Abstract
Aneuploidy, the most common chromosomal abnormality at birth and the main ascertained cause of pregnancy loss in humans, originates primarily from chromosome segregation errors during oogenesis. Here, we report that heterozygosity for a mutation in the mitotic checkpoint kinase gene, Bub1, induces aneuploidy in female germ cells of mice and that the effect increases with advancing maternal age. Analysis of Bub1 heterozygous oocytes showed that aneuploidy occurred primarily during the first meiotic division and involved premature sister chromatid separation. Furthermore, aneuploidy was inherited in zygotes and resulted in the loss of embryos after implantation. The incidence of aneuploidy in zygotes was sufficient to explain the reduced litter size in matings with Bub1 heterozygous females. No effects were seen in germ cells from heterozygous males. These findings show that Bub1 dysfunction is linked to inherited aneuploidy in female germ cells and may contribute to the maternal age-related increase in aneuploidy and pregnancy loss.
Keywords: fertility, maternal age, meiosis, oogenesis, spindle assembly checkpoint
Aneuploidy is the main ascertained cause of pregnancy loss in humans and the most common chromosomal abnormality at birth (1, 2). Aneuploidy in germ cells is primarily due to chromosomal segregation errors during the first meiotic division of oogenesis (1, 3). Numerous hypotheses and etiologies have been proposed for human aneuploidy, however, the only consistent finding remains its positive correlation with maternal age (2). Accurate chromosome segregation requires the coordinated interaction of many protein kinases and phosphatases, cellular organelles such as microtubules, motor proteins, centrosomes, kinetochores, and spindle checkpoint proteins. The spindle assembly checkpoint (SAC) is comprised of a set of highly conserved proteins, including Mad (mitotic arrest-deficient) and Bub (budding uninhibited by benzimidazole) proteins, and arrests cells during the mitotic metaphase to anaphase transition in response to kinetochores that are unattached to spindle microtubules or lack of tension from spindle fibers (3–6). The SAC prevents the precocious separation of sister chromatids and ensures accurate chromosomal segregation at anaphase by affecting the activity of the anaphase promoting complex/cyclosome (APC/C) (7). The APC/C ubiquitinates the securin component of the securin-separase complex in mitosis and tags it for degradation. Upon degradation of securin, separase is free to cleave the Scc1 subunit of the cohesin complex that holds sister chromatids together and allow their separation. In addition to its well-known role during mitosis, the SAC is active during mammalian oogenesis (8–10), and APC/C activity is required for homologous disjunction during the first meiotic division (11). Furthermore, meiosis in vertebrates requires separase-dependent depletion of Rec8 along chromosomal arms for subsequent chiasmata resolution (12). Lack of a functional spindle checkpoint because of defects in any of the SAC proteins may lead to chromosome missegregation and aneuploidy (8, 9, 13, 14).
The budding uninhibited by benzimidazole protein 1 (Bub1) is a protein kinase that localizes to kinetochores very early during mitosis and is required for the efficient kinetochore localization of other SAC proteins (15) and monitoring microtubule attachment to the kinetochore (16). In response to spindle damage, Bub1 phosphorylates Mad1 leading to the dissociation of the Mad1-Mad2 complex. Unbound Mad2 can then bind and inhibit Cdc20, an activator of APC/C (7, 17). Depletion of Bub1 in mammalian cells leads to misaligned chromosomes during mitosis (18) and to loss of chromatid cohesion (19–21). Conditional deletion of Bub1 in mice leads to developmental and proliferation defects in mouse embryonic fibroblasts (MEFs) and spermatogonial cells (21) whereas a hypomorphic mouse mutant model that expresses reduced levels of Bub1 shows an increased incidence of tumors (22). In mouse oocytes, Bub1 localizes to kinetochores (10, 23), and a dominant-negative form of Bub1 that perturbs the kinetochore localization activity of the wild-type (WT) protein leads to the acceleration of meiosis I, as has also been shown for Mad2 (9, 24, 25). It was recently shown that specific depletion of Bub1 in mouse oocytes results in chromosome missegregation at meiosis I and loss of cohesion between sister chromatids (26).
In this study, we analyzed the functional role(s) of Bub1 during embryogenesis and germ cell development in mice using a knockin gene-trap mutation within the Bub1 gene that leads to the generation of a truncated Bub1-βgeo fusion protein containing the first 269 aa of Bub1. The mutated Bub1 protein includes the Mad3-Bub1 domain and the kinetochore binding domain fused to β-galactosidase-neomycin (βgeo) reporter protein (Fig. 1 A–C and Fig. S1). We show that heterozygosity for this mutation results in female-specific, age-dependent increases in germ cell aneuploidy that is inherited in zygotes and affects the survival of the embryos. Our results suggest that heterozygous mutations in the human BUB1 gene may be an etiological cause of the known maternal-age effect for germ cell aneuploidy.
Fig. 1.
