Abstract
Rad51 is a core component of the eukaryotic homologous recombination machinery and is responsible for key mechanistic steps during strand invasion. Higher order oligomers of Rad51 display a remarkable degree of structural variation, forming rings, compressed filaments, and elongated filaments. It is unclear whether Rad51 can transition directly between these different oligomeric structures without disassembling first into monomers. We have used single-molecule microscopy to investigate the behavior of human Rad51 assembled on double-stranded DNA. Our results show that human Rad51 can form elongated nucleoprotein filaments on DNA, but ATP hydrolysis causes a decrease in their length without concomitant dissociation of protein. Compressed Rad51 filaments can re-elongate when presented with either ATP or the non-hydrolyzable analog AMP-PNP, and these cycles of elongation and compression are reversible. A Rad51 mutant deficient in ATP hydrolysis is locked into an extended conformation that is incapable of transitioning to a compressed filament. Similarly, wild-type Rad51 bound to DNA in the presence of AMP-PNP was trapped in the elongated state. Proteins incapable of transitioning to the compressed state were also highly resistant to dissociation from the DNA. Taken together, our results indicate that nucleotide hydrolysis by human Rad51 triggers a reversible structural transition leading to filaments with reduced helical pitch.
Keywords: DNA curtain, homologous recombination, single molecule imaging
Double-stranded DNA breaks (DSBs) are 1 of the most deleterious forms of DNA damage and can lead to cell death or oncogenic transformation. Homologous recombination (HR) is an evolutionarily conserved pathway used to repair DSBs, and is essential for maintaining genomic stability (1, 2). When a DSB occurs, the 5′ ends of the DNA are resected, yielding long 3′ single-stranded DNA (ssDNA) overhangs, which are the loading site for a DNA recombinase. The recombinase aligns the ssDNA with a homologous double stranded DNA (dsDNA) and then invades the duplex to form a D-loop. The invading end can then serve as a primer for the replication machinery, which uses the homologous duplex as a template, and the resulting products are resolved to restore the continuity of the chromosomes.
The DNA transactions that take place during HR are mediated by members of the RAD52 epistasis group of proteins (1–3). This includes the recombinase Rad51, which plays a central role in HR and assembles into a nucleoprotein filament on the ssDNA overhangs generated at the DSB (4, 5). This filament is responsible for catalyzing the pairing, alignment, and strand invasion steps during recombination (6). Rad51 is sufficient to catalyze these reactions in vitro (7), however, numerous accessory factors are required in vivo and their functions range from facilitating Rad51 loading at the outset of the reaction to promoting the disassembly of Rad51 upon completion of strand invasion (1, 3, 8, 9).
Human Rad51 has a flexible N-terminal domain and a central ATP-binding core closely related to bacterial RecA (10). Rad51 forms right-handed helical filaments that extend the bound DNA by approximately 50% relative to B-form DNA (11, 12). The structural parameters defining the geometry of the filaments are variable, and can even vary within the same filament. In general, filaments active for DNA strand exchange have a steeper helical pitch (≈90–130 Å) compared to inactive forms, which are more compressed (≈65–85 Å). All evidence suggests that the elongated filaments are in the ATP bound state, whereas compressed filaments represent the ADP bound state. Ring-like structures have also been observed, and these are comprised of 6–8 subunits with an internal pore large enough to allow passage of DNA (13–16). These ring-like forms have no known function, but may represent an inactive storage form of the protein. The relationship between the different recombinase structures remains unknown, and it is not clear whether 1 structural form can directly transition to another without first disassembling into monomeric units.
To evaluate the properties of Rad51 we have established a single-molecule assay that allows us to probe individual nucleoprotein filaments in real time. Here we use this assay to examine the behavior of human Rad51 assembled onto dsDNA. Our results demonstrate that human Rad51 can transition between an elongated nucleoprotein filament and a more compressed structure. These transitions are triggered by ATP hydrolysis, and the Rad51 filaments could reversibly interconvert between the elongated and compressed forms when nucleotide cofactor was replenished. We suggest that the transitions between these structural forms may be a target for regulation during homologous recombination.
Results
Visualizing Rad51 Nucleoprotein Filaments.
