with just 1 head attached to tubulin, a condition where intramolecular strain cannot develop.
What is needed to resolve this uncertainty is a method of monitoring the conformational state of a processively moving kinesin at the single-molecule level, with enough spatial and temporal resolution to unequivocally test the various models that have been proposed. In this study, we used single-molecule rhodamine fluorescence quenching (smFQ), which provides structurally sensitive information on the neck linkers with the temporal resolution needed to resolve the state of the kinesin motor through the course of a processive run. Importantly, smFQ allows us to monitor the conformation of kinesin during discrete phases of its mechanochemical cycle, which is not possible with other methods, such as fluorescence imaging with 1-nm accuracy (FIONA) or optical trapping (2, 13). In this study, we have combined the temporal accuracy of smFQ with the spatial accuracy of FIONA to monitor the interhead distance of single kinesin molecules moving processively on microtubules. These studies have allowed us to create a model that explains how kinesin can be so processive.
Results
Rhodamine Quenching Can Monitor the Effect of Microtubule and Nucleotide Binding on the Degree of Separation of the Two Neck Linkers.
Labeling a dimeric kinesin construct with tetramethylrhodamine-5 (TMR)-maleimide at position 333, within its neck linker, produces a preparation whose fluorescence emission senses the relative separation of the two neck linkers. The fluorescence of TMR-kinesin is strongly quenched, because the proximity of the two neck linkers to each other allows the rhodamine fluorophores to dimerize (6, 7). Forward stepping of kinesin on the microtubule separates the neck linkers and their attached rhodamines and consequently produces a large fluorescence enhancement. In this study, we have applied this methodology to examine the process of kinesin I stepping at both the ensemble and single-molecule levels by using 2 engineered kinesin dimers: K413, consisting of the first 413 amino-terminal residues, and K560, a longer 560-residue construct.
We first examined how binding of kinesin to the microtubule affects the fluorescence of the TMR probe. We generated 2 rhodamine-labeled K560 preparations with different labeling stoichiometries, one with 0.93 TMR/heavy chain, and the other with 0.05 TMR/heavy chain. As Fig. 1A shows, K560 with a single TMR probe per dimer (magenta spectrum) has a relative quantum yield that is ≈2.6-fold larger than doubly labeled TMR-K560 (red spectrum). Adding a 5-fold molar excess of microtubules in the absence of added nucleotide (Fig. 1A, blue spectrum) enhances the fluorescence of doubly labeled TMR-K560 by ≈40%. Adding ADP to 2 mM dissociates TMR-K560 from the microtubule and restores the fluorescence of doubly labeled TMR-K560 to that in the absence of microtubules (Fig. 1A, dark green spectrum). By contrast, adding microtubules to singly labeled TMR-K560 increased fluorescence emission by <5%.
Fig. 1.
Microtubule binding induces an equilibrium between two kinesin orientations. (A) Rhodamine fluorescence emission (λex = 545 nm) of doubly labeled TMR-K560 containing 0.5 μM rhodamine and 0.95 mol rhodamine/mol K560 heavy chain (red spectrum), singly labeled TMR-K560 containing 0.5 μM rhodamine and 0.05 mol rhodamine/mol K560 heavy chain (magenta spectrum), 0.5 μM doubly labeled TMR-K560 + 2.5 μM microtubules (blue spectrum), and 0.5 μM doubly labeled TMR-K560 + 2.5 μM microtubules + 2 mM ADP (green spectrum). (B) Mixing a complex of doubly labeled TMR-K560 + a 5-fold excess of microtubules with 2 mM ADP in the stopped flow produces a time-dependent decrease in fluorescence intensity (red data points) that fits a single exponential decay (yellow solid line) with rate constant of 492 ± 13 s−1. Conditions were 100 mM KCl, 25 mM Hepes, 2 mM MgCl2, 1 mM DTT, pH 7.50 at 20 °C.
