Abstract
Huntington's disease is linked to the insertion of glutamine (Q) in the protein huntingtin, resulting in polyglutamine (polyQ) expansions that self-associate to form aggregates. While polyQ aggregation has been the subject of intense study, a correspondingly thorough understanding of individual polyQ chains is lacking. Here we demonstrate a single molecule force-clamp technique that directly probes the mechanical properties of single polyQ chains. We have made polyQ constructs of varying lengths that span the length range of normal and diseased polyQ expansions. Each polyQ construct is flanked by the I27 titin module, providing a clear mechanical fingerprint of the molecule being pulled. Remarkably, under the application of force, no extension is observed for any of the polyQ constructs. This is in direct contrast with the random coil protein PEVK of titin, which readily extends under force. Our measurements suggest that polyQ chains form mechanically stable collapsed structures. We test this hypothesis by disrupting polyQ chains with insertions of proline residues and find that their mechanical extensibility is sensitive to the position of the proline interruption. These experiments demonstrate that polyQ chains collapse to form a heterogeneous ensemble of conformations that are mechanically resilient. We further use a heat-annealing molecular dynamics protocol to extensively search the conformation space and find that polyQ can exist in highly mechanically stable compact globular conformations. The mechanical rigidity of these collapsed structures may exceed the functional ability of eukaryotic proteasomes, resulting in the accumulation of undigested polyQ sequences in vivo.
Keywords: mechanical, polyglutamine, protein folding, single molecule
Homopolypeptide (HPP) repeats are regions within proteins that comprise a single tract of a particular amino acid. These regions exist in a wide variety of proteins with approximately 1.4% of all proteins containing HPP repeats of n > 6 amino acids (1). Recent studies have found that while HPP repeats are extremely rare in prokaryotes, they are very common in eukaryotes (1). Within eukarya, the most common uninterrupted repeats are glutamine (Q) and alanine (A). While their role is yet to be elucidated, uncontrolled genetic expansion of HPP regions has been shown to lead to the development of serious, debilitating human diseases (2, 3). Of all HPP repeats characterized to date, polyglutamine (polyQ) repeats are the most extensively studied. In particular, Huntington's disease (HD), an inherited genetic neurological disorder, has been linked to mutations in the protein huntingtin that lead to the insertion of many repeats of the amino acid Q (2). This increase in the number of Q is known as polyQ expansion and is currently associated with 8 other HPP neurodegenerative diseases (3).
In recent years, there has been a large effort to understand the functional role of HPP repeats (1) and their relation to human disease (4–6). However, the role of many HPP repeats remains obscure and it is likely that numerous diseases remain to be identified. Strikingly, HPP expansions in diseased proteins are thought to self associate to form aggregates and postmortem examinations of HD patients identify large aggregates deposited within brain neurons (7–9). However, the mechanism of HPP toxicity remains controversial (1). While much important work has been done to understand the ensemble properties of HPP aggregation (10–14) a correspondingly thorough understanding of individual HPP chains is lacking. Crucially, an understanding of the molecular properties of single HPP chains is the first step in characterizing HPP expansion diseases. Interestingly, recent experimental and computational studies have shown that polyQ chains form collapsed, disordered globules in aqueous solution, suggesting that water is a poor solvent for this HPP (15–17). However, the conformational stability of these collapsed structures has not been investigated to date. In recent years, significant advances have been made in our understanding of the conformational stability of proteins and the molecular architecture of transition states of protein folding by mechanically stretching proteins (18, 19). This approach has permitted the mechanical characterization of proteins that acquire a particular native fold and proteins which acquire random, intrinsically disordered structures (18–24). The next challenge is to use these techniques to understand the mechanical properties of HPP chains that do not possess a distinct native fold.
We have developed a strategy to generate an experimental fingerprint to unambiguously identify and characterize a single molecule containing specific HPP chains. We use single molecule force-clamp spectroscopy as a tool to examine the mechanical properties of a polyprotein under a constant stretching force (19). Force-clamp trajectories of polyproteins provide a characteristic staircase-like pattern that serves as a fingerprint of single molecules in experiments of mechanical unfolding and refolding (19). These experiments have been successful in probing the mechanical properties of a range of proteins, such as the Ig protein I27 found in human cardiac titin (20–22). Stretching a HPP chain would give a force-clamp trajectory that would be difficult to distinguish as a mechanical fingerprint and separate from the background of non-specific interactions which dominate these experiments (22). Therefore, we have used protein engineering techniques to assemble chimera polyproteins composed of identical repeats of I27-PP, the protein I27 linked to a particular polypeptide segment PP. This experimental design is based on the discovery that the unfolding of tandem protein modules follows their mechanical stability rather than their ordering within the protein (20). This engineered chimera, I27-PP, combines the uncertain structure of the polypeptide segment PP with the well-characterized I27 module, which can be readily identified by AFM (20). Hence we use the I27 module as an easily recognizable marker of the boundaries of the polypeptide segment. The ability to recognize single HPP chains offers a significant advantage in that the mechanical properties of individual chains can now be investigated. This level of experimental control and manipulation would not be possible in standard bulk measurements where the properties of an ensemble of HPP chains are characterized. Herein, we use a combination of polyprotein engineering and force-clamp spectroscopy to characterize HPP and heteropolypeptide chains revealing information on their mechanical properties and providing important insight into this field.