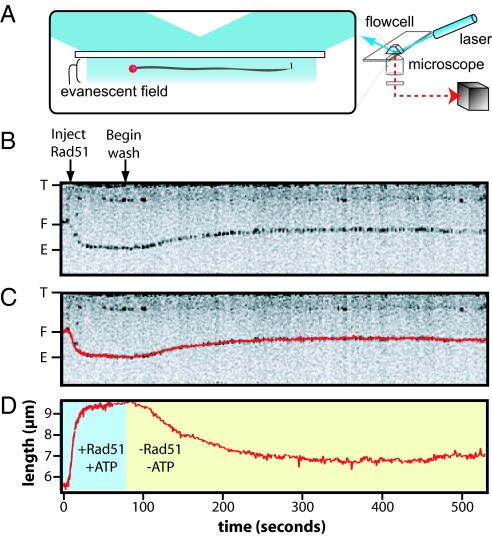
We have previously used total internal reflection fluorescence microscopy (TIRFM) to monitor the assembly of human Rad51 filaments on YOYO1-stained DNA curtains (17). However, Rad51 displaces YOYO1 from DNA, making it difficult to detect the fluorescence signal at high protein concentrations (17). To avoid this problem, we developed an assay that uses DNA substrates tagged at 1 end with a fluorescent quantum dot (QD; Fig. 1). Fig. 1 illustrates the procedure for visualizing Rad51 filaments with TIRFM. After locating a DNA curtain (17, 18), filament assembly was initiated by injecting 1 μM Rad51 in buffer containing 40 mM Tris (pH 7.8), 0.2 mg/mL BSA, 1 mM MgCl2, 1 mM DTT, and 1 mM ATP; unless otherwise stated, these conditions were used for all experiments reported below. Under these conditions, Rad51 completely covered the DNA as indicated by its length (Fig. 1). Fig. 1B shows an example of a kymogram depicting the change in the length of a single DNA molecule during the assembly of a Rad51 filament, and the subsequent decrease in the DNA length when free Rad51 and free ATP were flushed from the sample chamber.
Fig. 1.
Visualizing the behavior of human Rad51 nucleoprotein filaments. A shows a schematic of the TIRFM system used with a QD tagged DNA substrate. B shows a kymogram of a single DNA molecule and the black line demarks the QD tag at the end of the molecule. C shows the same kymogram superimposed with the particle-tracking data used to quantify the length of the DNA over time. The particle-tracking data alone are shown in D, and the assembly and chase phases of the reaction are denoted by blue and yellow background, respectively. When Rad51 is injected into the flowcell, assembly of the nucleoprotein filament causes lengthening of the DNA molecule and alters the position of the QD. Chasing the assembled filaments with wash buffer causes them to decrease in length. For the Top and Middle, “T” demarks the tethered end of the DNA molecule, “F” denotes the labeled end of the DNA in the absence of Rad51, and “E” denotes the location of the DNA after assembly of the extended nucleoprotein filament. In the Bottom, the length of the DNA is shown in microns (μm), and time is shown in seconds for all 3 panels.
Human Rad51 Can Undergo Structural Transitions Without Dissociating from DNA.
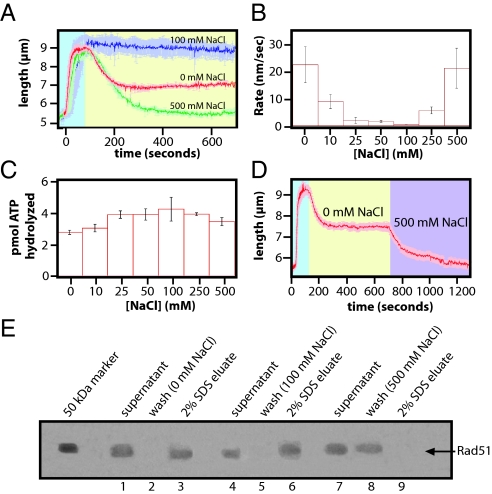
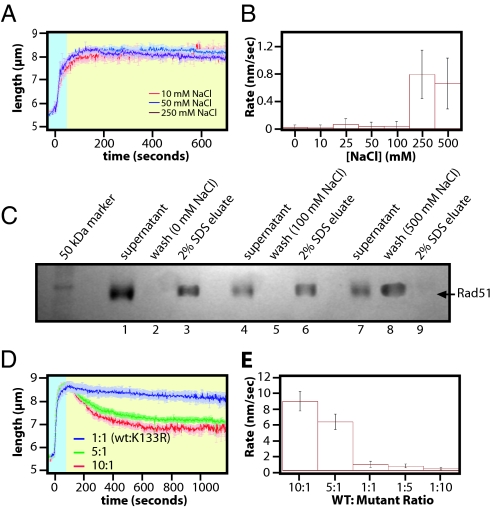
The DNA molecules bound by Rad51 filaments did not return to their original lengths when chased with buffer lacking both free Rad51 and free ATP (Fig. 1 B–D). There were 2 possible explanations for this observation, either some protein remained bound to the DNA, or Rad51 underwent a structural rearrangement to form a compressed filament. To help distinguish between these 2 possibilities we tested the effect of NaCl on the behavior of Rad51 during the chase phase of the assay. As shown in Fig. 2 A and B, in the absence of NaCl the Rad51 filaments decreased in length at a rate of 22.48 ± 6.59 nm/s (see Table S1) before plateauing at a final length of 7.09 ± 0.32 μm, which was approximately 25% longer than the naked DNA. As the concentration of NaCl was increased during the chase stage of the experiments, the rate at which the DNA molecules shorten decreased to just 0.614 ± 0.010 nm/s at 100 mM NaCl, and the molecules were still 8.78 ± 0.51 μm 10 min after initiating the chase. This indicated that little protein dissociated from the DNA and that the filaments remained in an elongated conformation. ATPase assays revealed that ATP hydrolysis was not inhibited at 100 mM NaCl (Fig. 2C). As the concentration of NaCl was increased further in the TIRFM assay the Rad51 filaments shortened more rapidly, with a measured rate of 21.12 ± 7.34 nm/s at 500 mM NaCl. More importantly, at this higher concentration of salt the DNA returned to near its original length with a final measured value of 5.91 ± 1.02 μm, indicating complete dissociation of Rad51.