These results suggest that microtubule binding induces an equilibrium distribution of two conformations in kinesin. We measured the rate of formation of this equilibrium by mixing a TMR-K560 + microtubule complex with 2 mM ADP in the stopped flow, which drives this equilibrium to favor the neck linker-associated, rhodamine-quenched conformation, and produces a fluorescence decrease characterized by a rate constant of 492 ± 13 s−1 (Fig. 1B).
Our measurements of single molecules of TMR-K560 support this model. We confirmed the stoichiometry of TMR labeling by demonstrating 1-step photobleaching for singly labeled TMR-K560 and 2-step photobleaching for doubly labeled TMR-K560. Fig. S1 illustrates traces of fluorescence versus time for single molecules of TMR-K560 that were attached to axonemes in the absence of ATP, imaged at 200 Hz, with 1 (A) or 2 (B) TMR per dimer. To compare the fluorescence of different TMR kinesin preparations at the single-molecule level, we created an intensity scale of arbitrary units (a.u.) where we define background as 0 a.u. and the mean fluorescence intensity of a singly labeled TMR kinesin as 1 a.u. (Fig. S1C). Using this scale, we found that the fluorescence output for doubly labeled TMR-K560 showed a broad distribution (Fig. S1B), with a mean intensity of 1.22 ± 0.96 a.u. (n = 50; Fig. S1D).
Mixing rhodamine-labeled kinesin I + microtubules with ATP in the stopped flow produces a rapid rise in fluorescence intensity, followed by a fall (refs. 6 and 7 and Fig. S2A). The rate constant of the rising phase (kobs), varies with ATP concentration, defining a maximum of 357 ± 23 s−1 at 20 °C (Fig. S2B, red curve). Adequate fitting of the falling phase requires a double exponential function. The more rapid component constitutes ≈80% of the total amplitude; and its rate constant, at 72 ± 9 s−1, shows little variation with ATP concentration (Fig. S2B, magenta). The rate constant of the slower component varies hyperbolically with ATP concentration, with an extrapolated maximum of 30 ± 3 s−1 (Fig. S2B, blue; shown on an expanded scale in Fig. S2C).
Rhodamine Quenching Can Be Used to Identify Individual Steps at the Single-Molecule Level.
We monitored the fluorescence intensity of individual TMR-labeled K413 dimers as they walked processively on a microtubule. We note that the velocities for K560 and K413 are essentially identical over a 100-fold range of ATP concentrations. At 10 μM ATP, we observed repetitive, alternating cycles of fluorescence enhancement and quenching as individual kinesin motors walked along axonemes (Fig. 2 A and B). The kinesin molecule in Fig. 2A underwent a displacement of 450 nm, corresponding to 55 steps, assuming a step size of 8.3 nm (10). During this same processive run, we observed 49 cycles of quenching and unquenching (Fig. 2B). In general, we found that there were 0.93 ± 0.06 (mean ± SEM) quenching events for every 8.2-nm step. A histogram of intensities during the processive run for this molecule (Fig. 2C) demonstrates two populations with peaks at ≈0 and 2 a.u. A histogram composed of the processive runs of 18 molecules at 5 μM ATP also shows this bimodal distribution (Fig. S3).
Fig. 2.
Single-molecule measurements of TMR-labeled kinesin displacement and fluorescence quenching/unquenching. (A) Displacement of the TMR-labeled kinesin at 10 μM ATP and imaged at 500 Hz is depicted in black. A linear fit was performed (red line) to estimate the velocity and deduce the number of steps. This particular kinesin-1 molecule took 55 steps. (B) Fluorescence intensity of the TMR probe during the processive run depicted in A. Rhodamine intensity is blue, and fitting is red. A total of 49 quenching events were observed. (Inset) An expanded portion of the trace, depicting fluorescence fluctuations between 1.6 and 2.6 s. Intensity counts were normalized by setting the quenched state to 0 a.u. Fluorescence intensity occasionally exceeded 2 a.u. because of the presence of other kinesin molecules rapidly landing and dissociating in the close proximity of molecule of interest. (C) The histogram for the intensity counts of all CCD frames throughout the movement shows 2 distinct peaks. (D) Dwell time histogram for 544 unquenched states. Red line shows the fit to a single exponential decay function τunquenched = 40.0 ± 1.0 ms (R2 = 0.99). (E) Dwell time histogram for 545 quenched states. Red line shows the fit to a single exponential decay R2 = 0.99. τunquenched = 34.0 ± 1.4 ms (R2 = 0.99).