Results and Discussion
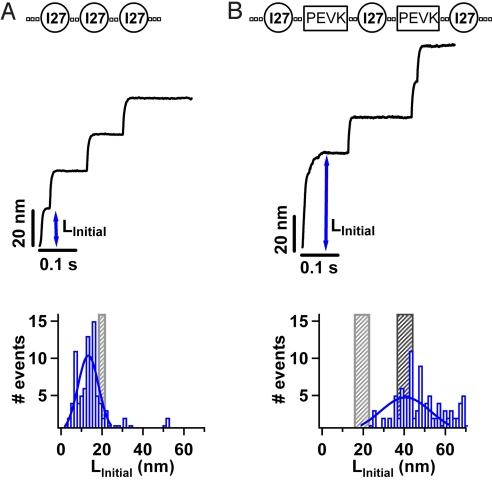
In our experiments, we design and construct polyproteins that display unique mechanical fingerprints. Polyproteins are multidomain proteins composed of identical repeats of proteins (20). Our first construct, (I27)3, is a polyprotein containing 3 repeats of the human cardiac titin Ig domain I27. The I27 protein is ideal for these experiments given that its mechanical properties have been well-characterized both experimentally (20–22) and also in silico using molecular dynamics techniques (23). The use of polyproteins is essential in that they provide a clear mechanical fingerprint to distinguish them against a background of spurious interactions and also provide a larger number of events per recording than otherwise possible with monomers (22). We measure the mechanical properties of the (I27)3 polyprotein by applying a constant force along a well defined direction, the end to end length of the protein. Under force-clamp conditions, stretching a polyprotein results in a well defined series of step increases in length, marking the unfolding and extension of the individual modules in the chain. Stretching a single (I27)3 polyprotein at a constant force of 180 pN, results in a series of step increases in length of 24 ± 0.4 nm (Fig. 1A). The size of the observed steps corresponds to the number of amino acids released by each unfolding event (24). A force-clamp trajectory showing 3 step increases in length (Fig. 1A) corresponds to the mechanical unfolding of 1 (I27)3 construct. In each of these single molecule trajectories, before unfolding of the I27 protein occurs, an initial extension in length (LInitial) is observed. This LInitial is the elongation of the 3 folded I27 proteins, the “handles” engineered on either side of the protein to facilitate protein pickup and the “linkers” connecting each of the I27 proteins [SI Text and Table S1]. In the example shown LInitial is approximately 21 nm. A histogram of LInitial is shown in Fig. 1A for trajectories containing staircases of 3 I27-unfolding events with steps of 24 nm. A Gaussian fit to the histogram (solid line in Fig. 1A) gives an average LInitial of 13.3 ± 7.1 nm. Since the construct has been designed using polyprotein engineering, we can calculate the expected extension, given that a folded I27 protein has a contour length L>I27 approximately 4.5 nm (25), the length of 1 aa residue LAA approximately 0.4 nm (24) and the number of amino acid residues in the handles and linkers NLink is well-defined (Table S1). Therefore, the contour length of the 3 folded I27 proteins (3 × LI27) and the handles and linkers (NLink× LAA) is approximately 21.5 nm. Using the worm-like chain (WLC) model of polymer elasticity (26), we calculate the expected initial extension, LWLC, at a force of 180 pN. We calculate LWLC for a range of different persistence lengths, p, (25, 26) (Table S1); when p = 0.2 nm, LWLC = 18.06 nm, and when p = 2.3 nm, LWLC is 21.07 nm. This allows us to define a region (gray shaded region in Fig. 1A) that represents the expected initial extension of the construct. We find that the average measured LInitial (histogram in Fig. 1A) is slightly lower than that expected LWLC (gray shaded region in Fig. 1A). The close agreement between LInitial and LWLC illustrates the effectiveness of this experimental tool in directly measuring the extension of amino acids contained within a polyprotein construct.
Fig. 1.
A single molecule protocol to probe the mechanical properties of polypeptide chains (A) We construct the polyprotein (I27)3 containing 3 identical repeats of the human cardiac titin Ig domain I27. Using force-clamp spectroscopy we apply a constant force of 180 pN to the construct resulting in a well defined series of step increases in length, marking the unfolding of each I27 protein in the chain. Complete extension of the construct is identified by the mechanical fingerprint of 3 step increases in length of 24 nm. An initial extension in length (LInitial) is observed, corresponding to the elongation of the folded I27 proteins and the amino acid linkers in the (I27)3 construct. A histogram of LInitial is shown for all force-clamp trajectories (n = 93) containing 3 I27 unfolding events. A Gaussian fit to the histogram gives an average LInitial = 13.30 ± 7.10 nm, in close agreement with the expected extension of 3 folded I27 proteins and the linkers in the construct (gray shaded area) (B) We construct the chimera (I27-PEVK-I27-PEVK-I27) containing the random coil protein PEVK in tandem with the I27 protein. We measure the mechanical properties by applying a constant force of 180 pN, resulting in a series of step increases in length. Each trajectory contains an initial extension LInitial corresponding to the elongation of the folded I27 proteins, PEVK and linkers in the construct. A histogram of LInitial is shown for force-clamp trajectories (n = 106) containing staircases of 3 I27 unfolding events with steps of 24 nm. A Gaussian fit to the histogram gives an average LInitial = 40.65 ± 17.26 nm, in agreement with the expected extension of 3 folded I27 proteins, PEVK and the amino acid linkers in the construct (black shaded area).