Fig. 2.
Reduction in the length of the human filaments is not caused solely by protein dissociation. Nucleoprotein filaments were assembled with 1 μM hRad51, 1 mM ATP, 1 mM MgCl2, 40 mM Tris (pH 7.8), 0.2 mg/mL BSA, and 1 mM DTT. All reactions were performed at 37 °C. After assembly, the filaments were chased with buffer that lacked free Rad51, lacked ATP, and contained the indicated amounts of NaCl. The assembly and chase phases of the reaction are denoted by blue and yellow background, respectively. The graph in A shows representative examples of traces at different concentrations of NaCl, and each trace represents the average of 3 separate reactions. Rates (nm/s) for filament shortening were derived from these traces by fitting the disassembly phase of the reaction to a sigmoidal function and are plotted in B, and the rates for all tested reaction conditions are summarized in Table S1. (C) shows the results of ATPase assays with Rad51 at different concentrations of NaCl. In D, human Rad51 nucleoprotein filaments were first assembled and then chased with buffer containing 1 mM MgCl2, 40 mM Tris (pH 7.8), 0.2 mg/mL BSA, and 1 mM DTT. A second chase with buffer containing 500 mM NaCl was initiated 10 min later. E shows an SDS/PAGE with the results for a magnetic bead pull down assay demonstrating that Rad51 remained bound to the DNA in reactions containing no salt (lanes 1–3) or 100 mM NaCl (lanes 4–6), whereas 500 mM NaCl removed Rad51 from the DNA (lanes 7 and 8).
To determine whether high salt would dissociate protein from the compressed filaments, a second chase was conducted with high salt (Fig. 2D). This second chase caused the DNA to return to its original length, further suggesting that the partially extended DNA molecules were still bound by Rad51 that had transitioned to a compressed filament. This conclusion that Rad51 remained bound to the DNA as a compressed filament was consistent with a recent study from Hilario et al., which also demonstrated that fluorescently tagged Rad51 could transition to a compressed helical filament upon hydrolysis of ATP (19).
We also performed pull-down experiments with streptavidin coated magnetic beads and biotinylated DNA substrates (20). In these assays, Rad51 was bound to the bead tethered DNA, the beads were pelleted to remove the free protein, then they were washed under the same buffer conditions used in the TIRFM experiments. Any protein remaining bound to the DNA was eluted by the addition of 2% SDS. Aliquots from each stage of the experiment were saved and analyzed by SDS/PAGE and Coomassie staining. As shown in Fig. 2E, Rad51 remained bound to the DNA when the washes lacked NaCl and also when the washes contained 100 mM NaCl. In contrast, Rad51 eluted in the wash fractions when the beads were rinsed with 500 mM NaCl. The results from the bulk and single molecule experiments suggested that (i) high salt provoked complete dissociation of human Rad51 from the DNA, (ii) low salt caused the elongated filaments to undergo a structural transition in the absence of ATP that led to a reduced helical pitch without loss of bound protein, whereas (iii) buffer conditions more closely resembling physiological salt concentrations stabilized the filaments in the elongated conformation.
ATP Hydrolysis Triggers Compression of Human Rad51 Filaments.
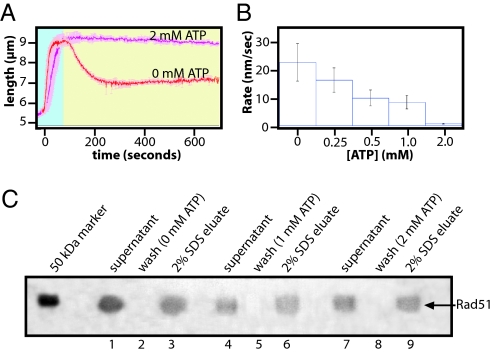
We next tested the effect of free ATP in the chase buffer. As shown in Fig. 3, increasing the concentration of ATP in the chase buffer caused a decrease in the rate at which the DNA shortened. When chased with 2 mM ATP, the filaments did not change appreciably in length during a 10-min observation (Fig. 3 A and B). These results demonstrated that free ATP stabilized the human Rad51 filaments in the elongated conformation. Results from the bead pull-down assays also confirmed that Rad51 remained bound to the DNA when ATP was included in the buffer wash (Fig. 3C).