If one cycle of quenching and unquenching corresponds to a single step, then the dwell time for the motor should be equal to the sum of quenched and unquenched durations. We found that the average velocity of kinesin molecules in the presence of 10 μM ATP was 109 ± 8 nm/s or 13.0 ± 1.2 steps/s (n = 91). The quenched (Fig. 2D) and unquenched (Fig. 2E) durations were determined by fitting to a single exponential decay and were found to be 34.0 ± 1.4 and 40.0 ± 1.0 ms, respectively (544 quenching events). The sum of these values defines a dwell time per step of 74.0 ± 1.7 ms, which corresponds to ≈13.5 steps per s (n = 43), in excellent agreement with our measured value. Likewise, at 22 μM ATP, the quenched and unquenched durations (580 quenching events) were found to be 31 ± 1.0 and 25 ± 1.3 ms, respectively, corresponding to a dwell time of 56 ± 1.6 ms and a velocity of ≈17.9 steps per s. We measured an average velocity of 18.6 ± 1.1 steps per s (n = 32), also in excellent agreement with our lifetime-derived velocity.
We performed several studies to rule out the possibility that the changes in rhodamine fluorescence are caused by fluorophore blinking from triplet-state decay. First, all of our experiments used oxygen scavengers to minimize oxygen-dependent triplet-state formation. Second, we repeated the same motility assay with K413 labeled with an average of <1 TMR fluorophore per kinesin dimer. We found that the fluorescence intensity of these singly labeled K413 molecules did not show any significant fluctuations (Fig. S4A) when using the same ATP concentration and imaging rate (500 Hz). Blinking events appeared as a minor peak on the fluorescence intensity histogram, corresponding to ≈5% of all counts (Fig. S4A, red histogram). Third, when the motor paused, the rhodamine fluorescence remained unquenched and did not vary in a way expected for random blinking (Fig. S4B). Fourth, doubling the laser power did not change the frequency of quenching events for doubly labeled TMR-K413.
Steady-State Rhodamine Fluorescence Intensity at the Single-Molecule Level Varies with ATP Concentration.
Individual quenching events at the single-molecule level became too rapid to observe at 50 μM [ATP]. To monitor how ATP concentration affects the distribution of bound states at the single-molecule level, we therefore imaged at 5 Hz, where we could measure an average fluorescence intensity during a processive run. Fig. 3A shows an example at 1 mM [ATP]. As expected, the average intensity for a singly labeled kinesin remains nearly invariant at ≈1 a.u. over a 100-fold range of ATP concentration (Fig. 3B, red circles). By contrast, the amplitude of doubly labeled TMR-kinesin varies with [ATP] hyperbolically, extrapolating to 0.87 ± 0.11 a.u. in the absence of ATP and 1.5 ± 0.23 a.u. at complete saturation. We correlated these results with measurements of rhodamine fluorescence at the ensemble level, by adding ATP to a complex of 1 μM doubly labeled TMR-K413 + 5 μM microtubules in the presence of an ATP-regenerating system. Fitting the steady-state intensity, measured in a fluorimeter, to a rectangular hyperbola and extrapolation of the fit to 0 [ATP] reveals a fluorescence emission ≈30% that seen in the presence of saturating ATP (Fig. 3B Inset).
Fig. 3.