To probe the mechanical properties of polypeptide chains that do not possess a well-defined native fold, we developed this approach to study polyprotein constructs containing specific polypeptide segments. In particular, we designed and engineered a polyprotein, (I27-PEVK), containing the I27 protein in tandem with the random coil protein PEVK (exon 161) (Materials and Methods). Specifically the construct is (I27-PEVK-I27-PEVK-I27) (Table S1). The PEVK domain is found in the giant muscle protein titin and has unknown secondary structure (27). Previous single molecule studies have shown that cardiac PEVK acts as an entropic spring with the properties of a random coil, exhibiting conformations of different flexibility and no mechanical stability, readily extending under the application of a mechanical force (28). The I27 proteins in the (I27-PEVK) construct provide a clear mechanical fingerprint for their unfolding, with step increases in length of 24 nm (28). Since the I27 protein and PEVK are constructed in an alternating pattern, if 3 I27 unfolding events are observed, then 2 PEVK segments must have been stretched. Using this approach, we can have certainty that the PEVK segments are stretched and that the mechanical response of PEVK is represented by the initial extension of the force-clamp trajectory, before any of the I27 modules unfold (28). We measured the mechanical properties of (I27-PEVK) by applying a constant force of 180 pN. Force-clamp trajectories showing 3 step increases in length (Fig. 1B) correspond to the full mechanical extension of 1 (I27-PEVK) construct (Fig S1). We verify that these trajectories indeed represent single molecules by measuring the unfolding rate of the I27 molecules in the (I27-PEVK) construct (Fig. S2). In each trajectory an initial extension in length (LInitial) is observed, corresponding to the elongation of the folded I27 proteins, the PEVK segments and the amino acid handles and linkers in the (I27-PEVK) construct (Table S1). A histogram of LInitial is shown in Fig. 1B for force-clamp trajectories containing staircases of 3 I27-unfolding events with steps of 24 nm. It is apparent that a distribution of LInitial is measured for the (I27-PEVK) construct (Fig. 1B), as was measured for the (I27)3 construct (Fig. 1A). This distribution of LInitial reflects the different attachment points between the construct and the cantilever tip and substrate (22). A Gaussian fit to the histogram gives an average LInitial of 40.65 ± 17.26 nm. The contour length of the construct includes 3 folded I27 proteins (3 × LI27), handles, and linkers (NLink× LAA) and the PEVK segments (Table S1). As before, using the WLC model we calculate the expected initial extension, LWLC for 2 different persistence lengths (black shaded region in Fig. 1B). For comparison we show the expected initial extension for only the folded I27 proteins and the amino acid handles and linkers (gray shaded region in Fig. 1B). It is apparent that the histogram of measured Linitial includes the extension of PEVK segments in the construct. The close agreement of LInitial with the expected value LWLC demonstrates the effectiveness of our experimental approach in directly measuring the extension of a random-coil protein PEVK contained within a polyprotein construct.
To probe the mechanical properties of HPP chains which do not possess a well-defined native fold, we used this approach to study polyprotein constructs containing specific polyQ segments. We designed and engineered a chimera protein, (I27-Q25), containing the I27 protein and 25 glutamine residues, Q25 (see Materials and Methods). Specifically, the construct was (I27-Q25-I27-Q25-I27). We measured the mechanical properties of (I27-Q25) by applying a constant force of 180 pN (Fig. 2B). A histogram of LInitial is shown in Fig. 2B for force-clamp trajectories containing staircases of 3 I27-unfolding events. A Gaussian fit to the histogram gives an average LInitial of 13.79 ± 9.63 nm. The contour length of the 3 folded I27 proteins, the handles, and linkers and the polyQ segments is approximately 43.1 nm (Table S1). Using the WLC model we calculate the expected initial extension, LWLC. We find that the average measured LInitial (histogram in Fig. 2B) is significantly lower than that expected LWLC (black shaded region in Fig. 2B). Remarkably, the average measured LInitial is instead close to the expected extension for only the folded I27 and amino acid handles, and linkers (gray shaded region in Fig. 2B). These measurements indicate that the polyQ chains in the (I27-Q25) construct are not extending under an applied force of 180 pN. This is in direct contrast with the behavior of the random-coil protein PEVK which readily extends under an applied force (Fig. 1B). The inextensibility of the polyQ chains in (I27-Q25) suggests that the HPP chain is forming highly compact structures which are mechanically resilient to an applied force of 180 pN. It is worth noting that many proteins which naturally fold can readily be extended under a constant applied force of 180 pN or lower; including I27 (21), ubiquitin (29), protein L (22), protein G (30), and ankryin (31, 32). It is therefore striking that a HPP, which does not have a well-defined fold, should exhibit such mechanical resilience.
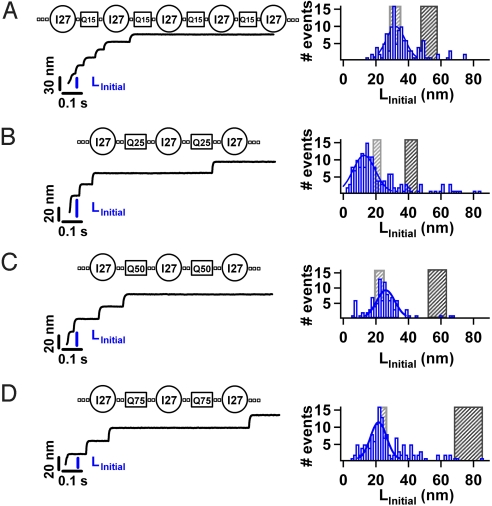
Fig. 2.
Probing the mechanical properties of homopolypeptide chains. We construct chimeras containing the I27 protein and polyglutamine chains of different length, namely Q15 (A), Q25 (B) Q50 (C), and Q75 (D). Applying a constant force of 180 pN, results in a series of step increases in length of 24 nm. We identify the full mechanical extension of a complete construct by the presence of 3 I27 unfolding steps for the Q25, Q50, and Q75 constructs and 5 I27 unfolding steps for the Q15 construct (Table S1). We measure the initial extension LInitial for each trajectory that satisfies these stringent criteria. A histogram of LInitial is shown for each of the constructs; Q15 (n = 121), Q25 (n = 165), Q50 (n = 100), and Q75 (n = 149). A Gaussian fit to the histograms (solid line) gives an average LInitial for each construct Q15 (LInitial = 31.16 ± 8.21 nm), Q25 (LInitial = 13.79 ± 9.63 nm), Q50 (LInitial = 26.09 ± 7.93 nm), Q75 (LInitial = 20.65 ± 6.98 nm), (QP)24 (LInitial = 42.3.1 ± 9.63 nm), and (Q11P)4 (LInitial = 70.00 ± 12.39 nm). For all polyglutamine chains, the measured LInitial is significantly shorter than that expected for full extension of the construct (black shaded area). Instead LInitial is in close agreement with the expected length extension of only the folded I27 proteins and linkers (gray shaded area).