Fig. 3.
ATP stabilizes the elongated Rad51 filaments. Rad51 filaments were assembled as described in Fig. 2, and then chased with buffer that contained varying concentrations of ATP but lacked free Rad51. A shows the effects of 0 mM and 2 mM ATP on human Rad51, and the bar graph in B shows the calculated shortening rates for 0, 0.25, 0.5, 1.0, and 2.0 mM ATP. The SDS/PAGE in C shows the magnetic bead pull-down assay performed with no ATP during the wash (lanes 1–3), or with 1 mM ATP (lanes 4–6) or 2 mM ATP (lanes 7–9) included in the wash buffers.
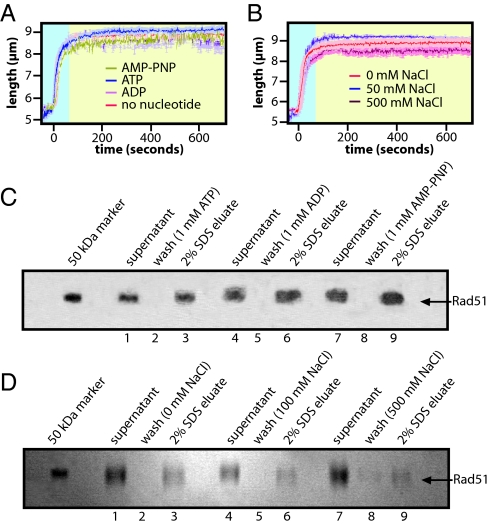
Calcium inhibits ATP hydrolysis by human Rad51 and stabilizes the protein in an elongated conformation proficient for strand exchange (21). However, we were unable to assess its effect in our assays because Ca2+ caused reversible adsorption of DNA to the lipid bilayer. As an alternative, we tested the effect of AMP-PNP, a non-hydrolyzable ATP analog, on filament stability. As previously shown, human Rad51 readily extends DNA in the TIRFM assay when AMP-PNP is used as the nucleotide cofactor (Fig. 4A) (17). These filaments remained elongated when chased with no nucleotide, 1 mM AMP-PNP, 1 mM ATP, or 1 mM ADP (Fig. 4A), suggesting that they did not release the bound AMP-PNP. In addition, Rad51 filaments made with AMP-PNP were highly resistant to dissociation even when chased with buffers containing up to 500 mM NaCl (Fig. 4B and Table S1). These results were corroborated with the magnetic bead assay (Fig. 4 C and D), and together these findings indicated that AMP-PNP locks human Rad51 into an extended configuration incapable of converting to a compressed filament.
Fig. 4.
Non-hydrolyzable nucleotide traps human Rad51 in an elongated conformation. Rad51 filaments were assembled as described in Fig. 3, but 1 mM ATP was replaced with 1 mM AMP-PNP. For the traces shown in A the nucleoprotein filaments were chased with buffer that contained either 1 mM AMP-PNP, ATP, or ADP. For traces in B the filaments were chased with buffer that lack nucleotide cofactor and contained varying concentrations of NaCl. C and D show the corresponding results from the magnetic bead pull-down experiments.
If ATP hydrolysis was necessary for the structural transition, then Rad51 mutants incapable of hydrolyzing ATP should be locked in the extended conformation. To test this hypothesis, we analyzed Rad51 K133R, which has a mutation within the Walker A motif that permits ATP binding, but prevents hydrolysis (22, 23). When filaments made with Rad51 K133R were chased with buffer lacking free protein and free ATP, they remained trapped in an extended conformation. The nucleoprotein filaments made with Rad51 K133R also displayed a 32-fold decrease in the dissociation rate relative to wild-type protein when chased with 500 mM NaCl (Fig. 5 A–C and Table S1). Control experiments showed that the formation of Rad51 K133R elongated filaments was dependent upon ATP, confirming that the mutation did not bypass the need for a nucleotide cofactor. These findings demonstrated that Rad51 K133R paralleled the behavior of filaments made with wild-type Rad51 plus AMP-PNP.
Fig. 5.
The ATPase-deficient mutant Rad51 K133R forms stable, elongated filaments on DNA. Rad51 nucleoprotein filaments were assembled as described in Fig. 2, using the ATPase deficient mutant human Rad51 K133R (A). The nucleoprotein filaments were then chased with buffer that lacked free Rad51, lacked ATP, and contained the indicated concentrations of NaCl and the rates at which the DNA decreased in length are shown in B. C shows the corresponding results from the magnetic bead pull-down experiments. D and E show TIRFM assays with nucleoprotein filaments assembled with mixtures of wild-type Rad51 and Rad51 K133R. The ratio of the 2 proteins was varied while maintaining a constant total concentration of protein at 1 μM. The corresponding rates for wild-type:mutant ratios of 10:1, 5:1, 1:1, 1:5, and 1:10 were 8.71 ± 1.22, 6.14 ± 0.97, 0.77 ± 0.40, 0.57 ± 0.24, and 0.23 ± 0.17 nm/s, respectively.