Effect of ATP concentration on single-molecule average intensity of doubly labeled TMR kinesin I. (A) Displacement (Upper, black line) and intensity (Lower, blue line) of a kinesin molecule moving in the presence of 1 mM ATP and imaged at 500 Hz is shown. The fluorescence intensity of TMR-labeled kinesin remains relatively constant at ≈2.0 a.u. in the presence of 1 mM ATP. (B) Variation of average fluorescence amplitude with ATP concentration for doubly labeled TMR kinesin (blue) and singly labeled TMR kinesin (red). Singly labeled TMR kinesin shows little variation, remaining at ≈1.0 a.u. (dotted red line). By contrast, the amplitude of doubly labeled TMR-kinesin varies in a hyperbolic manner, extrapolating to 0.87 ± 0.11 a.u. in the absence of ATP and 1.5 ± 0.23 a.u. at complete saturation. The extrapolated intensity at 0 ATP concentration would correspond to a value of k1/k-1 of 1.3. (Inset) ATP was added to a rigor complex consisting of 1 μM TMR-K413 + 5 μM microtubules in the presence of an ATP regenerating system (4). The resulting fluorescence intensity is plotted as a function of [ATP], after subtracting the fluorescence emission of TMR-K413 in the absence of microtubules. Fitting to a rectangular hyperbola and extrapolation of the fit to 0 ATP concentration reveals a fluorescence emission ≈30% that seen in the presence of saturating ATP.
FIONA Measurements Determine the Positions of the Two Heads with High Spatial and Temporal Resolution.
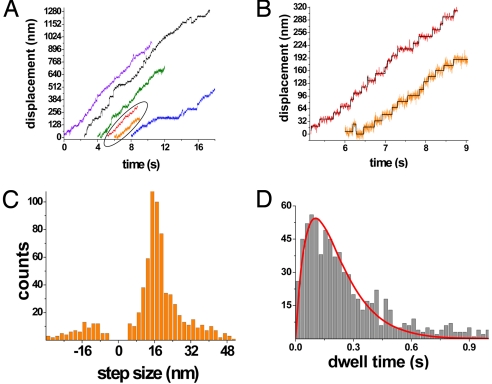
Although rhodamine smFQ provides information about the timing of structural changes in the neck linker, it does not provide information on the position of the heads. We therefore labeled K560 with quantum dots on the amino terminus of the motor domain at position 215 and imaged their position at 500 Hz by using FIONA. At 5 μM ATP, we observed abrupt ≈16-nm steps, consistent with a hand-over-hand mechanism (Fig. 4 A and B). The average step size was 16.3 ± 4.4 nm (Fig. 4C). Fitting the dwell time histogram required use of a probability function of the form P(t) = tk2e−kt (Fig. 4D), which implies that each dwell time contains 2 stepping events (2, 4), with a stepping rate constant (k) of 9.8 ± 0.3 s−1 (mean ± SEM). This result compares with a value of 8.7 ± 0.5 s−1, derived from the Michaelis–Menten dependence of velocity with [ATP]. By contrast, if only one stepping event occurred with each dwell time, the histogram would fit a single exponential decay [P(t) ∝ e−kt].
Fig. 4.
FIONA measurements of displacement at fast imaging rates. (A) Displacement traces of 6 different quantum dot-labeled kinesins in the presence of 5 μM ATP. Imaging rate is 500 Hz except for the red and black traces where it is at 250 Hz. Multiple consecutive 16-nm steps were observed for these molecules. All stepping events were instantaneous within our time resolution. (B) Zoomed images of selected traces from A. Black lines are computer-generated fits. (C) Step-size histogram of 770 steps from 30 kinesin molecules. The average step size is 16.3 ± 4.4 nm. Backward steps were occasionally observed (−15.6 ± 6.6 nm). (D) Dwell time histogram of 770 steps. The red line represents the best fit to the convolution function, P(t) = A·t·k2·exp(−k·t), with a stepping rate (k) of 9.76 s−1 (R2 = 0.97).
The standard deviation of the position measurement from quantum dot-labeled kinesin heads between consecutive steps was 6.8 nm (770 dwell periods; Fig. S5). This variation reflects the combined effects of vibration, imaging uncertainty, flexibility in the biotin-streptavidin link, and Brownian movements of the head (σtotal2 = σvibrations2 + σimaging2 + σhead2). In a control experiment, we simulated the noise contributed by vibration, imaging uncertainty, and biotin-streptavidin wobbling by attaching streptavidin coated quantum dots to a BSA-biotin-coated glass surface and imaging them at 500 Hz. The standard deviation of the position signal for 50 quantum dots (1,000 data points for each quantum dot) was 6.1 nm. We therefore conclude that Brownian movements of the head (σhead2) contribute to the total positional variation by ≈3 nm (σhead = ). Abrupt 16-nm steps and the reasonably small Brownian movement of the heads strongly suggest that the distance between kinesin heads remains at ≈8 nm while kinesin heads dwell on microtubules.