The number of extra residues in an uncontrolled genetic expansion of HPP regions is thought to be strongly correlated to the age of onset of a number of diseases (6). To further probe the mechanical properties of individual polyQ chains we designed chimera constructs containing different lengths of polyQ chains, namely (I27-Q15), (I27-Q50), and (I27-Q75) (Table S1). In each case the polyQ chains were sandwiched between I27 proteins that provided a clear mechanical fingerprint for the construct in our force-clamp protocol. We measured the mechanical properties of each polyQ construct by applying a constant force of 180 pN (Fig. 2 A–D). LInitial is measured for each force-clamp trajectory containing the full mechanical fingerprint of the I27 proteins, 3 I27 steps for (I27-Q50) and (I27-Q75) and 5 I27 steps for (I27-Q15). A longer construct is necessary for (I27-Q15) due to the shorter length of NPP = 15 (≈6 nm). A histogram of LInitial is shown in Fig. 2 A–D giving an average LInitial for each construct; Q15 (LInitial = 31.16 ± 8.21 nm), Q50 (LInitial = 26.09 ± 7.93 nm), and Q75 (LInitial = 20.65 ± 6.98 nm). Using the WLC model, we calculate LWLC for the constructs (Table S1) (28). For all 3 constructs, we find that the average measured LInitial (histograms in Fig. 2 A–D) are significantly lower than that expected LWLC (black shaded regions Fig. 2 A–D). Surprisingly, the average measured LInitial is instead close to the expected extension for only the folded I27 and the handles and linkers (gray shaded region in Fig. 2 A–D). These measurements indicate that polyQ chains, Q15, Q50, and Q75, are not extending under an applied force of 180 pN (Fig. 2B). It is noteworthy that LInitial is remarkably similar for all 3 constructs, which is surprising given the total number of Q amino acids increases from only NPP = 60 in Q15 to NPP = 150 in Q75 (Table S1). These measurements suggest that each polyQ chain is forming compact, disordered structures that are mechanically resilient, yet lack elongated structures such as β-sheets. Again, this is in direct contrast with the behavior of the random-coil protein PEVK which readily extends under an applied force (Fig. 1B). Crucially by combining force-clamp spectroscopy and polyprotein engineering, we can have certainty that single polyQ chains are being mechanically extended. This confidence results from obtaining an unambiguous mechanical fingerprint for the extension of 1 polyprotein construct (Fig. S1) and accurately measuring the unfolding rate of the I27 marker proteins in each construct (Fig. S2). Since the unfolding rate of a construct is highly sensitive to the way in which the force is distributed throughout the protein these measurements provide assurance that the mechanical properties of single molecules are measured. We propose that the inextensibility of single polyQ chains is due to the formation of compact and highly stable conformations. The presence of these conformations is independent of the number of Q in the chain, within the range NPP = 15 to 75. Therefore, while the number of Q is thought to correlate with age of onset in diseases, individual chains which span the length range of normal and diseased HPP proteins show no apparent evidence for a change in their mechanical properties.
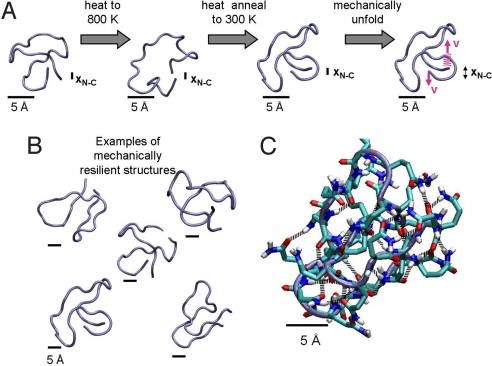
Interestingly, the observation of collapsed structures in HPP chains is not inconsistent with earlier experimental and theoretical studies (15–17, 33, 34). In particular, previous fluorescence correlation spectroscopy studies have demonstrated that polyQ chains of lengths n = 15–55 form collapsed, disordered globules in aqueous solution (15). Furthermore, computational studies have shown that collapsed polyQ globules are disordered and their dynamics of their conformational relaxation has signatures of glassy behavior (16, 17). However, in addition to collapsed structures, molecular dynamics simulations on monomeric polyQ have identified a wide range of different structures including; random coils (35), β-helices (35, 36), steric zipper motifs (37) and collapsed β-sheet structures (37). To gain insight into the relation between polyQ's molecular architecture and its observed extreme mechanical stability we use heat annealing (38, 39) to extensively search the conformation space of polyQ. Since in our AFM experiments an extended polyQ state was never observed (Fig. 2) we constrained the end-to-end length of the polyQ chains (of length NPP = 25) to be small in a heat annealing protocol and then ran steered molecular dynamics (SMD) simulations (23) on the annealed conformations. Interestingly, we found that the end-to-end distance constraint is unnecessary since heat annealing of unconstrained chains generated similar collapsed conformations to the constrained systems. The SMD simulations involve the application of external forces to polyQ chains by pulling their terminal residues in opposite directions (see SI Text for details), and complement the AFM observations by providing a detailed atomic picture of stretching and unfolding of individual protein domains (23). Thus we performed simulations of polyQ chains of length NPP = 25 with and without end-to-end length constraints by first heating the chains to a high temperature and then annealing (38, 39) them to obtain 10,000 collapsed structures (Fig. 3). Each collapsed structure was then pulled at a constant rate to trigger unraveling (see SI Text). In the majority of annealed structures generated from both constrained and unconstrained simulations the 2 terminal residues were found to be hydrogen bonded to each other. Upon pulling, we measured a wide range of unraveling forces for the ensemble of collapsed structures. Interestingly, a number of these configurations exhibited very high mechanical stability with rupture forces of more than 900 pN and up to 1,500 pN (Figs. 3B and Fig. S3). Significantly, these forces are much higher than that required for the unfolding of I27, using the same SMD procedure (see SI Text for detail), and many other proteins (19). Remarkably, these representative structures all share similar structural and mechanical characteristics (Fig. 3B). Upon close examination of the force induced unraveling of these mechanically resilient structures we find that up to 17 hydrogen bonds must be ruptured before unraveling occurs (Figs. 3C and Fig. S4). Furthermore, each structure exhibits a high propensity for multiple hydrogen bonds between and around the N and C termini of the collapsed polyQ chain (Fig. 3C). Given that a protein such as I27 easily unfolds at a pulling force of 180 pN and 6 key hydrogen bonds are broken (23) we can speculate that the simultaneous rupture of a greater number of hydrogen bonds would require a higher rupture force, as indeed seems to be the case. By using a heat annealing approach, a wide region of conformational space has thus been extensively probed. While only a fraction of the configurations possess high mechanical resilience, it is likely that with time this population of mechanically resilient structures would increase as the collapsed ensemble evolved toward more stable structures. Indeed, future studies using SMD simulations for extended equilibration times will probe the evolution of the ensemble of collapsed polyQ structures. More significantly, the plausible structures obtained from SMD simulations of polyQ in water (Fig. 3B) and the high mechanical stability shared by these structures provide support for the existence of compact mechanically resilient structures.