Rad51 K133R is a dominant-negative mutant when expressed at normal levels in a wild-type background (22). This suggested that mixed filaments comprised of both proteins would display behaviors comparable to the K133R mutant. To test this hypothesis we assembled mixed filaments comprised of varying ratios of wild-type and mutant proteins. As shown in Fig. 5 D and E, a 10:1 ratio of wild-type to mutant slowed the rate at which the DNA shortened by a factor of 2.6-fold compared with wild-type Rad51 alone (Fig. 3 and Table S1). At a 1:1 ratio, this effect was more pronounced, revealing a rate 30-fold lower than the wild-type protein alone. There were 2 potential explanations for these observations, either the mutant protein bound DNA faster than wild-type Rad51 leading to filaments comprised mostly of Rad51 K133R, or the presence of the mutant protein prevented the wild-type protein from undergoing the transition to the compressed structure. We previously demonstrated that Rad51 K133R binds dsDNA 2-fold more slowly than wild-type Rad51 (17), and these findings have been confirmed in a separate study using gel-shift experiments (23). This suggests that Rad51 K133R should be underrepresented in the mixed filaments, and yet could still exert an influence on the behavior of the wild-type Rad51. We concluded that the presence of the ATPase deficient mutant Rad51 K133R could effectively slow or stop the transition of the wild-type Rad51 from the elongated to the compressed state, and most likely exerted this effect through allosteric interactions between wild-type and mutant monomers within the filament.
Reversible Extension and Compression of Human Rad51 Nucleoprotein Filaments.
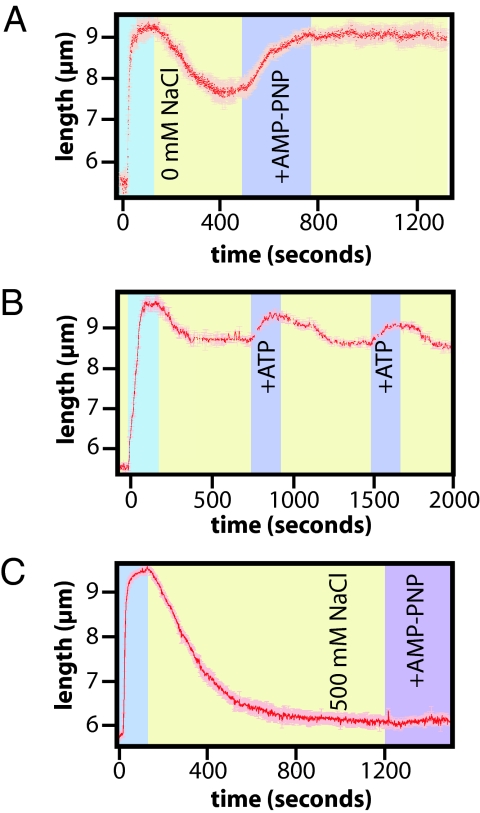
If Rad51 transitioned to a compressed filament, then replenishing the nucleotide cofactor might enable the filament to transition back to the elongated conformation. To test this prediction we assembled elongated Rad51 filaments with ATP (Fig. 6A). These filaments were chased with buffer lacking both free Rad51 and ATP to trigger formation of compressed filaments. The compressed filaments were chased a second time with assembly buffer containing 1 mM AMP-PNP as the nucleotide cofactor. As shown here, when the compressed filaments were chased with AMP-PNP the DNA molecules increased in length (Fig. 6A).
Fig. 6.
Reversible transitions between elongated and compressed filaments. In A, Rad51 nucleoprotein filaments were assembled with ATP, and then chased with buffer lacking nucleotide cofactor leading to formation of compressed filaments. One millimolar AMP-PNP was then injected into the sample chamber at the indicated time point revealing re-elongation of the compressed filaments. B shows cycles of successive injections of 2 mM ATP each followed by washes with buffer lacking nucleotide cofactor. C shows a control experiment in which Rad51 was stripped from DNA with 500 mM NaCl, followed by the injection of 1 mM AMP-PNP.
Re-extension of the DNA was also observed when the compressed filaments were chased with 2 mM ATP (Fig. 6B), and the filaments could be cycled through multiple rounds of elongation and compression when alternately chased with buffer ±ATP (Fig. 6B). These results confirmed that not only was Rad51 still bound to the DNA in a compressed conformation, but also that the bound protein was able to re-stretch the DNA when nucleotide cofactor was replenished. The re-elongated filaments did not attain the original length seen during the initial assembly reaction, but bead pull-down assays suggested there was minimal loss of protein from the DNA. This suggested 2 possibilities. The filament may have uniformly elongated, but failed to attain the same degree of extension observed for the original filaments. Alternatively, elongation may not have been uniform throughout the filament, with some regions remaining compressed while others elongated. Future studies will be necessary to distinguish between these scenarios.