Discussion
In this study, we have used the fluorescence quenching that occurs with rhodamine dimer formation at both the ensemble and single-molecule levels to generate a model that addresses several unresolved questions regarding kinesin's stepping mechanism and is summarized schematically in Fig. 5. We enter the model with one head of the kinesin dimer bound to the microtubule in rigor. The other head, referred to as tethered, has ADP in its catalytic site (14). Although this state has been thought of as a discrete conformation (5, 15), our data show instead that it is an equilibrium mixture of 2 conformations, corresponding to quenched and unquenched, and characterized by rate constants k1 and k-1. This mixture produces an intermediate degree of fluorescence emission in the absence of ATP at both the ensemble (Fig. 1) and single-molecule (Fig. S1) levels, which implies that both conformations are appreciably populated when bound to the microtubule. Although the rhodamine fluorescence studies do not tell us where the tethered head is in relation to the rigor head, our results with FIONA (Fig. 4) imply that the two heads remain separated by ≈8 nm in both the quenched and unquenched conformations, because the distance between kinesin heads remains at ≈8 nm throughout a processive run. These studies would therefore predict that because the leftmost image in Fig. 5 is unquenched, the lifetime of the unquenched state should decrease with increasing [ATP]. This prediction is confirmed by our direct measures of unquenched lifetimes at 10 and 22 μM ATP (Fig. 2 D and E) and is consistent with prior kinetic models (9, 10).
Fig. 5.
Schematic depiction of the kinesin mechanochemical cycle. Four kinesin dimers are depicted and are symbolized by the two triangular heads, connected to a coiled coil (purple) via the neck linkers (blue line). The two heads are labeled magenta and yellow to distinguish them from each other. The microtubule is depicted as the gray structure, the + and − ends are indicated in black, and the direction of movement of the kinesin molecules is indicated by the arrow at the bottom. Binding of kinesin to the microtubule generates a rapid equilibrium between two conformations, whose rate of formation, k1 + k-1 = 492 ± 13 s−1. These conformations are distinguished by the positions of the neck linkers and TMR probes (red discs), separated and unquenched (illustrated by the rays emanating from the leftmost kinesin) and closely associated and quenched (second kinesin from left). Nucleotide can only bind to the lead, yellow head when the two neck linkers are in close proximity. Binding of ATP (blue sphere) leads to forward stepping (third kinesin from left), separation of the neck linkers, and unquenching of the TMR probes. This step occurs with apparent second-order rate constant of k2·KT, where 1/KT is the dissociation constant of ATP to kinesin (25 ± 6 μM), and k2 is the rate constant for forward stepping (≫1,000 s−1). ATP hydrolysis on the new trailing head, occurring with rate constant of kh, and ADP dissociation from the lead head, occurring with rate constant kd, follow, generating the rightmost kinesin conformation. Both of these steps are irreversible and can be described by a composite rate constant k3 = kh·kd/kh + kd. Phosphate release follows with rate constant k4 to complete the cycle. The relative reaction rates for the various steps are symbolized by the widths of the reaction arrows, with the widest arrow corresponding to the fastest rate.