Fig. 3.
Representative structures of polyglutamine chains display extreme mechanical stability (A) SMD simulations provide a detailed atomic picture of stretching individual polyQ chains. In the simulations, polyQ chains of length NPP = 25 were first heated to a high temperature and then heat annealed to obtain collapsed structures. Each collapsed structure (N ≈10,000) was then pulled at a constant rate to trigger unraveling (see SI Text). Upon pulling, a number of configurations exhibit very high mechanical stability with rupture forces of more than 900 pN. Examples of these mechanically resilient structures are shown in (B and C) Upon close examination of the force induced unraveling of these mechanically resilient structures we find that a large number of hydrogen bonds must rupture simultaneously before unraveling occurs.
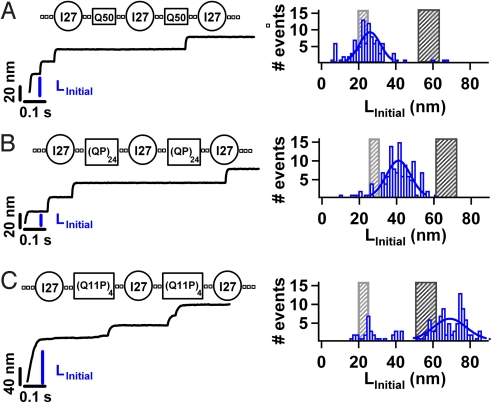
To test whether mechanical resilience is a unique property of HPP chains containing Q we designed and engineered a construct containing a different HPP sequence. We made the chimera construct (I27-A25), containing both I27 and 25 alanine residues (see Materials and Methods), an amino acid that is also been linked to HPP expansion diseases (40). Interestingly, force-clamp measurements demonstrate polyalanine chains, also form compact structures which are insensitive to an applied force of 180 pN (Fig. S5). These measurements suggest that mechanical resilience may be a common feature of HPP chains, in contrast with heteropolypeptide chains such as PEVK. To test this hypothesis, we made constructs with polypeptide segments containing Q repeats interrupted by proline residues [I27-(QP)24] and [I27-(Q11P)4] (Table S1). Importantly, these constructs are similar in length to the HPP segment in the (I27-Q50) construct allowing us to directly compare HPP and heteropolypeptide chains of similar length (SI Text; Fig. 4). We measured the mechanical properties of these constructs by applying a constant force of 180 pN (Fig. 4 B and C). A histogram of LInitial is shown in Fig. 4 B and C with an average LInitial of 42.3.1 ± 9.63 nm for [I27-(QP)24] and 70.1 ± 9.63 nm for [I27-(Q11P)4]. Upon the addition of prolines within the polyQ chain the mechanical extensibility of the chain changes significantly. Interestingly, the mechanical extensibility is very sensitive to the position of the proline interruption within the chain (Fig. 4 B and C and Fig. S6). While interrupting the polyQ chain with alternating proline residues increases the mechanical extensibility marginally (Fig. 4B), interrupting polyQ after 11 Q repeats modifies the mechanical extensibility significantly (Fig. 4C).
Fig. 4.
Probing the mechanical properties of heteropolypeptide chains. We construct chimeras containing the I27 protein and a heteropolypeptide chain containing glutamines interrupted by a proline residues, namely [I27-(QP)24] and [I27-(Q11P)4] (Table S1). Using force-clamp spectroscopy we apply a constant force of 180 pN along the end to end length of the constructs. We identify the full mechanical extension of a complete construct by the presence of 3 I27 unfolding steps for the (QP)24 and (Q11P)4 constructs (Table S1) where each I27 unfolding step is 24 nm in length. We measure the initial extension LInitial for each trajectory that satisfies these stringent criteria. A histogram of LInitial is shown for each of the constructs; (A) Q50 (n = 100) for comparison, (B) (QP)24 (n = 108), and (C) (Q11P)4 (n = 133). A Gaussian fit to the histograms (solid line) gives an average LInitial for each construct (QP)24 (LInitial = 42.3.1 ± 9.63 nm) and (Q11P)4 (LInitial = 70.00 ± 12.39 nm). The measured LInitial for the heteropolpeptide chain Q11P, is in agreement with the full extension of the folded I27 proteins, polypeptides, and linkers. Interestingly, LInitial for the (QP)24 construct lies between the gray and black shaded areas.