An alternative explanation for these results was that free Rad51 was not effectively removed from the sample chamber during the buffer washes. To rule out this possibility, Rad51 filaments were assembled with ATP, and then chased with buffer containing 500 mM NaCl. As expected, this caused dissociation of Rad51 and the DNA returned to its original length. Subsequent addition of assembly buffer containing 1 mM AMP-PNP did not cause any increase in the length of the DNA (Fig. 6C). This control confirmed that free protein was efficiently removed from the sample chamber during the buffer washes and provided conclusive evidence that the compressed filaments re-elongated when chased with AMP-PNP.
Discussion
This work demonstrates that Rad51 can undergo reversible transitions between filaments with elongated and compressed helical geometries. Our results implicate ATP hydrolysis and allosteric interactions between neighboring Rad51 monomers as key determinants in modulating the dynamics of the filaments. These structural transitions take place without an intervening step requiring dissociation of Rad51 into monomers. Importantly, a recent single molecule study has demonstrated that bacterial RecA bound to ssDNA can also undergo repeated cycles of elongation and compression (24), and another study found that human Rad51 can transition from an extended to a compressed filament (19). Thus, reversible structural transitions may be a common attribute of this family of DNA recombinases. Our work also reveals a correlation between the ability of Rad51 to transition between an elongated filament and a compressed filament and the ability of the protein to dissociate from DNA. Conditions that prevented the elongated to compressed structural transition, such as wild-type Rad51 plus AMP-PNP or the ATPase deficient Rad51 K133R mutant, also prevented the protein from dissociating from DNA even when chased with high salt buffer. These results imply that dissociation of Rad51 from DNA is preceded by conversion from an elongated to a compressed state, a conclusion supported by 2 recent single molecule studies (19, 25).
The Role of ATP Hydrolysis and Allostery in Mediating Structural Transitions and Disassembly of Rad51 Filaments.
Rad51 K133R acts as a dominant-negative suppressor of normal Rad51 function in mouse ES cells (22). This is despite the fact that Rad51 K133R binds DNA and can perform strand exchange in vitro (17, 23). Our data show that Rad51 K133R is locked in an elongated conformation, and was resistant to dissociation when chased with high salt buffer. Moreover, Rad51 K133R exerts a dominant effect when assembled into mixed filaments comprised of both wild-type Rad51 and Rad51 K133R. These findings suggest that communication between Rad51 monomers within the filaments was involved in triggering the observed structural transitions, and that Rad51 K133R inhibited transition of the filament to the compressed state. This argues that allosteric interactions within the filament allow adjacent monomers to sense and respond to the nucleotide bound status of their immediate neighbors and respond accordingly. The dominant-negative effect of Rad51 K133R likely inhibits dissociation of mixed Rad51 filaments from undamaged chromatin in vivo before HR, and may also prevent removal of Rad51 from the double-stranded products of the strand invasion reactions. Either scenario could lead to an accumulation of lethal recombination intermediates that compromise genome integrity.
Near physiological salt concentrations also lock the wild-type Rad51 filaments into an elongated conformation. Based on these findings, we initially presumed that filaments locked into the elongated conformation through addition of 100 mM NaCl would display little or no ATP hydrolysis activity. However, ATP hydrolysis assays demonstrated that these buffer conditions also supported ATPase activity, indicating that the elongated Rad51 filaments were proficient at hydrolyzing and releasing bound ATP. In addition, the structural transitions for human Rad51 were not seen when high concentrations of ATP were maintained in the chase buffer. These findings argue that there is a rate-limiting step following ATP hydrolysis and ADP+Pi release that precedes conversion to the compressed state. This proposed rate-limiting step is slower than ATP rebinding, thus abrogating the Rad51 structural transition at high concentrations of free nucleotide cofactor. Based on our findings with Rad51 K133R, we propose that the rate-limiting step involves communication between neighboring subunits, and that conversion to the compressed state requires several neighboring subunits within a contiguous allosterically coupled patch of Rad51 to be in the ADP or nucleotide free state.
Nucleoprotein Filament Architecture as a Regulatory Component during HR.
Accumulating evidence supports the hypothesis that conversion of elongated filaments to compressed structures, and vice versa, involves conformational shifts in the N-terminal domain of Rad51 (11, 26). Details of this conformational change and the factors involved in its regulation remain unexplored, but recent studies have implicated this N-terminal region as a prime target for regulatory factors. For example, recent work has shown that BRC3 peptides derived from Brca2 bind the N-terminal domain of Rad51, and preferentially interact with regions of the filaments that are highly elongated (27). Qualitatively similar findings have been made for other Brca2-derived peptides, suggesting a common regulatory mechanism (28, 29). In addition, a small molecule called RS-1 has recently been identified from a chemical library as an allosteric effector of human Rad51 (30). This compound stimulates strand invasion by stabilizing the filaments in an active elongated state (30). This same principle could hold true for any protein factor, or even small molecules, that could selectively bind to Rad51 while it was in the elongated conformation. Similarly, proteins or small molecules that preferentially interact with Rad51 filaments having a more compressed geometry would be expected to inhibit strand exchange by stabilizing an inactive conformation, and may possibly provoke more rapid dissociation of the protein from DNA. It will be of interest to determine how the structural transitions observed for Rad51 are influenced by protein cofactors known to mediate recombination by impacting the activity of Rad51 and related recombinases.