Our study supports and extends earlier models of kinesin stepping (6–8, 16), in which rearward strain on the forward head gates nucleotide binding. We now propose that this gating mechanism applies as well to those kinesin molecules that are “waiting” to bind ATP (two leftmost molecules in Fig. 5). We propose that ATP binding to the lead head is inhibited in any conformation of kinesin where the neck linkers are separated, and it allows us to make several predictions. First, binding of ATP to the catalytically empty head of kinesin on the microtubule (leftmost image in Fig. 5) should not occur any faster than k1. By contrast, in the absence of microtubules, the neck linkers of kinesin are in close proximity to each other, and ATP binding should not be rate limited by k1. In fact, we had found (7) that although the maximum rate of binding 2′ deoxy 3′ mant ATP to nucleotide-free kinesin is >1,000 s−1, the corresponding value for a complex of kinesin + microtubules is 457 ± 56 s−1. Second, our model predicts that like ATP binding, the observed rate constant for the forward step in the mechanochemical cycle (kobs in Fig. S2B) should also be rate-limited by k1 and should be much less than k2, the actual rate constant for the forward step. According to the model in Fig. 5,
 |
where
Note that k¯2 ≈ k2 at [ATP] ≫ KT, and KT is the binding constant for ATP. Given that k1 + k-1 is ≈500 s−1 (Fig. 1B), and that k1/k-1 ≈1.3–1.5 (Fig. 3B), we conclude that k1 ≈300 s−1, which is fairly close to our measured value of kobs of 357 ± 23 s−1. This implies that k2 ≫ k1, and, as predicted from our model, that kobs ≪ k2. This conclusion is also consistent with a previous study, which proposed that forward stepping occurs on a microsecond time scale (17).
Our ensemble kinetic data, summarized in Fig. S2, indicate that the return to the quenched state can occur via 2 separate pathways. The rate constant for the first, associated with 80% of the signal amplitude (Fig. S2B, magenta data points), does not vary with [ATP] and it averages 72 ± 9 s−1, similar to the rate of phosphate release (6, 7). We propose that this reaction occurs after kinesin takes a forward step (k2 in Fig. 5), is associated with phosphate release, and reflects the dissociation of the rear head of a doubly attached, internally strained kinesin. This is depicted in Fig. 5 by the curved arrow with rate constant k4. However, the rate constant for the smaller, 20% amplitude component (Fig. S2B, blue data points) does vary with [ATP]. We propose that this reflects an alternative reaction, where the strongly bound head of kinesin binds and hydrolyzes ATP and dissociates from the microtubule before it can step forward and generate internal strain. This reaction is not depicted in Fig. 5, because it is off of the main processive pathway. The rate constant for this reaction, at 30 ± 3 s−1, therefore describes the rate of release of an unstrained head. Thus, although we agree with a previous report that forward strain can accelerate the rate of trailing head release (18), our data would suggest that the ≈2.5-fold difference we observe is too small to play any significant role in the process of gating.
Our model proposes that only one conformation is competent to bind ATP, regardless of ATP concentration. This would correspond to the quenched species in Fig. 5 (second from left). In this regard, we agree with the model of Alonso et al. (12). However, their conclusion that the tethered head is held in a forward position while the motor is waiting to bind ATP is not consistent with our FIONA results (Fig. 4). These results demonstrate steps of ≈16 and ≈0 nm, whereas the model of Alonso et al. (12) would predict step sizes of significantly <16 nm and significantly >0 nm. Furthermore, our model depends on intramolecular strain to mediate gating, and thus fundamentally disagrees with their proposal that gating occurs from a one head bound, unstrained state. In a more recent study, Mori et al. (15) proposed that the nature of the ATP waiting state varies with ATP concentration, with one head bound at low ATP concentration and both heads bound at high ATP concentration. We agree that the nature of the predominant bound intermediate varies with ATP concentration. However, our results imply that gating occurs predominantly on the lead head when the neck linkers are separated and the system is under strain. This conclusion is consistent as well with prior studies (6–8) that have clearly demonstrated that ATP cannot bind to the lead head when it is experiencing rearward strain.