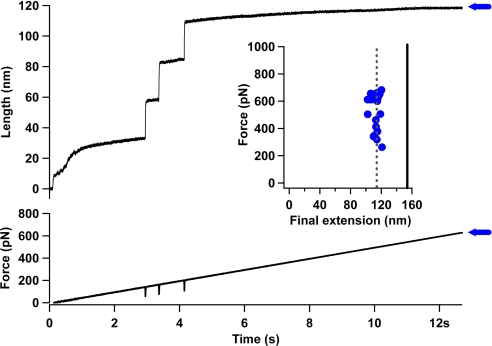
We propose that the inextensibility of HPP chains results from the formation of an ensemble of collapsed and mechanically resilient structures. This extreme mechanical resilience may be a common feature of HPP chains, in contrast with heteropolypeptide chains, such as PEVK and Q11P. To test this hypothesis, we make use of a force protocol that greatly extends the force range with which we probe the HPP chains (41). We used a force-ramp protocol to apply a linearly increasing force to the (I27-Q50) construct. In this force protocol we ramp the force from 0 pN to 1,000 pN over a time period of 15 s. Extension of the (I27-Q50) construct was identified by the presence of 3 I27 unfolding steps in the length trajectory (Fig. 5). Unfolding of the I27 protein occurred between a force of 150 pN and 200 pN, in accord with the known mechanical properties of this protein (20). Using this protocol, extension of the polyQ chain would be identified by a marked increase in length of approximately 40 nm (NPP = 100, LAA = 0.4 nm). We measured the final extension (blue horizontal arrow in Fig. 5) of the (I27-Q50) construct for each trajectory where the full I27 unfolding fingerprint could be identified. The average final extension 116 nm was much lower than that expected for the complete extension of 3 unfolded I27 proteins (89 amino acids × LAA), the glutamines (NPP× LAA) and the linkers (NLink× LAA) (solid black line). Instead, the final extension was in close agreement with the extension of only the unfolded I27 proteins and linkers (118 nm). Thus, no evidence for the extension of the polyQ chain was observed, even at forces as high as 800 pN, suggesting that the polyQ chains possess extreme mechanical stability. These results support the view that HPP chains display extreme mechanical resilience under the application of a high force. The behavior of a disordered globule under a stretching force has previously been addressed theoretically and numerically (42–46). These studies demonstrated that stretching of globular polymers shares some similarities with a first order transition that corresponds to the ‘bursting’ of the collapsed globule. In particular, below a critical applied force the globule remains compact, while above a critical applied force the globule unwinds into a stretched chain. Interestingly, one recent model for unwinding globules under tension (44) yields a critical force of approximately 46 pN for a chain of 50 monomers, a value far below the experimentally measured value of 800 pN we find for a polyQ chain of length n = 50.
Fig. 5.
Homopolypeptide chains display extreme mechanical resilience under the application of a high force (A) To further examine the mechanical properties of the polyglutamine chain we extended the force regime with which we probed the system. We used a force-ramp protocol to apply a linearly increasing force to the (I27-Q50) construct. In this force protocol we ramp the force from 0 pN to 1000 pN over a time period of 15 seconds. Extension of the (I27-Q50) construct was identified by the presence of 3 I27-unfolding steps in the length trajectory. Unfolding of the I27 protein occurred between a force of 150 pN and 200 pN, in accord with the known mechanical properties of this protein. Using this protocol, extension of the polyglutamine chain would be identified by a marked increase in length of approximately 40 nm (NPP x LAA) (B) We measured the final extension of the (I27-Q50) construct for each trajectory where the full I27-unfolding fingerprint could be identified. The average final extension approximately116 nm was much lower than that expected for the complete extension of 3 unfolded I27 proteins (89 × LAA), 100 glutamines (NPP× LAA) and the linkers (NLink× LAA) (solid black line). Instead, the final extension was in close agreement with the extension of only the unfolded I27 proteins and linkers. Thus, no evidence for the extension of the polyglutamine chain was observed, even at force as high as 800 pN suggesting that the polyglutamine chains possess extreme mechanical stability.
What properties of a polypeptide chain determine its ability to form stable collapsed structures that are mechanically resilient? In the case of heteropolypeptide chains 2 different scenarios have now been identified. In the first case, some sequences such as PEVK and Q11P form disordered collapsed structures which exhibit no mechanical stability (28). In the second case, proteins which fold such as I27, form an ensemble of collapsed structures exhibiting weak mechanical stability which then progress to a well-defined native fold that is more mechanically stable (47). These initial minimum energy collapsed structures (MECS) are thought to play a key role in reducing the dimensionality of the search for the native basin for a folding protein (47–50). The existence of MECS may be a general feature of proteins that are naturally designed through evolution to fold on biological timescales (48). Indeed, recent experiments have demonstrated the existence of MECS in the proteins ubiquitin and I27 and the key role they play in allowing the protein to reach a well-defined and functional native conformation (47). While the existence of MECS has long been predicted for folding proteins and hetereopolymers (49, 50) the structures we have identified for HPP chains of polyQ and polyA are clearly distinct. These HPP collapsed structures are very compact (Fig. 2) and exhibit extreme mechanical stability (Fig. 5). Furthermore, these structures do not appear to progress toward a well-defined native fold (Fig. 3B). Instead, these collapsed structures may represent the lowest energy structures for these molecules. Significantly, the number of structures in this ensemble and their relative stability are thought to be sensitive to the properties of the amino acids in the polypeptide chain. In particular, polar polypeptide chains are predicted to have a significantly higher number of MECS than non-polar chains (48), and HPP chains are predicted to have more stable collapsed structures than heteropolypeptide chains (51). Furthermore, recent analysis of order parameters derived from theories for flexible polymers has shown that water is a poor solvent for generic polypeptide backbones suggesting that backbone-solvent interactions are a crucial determinant of the conformational ensemble (52). From an evolutionary point of view, these highly stable MECS may not have evolved efficiently. Indeed, it has been found that while HPPs are extremely rare in prokaryotes, several eukaryote putative homologs of prokaryote proteins containing HPP's retain the repetitive region, suggesting an ancient origin for certain repeats (1). A recent global genome survey has identified that the majority of eukaryote proteins which contain HPP repeats perform roles in processes that require the assembly of large multiprotein or protein/nucleic acid complexes (1). The observation that HPP chains form collapsed structures that are mechanical resilient suggests that mechanical stability may have an important role in these assembly processes (53). Indeed, it is interesting to consider what physiological force or conditions could lead to the unraveling of HPP chains. A recent study has shown that eukaryotic proteasomes cannot digest polyQ sequences and instead releases them during degradation of polyQ containing proteins (54, 55). This inability to digest polyQ sequences may reflect the inextensibility of these highly stable MECS in vivo. One could speculate that with time the accumulation of unsuccessfully degraded polyQ leads to aggregation and the accumulation of larger deposits within brain neurons (7–9).