Materials and Methods
Proteins, DNA, and Quantum Dots.
Human Rad51 was overexpressed in E. coli HMS174(DE3)pLysS and purified as previously described using a combination of ammonium sulfate precipitation and Ni-chelating chromatography (17). The DNA substrate used here was a 23-kb segment of the human β-globin locus generated using PCR from whole human genomic DNA using an Expand 20-kb PCR kit (Roche) using the following primers: 5′-biotin-TEG-CAC AAG GGC TAC TGG TTG CCG ATT-3′ and 5′-digoxigenin-AGC TTC CCA ACG TGA TCG CCT TTC TCC CAT-3′. Primers were obtained from Operon Technologies and gel purified before use. PCR products were purified over a MicroSpin™ S-400 HR column (GE Healthcare) pre-equilibrated in TE. Once assembled into a DNA curtain, a solution containing 1 nM anti-DIG labeled QDs was injected to bind the digoxigenin-labeled end of the DNA substrate and free QDs were rinsed from the flowcell. QDs (QD 705; Invitrogen) were labeled with polyclonal sheep anti-digoxigenin (anti-DIG) Fab fragments (Roche) as per the manufacturer's instructions.
Data Collection and Analysis.
The microscope used in this study has been previously described (17). The system uses a Nikon TE2000U inverted microscope and a back-illuminated EMCCD. Illumination was provided by a 488 nm diode-pumped solid-state laser focused through a fused silica prism onto the microfluidic sample chamber. Unless otherwise stated the assembly reactions used a 700-μL injection loop containing 1 μM human Rad51, a flow rate of 400 μL/min in buffer containing 40 mM Tris (pH 7.8), 0.2 mg/mL BSA, 1 mM MgCl2, 1 mM DTT, and 1 mM ATP or 1 mM AMP-PNP. The chase reactions used the same buffer conditions with the exception of the indicated variables. The flow cells were mounted in a custom designed heating unit and all reactions were performed at 37 °C. Data collection and analysis has been previously described (17, 31).
Magnetic Bead and ATPase Assays.
The magnetic bead assay was adapted from a previously described method (20). In brief, a 60-mer dsDNA oligonucleotide, modified with biotin on 1 5′ end was bound to streptavidin-coated magnetic beads. In a total volume of 10 μL, Rad51 was bound to the oligonucleotide in reaction buffer containing 40 mM Tris-HCl (pH 7.8), 1 mM DTT, 1 mM MgCl2, 1 mM ATP or AMP-PNP, and 0.2 mg/mL BSA (BSA). Rad51 was allowed to bind for 2 min at 37 °C. A magnet was used to pellet the beads, allowing for the removal of the supernatant buffer and the excess, unbound Rad51. The beads were then washed 2 times, each for 10 min at 37 °C, under the condition of interest. For each wash, the beads were pelleted as previously and the buffer was exchanged. Once the reaction was complete, remaining Rad51 bound to the oligonucleotide was eluted with reaction buffer plus 2% SDS. Samples of supernatant, the pooled washes, and eluate were then separated on an SDS/PAGE gel and detected with SimplyBlue™ SafeStain (Invitrogen).
ATPase reactions (10 μL) were carried out in 40 mM Tris pH 7.8, 1 mM MgCl2, 0.2 mg/mL BSA, and 1 mM DTT with the indicated concentration of NaCl. Reactions contained 1 μM Rad51 and 2 μM ssDNA 60-mer oligonucleotide. The reactions were initiated by adding 1 μL of 1.3 μM [γ-32P] ATP. Reactions were incubated at 37 °C and 2-μL aliquots were removed after 2, 5, 10, and 30 min and terminated with the addition of 2 μL of 500 mM EDTA. Samples were separated on TLC plates and analyzed with a phospho imager.
Supplementary Material
Acknowledgments.
This work was supported in part by a grant from Susan G. Komen grant (to E.C.G.) and National Institutes of Health (NIH) Grants GM074739 (to E.C.G), ES07061 (to P.S.), and GM53738 (to H.K.). R.B.R. was supported by a NIH Biophysics training grant (5T32GM008281). D.N.M. was supported by a NRSA fellowship from the NIH (GM084587). We thank members of this laboratory for comments on the manuscript. We thank Luke Kaplan for assistance with data analysis, and we thank members of our laboratories for discussion and comments on the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811465106/DCSupplemental.