Four features of our model help explain how kinesin I can be so highly processive. First, fitting our single-molecule average intensity measurements as a function of ATP concentration (Fig. 3B) reveals a maximum of 1.5 ± 0.23 a.u. Because complete unquenching corresponds to 2 a.u., we conclude that under steady-state, processive conditions, ≈75% of the kinesin molecules are unquenched, cannot bind ATP to the lead head, and are therefore resistant to dissociation from the microtubule. Second, our results confirm that the process of forward stepping is very fast (17). This reaction converts the quenched, singly attached kinesin conformation (second from left in Fig. 5), which is susceptible to detachment, to an unquenched, doubly attached species (third from left in Fig. 5), and its speed further reduces the probability of premature detachment. Third, while kinesin is in an ATP waiting state (second image from left in Fig. 5), its tethered head alternates between two positions that are both appreciably populated (because k1/k-1 ≈ 1; Fig. 3). Finally, our FIONA results demonstrate that in the ATP waiting state, the ADP-containing head of kinesin is located 8 nm behind the attached head, in a location where it can at least intermittently interact with the microtubule lattice. Thus, if the tethered head can interact with the microtubule surface enzymatically (19), it may also be able to do so mechanically, generating sufficient strain to gate ATP binding to the rigor head and providing a further safety mechanism to prevent premature detachment.
Thus, our combined approach, using solution kinetics and single-molecule studies with a structurally sensitive fluorescent reporter on the neck linker, provides a highly sensitive method of monitoring the key conformations of the kinesin motor domain in real time.
Materials and Methods
TMR-Labeled Kinesin.
K413W340F and K560W340F mutants were purified and labeled with TMR dyes as described (6).
Quantum Dot-Labeled Kinesin Construct.
The K560GFP-215C construct was purified by following the protocol by Yildiz et al. (4). Cysteines on position 215 (on the head) were biotinylated (20). Biotinylated kinesin molecules were labeled with streptavidin-coated quantum dots (QS655; Invitrogen). This labeling was done while kinesins were attached to the axonemes to prevent formation of multikinesin-attached quantum dots.
In Vitro Motility Assay.
We used the motility assay that has been described (4). An imaging buffer [BRB80 buffer (80 mM Pipes, 1 mM MgCl2, 1 mM EGTA, pH 6.8) + 1% β-mercaptoethanol (BME), 1% GLOXY, 0.4% glucose, 0.01M DTT] was used in all our experiments to minimize blinking events (2).
Imaging.
TMR dyes or quantum dots were excited with a green (532 nm) laser by using a home-built objective-type total internal reflection fluorescence microscope (IX-71; Olympus) (3). Images were captured by a CCD camera (Andor iXon DU-860) and used to track the intensity counts of individual TMR-labeled kinesin molecules.
Data Analysis for Quenching Experiments.
CCD images were used for both intensity counting and localization purposes. Kinesin molecules that moved >150 nm were chosen for analysis. Images were acquired at 500 frames per s. Intensity counts within a region of interest window were summed and background-corrected by using a custom Matlab code. Displacements of TMR-labeled kinesin molecules were calculated by centroid tracking, yielding subpixel localization accuracy. We fit a line to the displacement curve of kinesin molecules to estimate the total displacement. The number of steps was determined by dividing the total displacement by the average step size of kinesin (8.3 nm) (13). Quenching events were scored by using a custom IDL code based on Student's t test (17). In this algorithm, an event is scored if, and only if, kinesin dwells for 3 time points (6 ms) and the intensity change is >50% of the average intensity level of unquenched dyes.
Intensity Counting Experiments with TMR-Labeled Kinesins.
We used the same imaging (5 Hz) and excitation settings (1-mW laser power) at all different ATP concentrations. Total intensity within a 0.49-μm2 region of interest was calculated and background-corrected. Displacements were calculated by fitting a 2D Gaussian function to the CCD images of TMR dyes, and velocities were calculated by fitting a line to the displacement trajectory (2). We used the average fluorescence intensity level of singly labeled TMR kinesin molecules.
Localizing Quantum Dots with FIONA.
CCD images of quantum dots (QS655; Invitrogen) were fit to 2D Gaussian function for tracking quantum dots with nanometer accuracy (2). Quantum dot-labeled kinesin molecules were acquired at 500 frames per s. Although blinking of quantum dots was minimized by oxygen scavengers and BME, ≈2% of the frames were discarded because our computer program occasionally failed to localize the quantum dots because of blinking events. Displacement traces were never smoothed or filtered. Stepping events were scored by a computer program based on a published algorithm (21).
Noise Analysis for the Quantum Dot-Labeled Head's Position.