Materials and Methods
Protein Engineering and Purification.
The details of the I27 (20) and PEVK (28) polyprotein engineering and purification have been reported previously. For the I27 and polypeptide segments chimera constructs we used successive cloning in pT7Blue vectors and then expressed the gene using vector pQE80L in Escherichia coli strain BLR(DE3).
Single Molecule-force Spectroscopy.
We used a custom-built atomic force microscope equipped with a PicoCube P363.3-CD piezoelectric translator (Physik Instrumente) controlled by an analog PID feedback system that has been described previously (29).
Supplementary Material
Acknowledgments.
This work has been supported by National Institutes of Health grants to J.M.F. (HL66030 and HL61228) and B.J.Berne (GM43340). We thank members of the Fernandez lab for helpful discussions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900678106/DCSupplemental.
References
- 1.Faux NG, et al. Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res. 2005;15:537–551. doi: 10.1101/gr.3096505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates GP, Benn C. In Huntington's Diseases. Oxford: Oxford Univ Press; 2002. The polyglutamine diseases. [Google Scholar]
- 3.Cummings CJ, Zoghbi HY. Trinucleotide repeats: Mechanisms and pathophysiology. Annu Rev Genomics Hum Genet. 2000;1:281–328. doi: 10.1146/annurev.genom.1.1.281. [DOI] [PubMed] [Google Scholar]
- 4.Truant R, Atwal RS, Desmond C, Munsie L, Tran T. Huntington's disease: Revisiting the aggregation hypothesis in polyglutamine neurodegenerative diseases. Febs Journal. 2008;275:4252–4262. doi: 10.1111/j.1742-4658.2008.06561.x. [DOI] [PubMed] [Google Scholar]
- 5.Wetzel R. Polyglutamine folding and aggregation in Huntington's disease. Biophys J. 2004;86:166A–166A. [Google Scholar]
- 6.Chen SM, Ferrone FA, Wetzel R. Huntington's disease age-of-onset linked to polyglutamine aggregation nucleation. Proc Natl Acad Sci USA. 2002;99:11884–11889. doi: 10.1073/pnas.182276099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perutz MF. Glutamine repeats and inherited neurodegenerative diseases: Molecular aspects. Curr Opin Struct Biol. 1996;6:848–858. doi: 10.1016/s0959-440x(96)80016-9. [DOI] [PubMed] [Google Scholar]
- 8.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 9.Osmand AP, Berthelier V, Wetzel R. Imaging polyglutamine deposits in brain tissue. Amyloid, Prions, Other Protein Aggregates. 2006;412(Pt B):106–122. doi: 10.1016/S0076-6879(06)12008-X. [DOI] [PubMed] [Google Scholar]
- 10.Sharma D, Shinchuk LM, Inouye H, Wetzel R, Kirschner DA. Polyglutamine homopolymers having 8–45 residues form slablike beta-crystallite assemblies. Proteins-Structure Funct Bioinformat. 2005;61:398–411. doi: 10.1002/prot.20602. [DOI] [PubMed] [Google Scholar]
- 11.Giri K, Ghosh U, Bhattacharyya NP, Basak S. Caspase 8 mediated apoptotic cell death induced by beta-sheet forming polyalanine peptides. FEBS Lett. 2003;555:380–384. doi: 10.1016/s0014-5793(03)01294-8. [DOI] [PubMed] [Google Scholar]
- 12.Wetzel R. Ideas of order for amyloid fibril structure. Structure. 2002;10:1031–1036. doi: 10.1016/s0969-2126(02)00809-2. [DOI] [PubMed] [Google Scholar]
- 13.Slepko N, et al. Normal-repeat-length polyglutamine peptides accelerate aggregation nucleation and cytotoxicity of expanded polyglutamine proteins. Proc Natl Acad Sci USA. 2006;103:14367–14372. doi: 10.1073/pnas.0602348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thakur AK, Wetzel R. Mutational analysis of the structural organization of polyglutamine aggregates. Proc Natl Acad Sci USA. 2002;99:17014–17019. doi: 10.1073/pnas.252523899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crick SL, Jayaraman M, Frieden C, Wetzel R, Pappu RV. Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions. Proc Natl Acad Sci USA. 2006;103:16764–16769. doi: 10.1073/pnas.0608175103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitalis A, Wang X, Pappu RV. Atomistic simulations of the effects of polyglutamine chain length and solvent quality on conformational equilibria and spontaneous homodimerization. J Mol Biol. 2008;384:279–297. doi: 10.1016/j.jmb.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vitalis A, Wang X, Pappu RV. Quantitative characterization of intrinsic disorder in polyglutamine: insights from analysis based on polymer theories. Biophys J. 2008;93:1923–1937. doi: 10.1529/biophysj.107.110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sulkowska JI, Cieplak M. Mechanical stretching of proteins - a theoretical survey of the Protein Data Bank. J Phys Condens Matter. 2007;19:283201–283261. [Google Scholar]
- 19.Fernandez JM, Li H. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science. 2004;303:1674–1678. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
- 20.Li HB, et al. Reverse engineering of the giant muscle protein titin. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- 21.Dougan L, Fang G, Lu H, Fernandez JM. Solvent molecules bridge the mechanical unfolding transition state of a protein. Proc Natl Acad Sci USA. 2008;105:3185–3190. doi: 10.1073/pnas.0706075105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Garcia-Manyes S, Brujic J, Fernandez JM. Force-clamp spectroscopy of single protein monomers reveals the individual unfolding and folding pathways of 127 and ubiquitin. Biophys J. 2007;93:2436–2446. doi: 10.1529/biophysj.107.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu H, Schulten K. The key event in force-induced unfolding of titin's immunoglobulin domains. Biophys J. 2000;79:51–65. doi: 10.1016/S0006-3495(00)76273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koti ASR, et al. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys J. 2007;92:225–233. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrion-Vazquez M, Marszalek PE, Oberhauser AF, Fernandez JM. Atomic force microscopy captures length phenotypes in single proteins. Proc Natl Acad Sci USA. 1999;96:11288–11292. doi: 10.1073/pnas.96.20.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 27.Linke WA, et al. PEVK domain of titin: An entropic spring with actin-binding properties. J Struct Biol. 2002;137:194–205. doi: 10.1006/jsbi.2002.4468. [DOI] [PubMed] [Google Scholar]