References
- 1.Symington LS. Role of RAD52 Epistasis group of genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 3.West SC. Molecular views of recombination proteins and their control. Nat Rev. 2003;4:1–11. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 4.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J Biol Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 5.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 6.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: A biochemical and physical comparison. Front Biosci. 1998;3:d530–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 7.Eggler A, Inman R, Cox M. Oct 18) The Rad51-dependent pairing of long DNA substrates is stabilized by replication protein A. J Biol Chem. 2002;277:39280–39288. doi: 10.1074/jbc.M204328200. [DOI] [PubMed] [Google Scholar]
- 8.Lisby M, Rothstein R. DNA damage checkpoint and repair centers. Curr Opin Cell Biol. 2004 Jun;16:328–334. doi: 10.1016/j.ceb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Symington LS, Heyer WD. Some disassembly required: Role of DNA translocases in the disruption of recombination intermediates and dead-end complexes. Genes Dev. 2006;20:2479–2486. doi: 10.1101/gad.1477106. [DOI] [PubMed] [Google Scholar]
- 10.Baumann P, West SC. Role of the human Rad51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 11.Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc Natl Acad Sci USA. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheridan SD, et al. A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments. Nucleic Acids Res. 2008;36:4057–4066. doi: 10.1093/nar/gkn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinebuchi T, et al. Structural basis for octomeric ring formation and DNA interaction of the human homologous-pairing protein Dmc1. Mol Cell. 2004;14:363–374. doi: 10.1016/s1097-2765(04)00218-7. [DOI] [PubMed] [Google Scholar]
- 14.Passy SI, Yu X, Li Z, Radding CM, Masson J-Y, West SC, Egelman EH. Human Dmc1 protein binds DNA as an octomeric ring. Proc Natl Acad Sci USA. 1999;96:10684–10688. doi: 10.1073/pnas.96.19.10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X, Egelman EH. The RecA hexamer is a structural homologue of ring helicases. Nat Struct Biol. 1997;4:101–104. doi: 10.1038/nsb0297-101. [DOI] [PubMed] [Google Scholar]
- 16.Yang S, Yu X, Seitz EM, Kowalczykowski SC, Egelman EH. Archaeal RadA protein binds DNA as both helical filaments and octameric rings. J Mol Biol. 2001;314:1077–1085. doi: 10.1006/jmbi.2000.5213. [DOI] [PubMed] [Google Scholar]
- 17.Prasad TK, Yeykal CC, Greene EC. Visualizing the assembly of human Rad51 filaments on double-stranded DNA. J Mol Biol. 2006;363:713–728. doi: 10.1016/j.jmb.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 18.Graneli A, Yeykal CC, Prasad TK, Greene EC. Organized arrays of individual DNA molecules tethered to supported lipid bilayers. Langmuir. 2006;22:292–299. doi: 10.1021/la051944a. [DOI] [PubMed] [Google Scholar]
- 19.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc Natl Acad Sci USA. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Busygina V, Sehorn MG, Shi IY, Tsubouchi H, Roeder GS, Sung P. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 2008;22:786–795. doi: 10.1101/gad.1638708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stark JM, Hu P, Pierce AJ, Moynahan ME, Ellis N, Jasin M. ATP hydrolysis by mammalian Rad51 has a key role during homology-directed DNA repair. J Biol Chem. 2002;277:20185–20194. doi: 10.1074/jbc.M112132200. [DOI] [PubMed] [Google Scholar]
- 23.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair. 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Nishinaka T, Doi Y, Hara R, Yashima E. Elastic behavior of RecA-DNA helical filaments. J Mol Biol. 2007;370:837–845. doi: 10.1016/j.jmb.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 25.van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJ, Wuite GJ. Counting Rad51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galkin VE, et al. The Rad51/RadA N-terminal domain activates nucleoprotein filament ATPase activity. Structure. 2006;14:983–992. doi: 10.1016/j.str.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Galkin VE, Esashi F, Yu X, Yang S, West SC, Egelman EH. BRCA2 BRC motifs bind Rad51-DNA filaments. Proc Natl Acad Sci USA. 2005;102:8537–8542. doi: 10.1073/pnas.0407266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 29.Petalcorin MI, Galkin VE, Yu X, Egelman EH, Boulton SJ. Stabilization of RAD-51-DNA filaments via an interaction domain in Caenorhabditis elegans BRCA2. Proc Natl Acad Sci USA. 2007;104:8299–8304. doi: 10.1073/pnas.0702805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayathilaka K, et al. A chemical compound that stimulates the human homologous recombination protein RAD51. Proc Natl Acad Sci USA. 2008;105:15848–15853. doi: 10.1073/pnas.0808046105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad TK, Robertson RB, Visnapuu ML, Chi P, Sung P, Greene EC. A DNA-translocating Snf2 molecular motor: Saccharomyces cerevisiae Rdh54 displays processive translocation and extrudes DNA loops. J Mol Biol. 2007;369:940–953. doi: 10.1016/j.jmb.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.