We did a control experiment to measure the contribution of the noise sources other than Brownian movements of the head itself. We incubated a glass surface with 0.67 mg/mL BSA-biotin solution (in BRB80) for 5 min and washing with BRB80 buffer. This BSA-biotin coated surface was then incubated with 50 pM quantum dots in BRB80 for 5 min. Finally, we washed the chamber with 50 μL of imaging buffer (1% BME, 1% GLOXY, 0.4% glucose in BRB80). Quantum dots were imaged with 500-Hz imaging rate and localized by using FIONA.
Supplementary Material
Acknowledgments.
We thank Marileen Dogterom for sharing the step finder program; Ben Blehm, Comert Kural, Evan Graves, Hasan Yardimci, Sultan Doganay, Sheyum Syed, and Tyler Lougheed for helpful discussions; and Trine Giaever for producing Fig. 5. This work was supported by National Institutes of Health Grants GM068625 (to P.R.S.) and AR048565 (to S.S.R.) and National Science Foundation Frontiers Grant 0822613 (to P.R.S).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808396106/DCSupplemental.
References
- 1.Okten Z, Churchman LS, Rock RS, Spudich JA. Myosin VI walks hand-over-hand along actin. Nat Struct Mol Biol. 2004;11:884–887. doi: 10.1038/nsmb815. [DOI] [PubMed] [Google Scholar]
- 2.Yildiz A, et al. Myosin V walks hand-over-hand: Single fluorophoree imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 3.Yildiz A, et al. Myosin VI steps via a hand-over-hand mechanism with its lever arm undergoing fluctuations when attached to actin. J Biol Chem. 2004;279:37223–37226. doi: 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
- 4.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004b;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 5.Guydosh NR, Block SM. Backsteps induced by nucleotide analogs suggest the front head of kinesins gated by strain. Proc Natl Acad Sci USA. 2006;103:8054–8059. doi: 10.1073/pnas.0600931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenfeld SS, Xing J, Jefferson GM, Cheung HC, King PH. Measuring kinesin's first step. J Biol Chem. 2002;277:36731–36739. doi: 10.1074/jbc.M205261200. [DOI] [PubMed] [Google Scholar]
- 7.Rosenfeld SS, Fordyce PM, Jefferson GM, King PH, Block SM. Stepping and stretching: How kinesin uses internal strain to walk processively. J Biol Chem. 2003;278:18550–18556. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uemura S, Ishiwata S. Loading direction regulates the affinity of ADP for kinesin. Nat Struct Biol. 2003;10:308–311. doi: 10.1038/nsb911. [DOI] [PubMed] [Google Scholar]
- 9.Ma YZ, Taylor EW. Interacting head mechanism of microtubule-kinesin ATPase. J Biol Chem. 1997;272:724–730. doi: 10.1074/jbc.272.2.724. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert SP, Moyer ML, Johnson KA. Alternating site mechanism of the kinesin ATPase. Biochemistry. 1998;37:792–799. doi: 10.1021/bi971117b. [DOI] [PubMed] [Google Scholar]
- 11.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 12.Alonso MC, et al. An ATP gate controls tubulin binding by the tethered head of kinesin-1. Science. 2007;316:120–123. doi: 10.1126/science.1136985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hackney DD. Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc Natl Acad Sci USA. 1994;91:6865–6869. doi: 10.1073/pnas.91.15.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori T, Vale RD, Tomishige M. How kinesin waits between steps. Nature. 2007;450:750–754. doi: 10.1038/nature06346. [DOI] [PubMed] [Google Scholar]
- 16.Klumpp LM, Hoenger A, Gilbert SP. Kinesin's second step. Proc Natl Acad Sci USA. 2004;101:3444–3449. doi: 10.1073/pnas.0307691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter NJ, Cross RA. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 18.Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hackney DD. The tethered motor domain of a kinesin–microtubule complex catalyzes reversible synthesis of bound ATP. Proc Natl Acad Sci USA. 2005;102:18338–18343. doi: 10.1073/pnas.0505288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reck-Peterson SL, et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerssemakers JW, et al. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.