- 28.Li HB, et al. Multiple conformations of PEVK proteins detected by single-molecule techniques. Proc Natl Acad Sci USA. 2001;98:10682–10686. doi: 10.1073/pnas.191189098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlierf M, Li H, Fernandez JM. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc Natl Acad Sci USA. 2004;101:7299–7304. doi: 10.1073/pnas.0400033101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao Y, Kuske R, Li HB. Direct observation of Markovian behavior of the mechanical unfolding of individual proteins. Biophys J. 2008;95:782–788. doi: 10.1529/biophysj.107.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee G, et al. Nanospring behaviour of ankyrin repeats. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 32.Li LW, Wetzel S, Pluckthun A, Fernandez JM. Stepwise unfolding of ankyrin repeats in a single protein revealed by atomic force microscopy. Biophys J. 2006;90:L30–L32. doi: 10.1529/biophysj.105.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dima RI, Thirumalai D. Asymmetry in the shapes of folded and denatured states of proteins. J Phys Chem B. 2004;108:6564–6570. [Google Scholar]
- 34.Wang XL, Vitalis A, Wyczalkowski MA, Pappu RV. Characterizing the conformational ensemble of monomeric polyglutamine. Proteins-Structure Funct Bioinformat. 2006;63:297–311. doi: 10.1002/prot.20761. [DOI] [PubMed] [Google Scholar]
- 35.Marchut AJ, Hall CK. Effects of chain length on the aggregation of model polyglutamine peptides: Molecular dynamics simulations. Proteins-Structure Funct Bioinformat. 2007;66:96–109. doi: 10.1002/prot.21132. [DOI] [PubMed] [Google Scholar]
- 36.Khare SD, et al. Molecular origin of polyglutamine aggregation in neurodegenerative diseases. PLoS Comput Biol. 2005;1:230–235. doi: 10.1371/journal.pcbi.0010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esposito L, et al. Insights into structure, stability, and toxicity of monomeric and aggregated polyglutamine models from molecular dynamics simulations. Biophys J. 2008;94:4031–4040. doi: 10.1529/biophysj.107.118935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chou KC, Carlacci L. Simulated Annealing Approach to the Study of Protein Structures. Prot Eng. 1991;4:661–667. doi: 10.1093/protein/4.6.661. [DOI] [PubMed] [Google Scholar]
- 39.Villani V, Tamburro AM. Simulated annealing and molecular dynamics of an elastin-related tetrapeptide in aqueous solution. J Mol Struct-Theochem. 1998;431:205–218. [Google Scholar]
- 40.Brown LY, Brown SA. Alanine tracts: The expanding story of human illness and trinucleotide repeats. Trends Genet. 2004;20:51–58. doi: 10.1016/j.tig.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Walther K, et al. Signatures of hydrophobic collapse in the extended proteins captured with force spectroscopy. Proc Natl Acad Sci USA. 2007;104:7916–7921. doi: 10.1073/pnas.0702179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams C, Brochard F, Frisch H. Polymer collapse. Annu Rev Phys Chem. 1981;32:433–451. [Google Scholar]
- 43.Craig A, Terentjev EM. Stretching globular protein. I. Single chains. J Chem Phys. 2005;122:194901–194906. doi: 10.1063/1.1898213. [DOI] [PubMed] [Google Scholar]
- 44.Frisch T, Verga A. Unwinding globules under tension and polymer collapse. Phys Rev E. 2002;65:41801–41807. doi: 10.1103/PhysRevE.65.041801. [DOI] [PubMed] [Google Scholar]
- 45.Halperin A, Zhulina EB. On the deformation behavior of collapsed polymers. Europhys Lett. 1991;6155:417–421. [Google Scholar]
- 46.Ziv G, Thirumalai D, Haran G. Collapse transitions in proteins. Phys Chem Chem Phys. 2009;11:83–93. doi: 10.1039/b813961j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Manyes S, Dougan L, Badilla CM, Brujic J, Fernandez JM. Direct observation of an ensemble of stable collapsed states in the mechanical folding of ubiquitin. Proc Natl Acad SCI USA. 2009 doi: 10.1073/pnas.0901213106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Camacho CJ, Thirumalai D. Minimum energy compact structures of random sequences of heteropolymers. Phys Rev Lett. 1993;71:2505–2508. doi: 10.1103/PhysRevLett.71.2505. [DOI] [PubMed] [Google Scholar]
- 49.Sali A, Shakhnovich E, Karplus M. How does a protein fold? Nature. 1994;369:248–251. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- 50.Yue K, Dill KA. Forces of tertiary structural organization in globular proteins. Proc Natl Acad Sci USA. 1995;92:146–150. doi: 10.1073/pnas.92.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel BA, Debenedetti PG, Stillinger FH, Rossky PJ. The effect of sequence on the conformational stability of a model heteropolymer in explicit water. J Chem Phys. 2008;128:175102. doi: 10.1063/1.2909974. [DOI] [PubMed] [Google Scholar]
- 52.Tran HT, Mao A, Pappu RV. Role of backbone-solvent interactions in determining conformational equilibria of intrinsically disordered proteins. J Am Chem Soc. 2008;130:7380–7392. doi: 10.1021/ja710446s. [DOI] [PubMed] [Google Scholar]
- 53.Colby DW, Cassady JP, Lin GC, Ingram VM, Wittrup KD. Stochastic kinetics of intracellular huntingtin aggregate formation. Nat Chem Biol. 2006;2:319–323. doi: 10.1038/nchembio792. [DOI] [PubMed] [Google Scholar]
- 54.Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- 55.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.