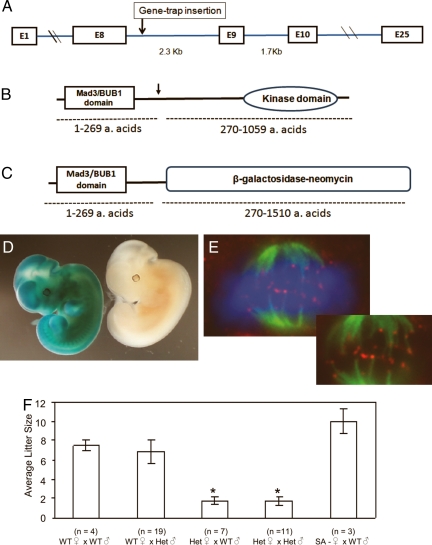
Bub1 mutant females are subfertile. (A) Gene structure of the murine Bub1. The relative insertion site is indicated by an arrow. (B) Bub1 protein organization. The functional domains within the Bub1 protein are shown, and the relative trap insertion site is indicated by an arrow. (C) Bub1-βgeo mutant protein organization. The replacement of the C-terminal amino acids by the β-galactosidase-neomycin (β-geo) fusion peptide is shown. (D) Expression of Bub1 in 10.5 dpc embryos using the β-geo reporter mediated conversion of X-gal substrate. Photograph shows a Bub1 heterozygote embryo (stained blue) and a control WT littermate embryo obtained from a WT female mouse crossed to a Bub+/m male. (E) Localization of the mutant Bub1-β-geo fusion peptide to kinetochores in Bub+/m mouse embryonic fibroblasts (MEFs). Immunofluorescence analysis of a metaphase cell stained with FITC-conjugated α-tubulin antibodies to visualize microtubules (green), anti-β-galactosidase antibodies bound to Texas-red conjugated secondary antibodies (red), and DAPI stained as blue to visualize DNA is shown above. Note the dotted appearance of the Bub1-β-geo fusion protein signals at the chromosome-microtubule junctions that indicate kinetochore localization. Inset shows the same image without the DAPI-stained DNA for better visualization of the kinetochore localization of the Bub1-β-galactosidase fusion protein. (F) The average litter size of various intercrosses obtained from Bub1+/m and WT mice are shown. The bars indicate the standard error within the dataset obtained from each intercross and n refers to the total number of crosses from each intercross. The differences between the litter sizes obtained from the heterozygous females were significantly different from the intercrosses obtained from the WT females. ∗, P < 0.0001, Student t test.
Results
Generation of Bub1 Mutant Mice.
To generate mutant mice that are homozygous for the Bub1 mutant allele (Bub1m/m), we intercrossed F1 heterozygous offspring (Bub1+/m) obtained from Bub1 chimeric males. Intercrosses of the F1 heterozygotes yielded no Bub1m/m offspring, confirming that the Bub1 homozygous mutation leads to embryonic lethality in mice (22). The essential role for Bub1 in embryonic development is consistent with studies that have shown a similar role for other SAC proteins (27–29). Intercrosses between heterozygous males and WT females yielded the expected ratio of heterozygous and WT offspring (60 Bub1+/m and 70 Bub1+/+ offspring from 19 matings) indicating that heterozygosity for Bub1 did not affect embryonic development. Expression analysis of the surrogate marker (β-gal) driven by the Bub1 promoter in E10.5 mutant heterozygous embryos showed that Bub1 was ubiquitously expressed in the embryo (Fig. 1D). In adult mouse tissues, Bub1 expression was high in thymus, ovary, and testis. However, much lower Bub1 expression levels were noted in tissues with differentiated cells that have a low proliferation potential (Fig. S2). Expression analysis of the Bub1-βgeo fusion protein in metaphase mouse embryonic fibroblasts (MEFs) showed that the fusion protein localizes to the kinetochores suggesting a putative dominant negative function by competing for kinetochore binding with the WT protein with intact kinase activity or preventing interactions with other SAC components (Fig. 1E).
Bub1 Heterozygous Female Mice Are Subfertile.
During the breedings aimed at generating homozygous mutants, we noted that the heterozygous intercrosses yielded abnormally low litter sizes (22 pups from 12 matings) that were not accounted for by the embryonic lethality of Bub1m/m embryos. To further investigate this phenotype, we generated offspring from intercrosses between Bub1+/m and WT mice (Fig. 1F). Crosses between WT females and heterozygous males, as well as WT females and WT males, yielded similar litter sizes (6.8 ± 1.2 versus 7.5 ± 0.5 SE). However, intercrosses involving Bub1+/m females (with WT males or heterozygous males) yielded a significantly lower number of offspring (1.7 ± 0.4 and 1.7 ± 0.5, respectively). To confirm that the phenotype was directly related to disruption of the Bub1 gene, we induced deletion of the splice acceptor (SA) sequence from the gene trap by mating Bub1 mutant mice with transgenic mice that expressed Cre-recombinase under the control of mouse protamine 1 promoter. The deletion of the SA sequence is expected to result in the reexpression of the WT Bub1 protein from the gene-trap allele (Fig. S1). When females heterozygous for the gene trap that lacked the splice acceptor (SA- females) were mated with WT males, the litter size was comparable to WT crosses, indicating that the SA deletion had reversed the subfertility defect (Fig. 1F).
We next determined whether the reduced litter size in Bub1+/m females was due to implantation defects. We analyzed uterine content in pregnant females between 6.5 and 9.5 days post coitum (dpc) and found a slight reduction (≈30%) in the number of implantation sites between WT and Bub1+/m females (Table 1). Analysis of the harvested embryos showed arrested development of a majority of the embryos at various stages that ranged from E6.5 through E8.0 (Table 1 and Fig. S3). Furthermore, the data also suggested that the Bub1+/m females were unable to support the normal development of embryos irrespective of their genotype as ≈80% of the implanted embryos were either arrested or dead even in crosses between Bub1+/m females and WT males that were expected to yield an equal proportion of WT and heterozygous embryos. To determine whether the subfertile phenotype of the Bub1+/m females was due to defects in follicular or uterine development, we performed histological analysis of serial sections of ovaries and uteri harvested from age matched mutant and WT mice (ages 7, 13, and 28 weeks). No significant differences in the various stages of follicular or uterine development between the 2 groups were seen; however, Bub1+/m females exhibited slightly increased levels of epithelial apoptosis, multifocal superficial stromal necrosis, and neutrophilic endometrial infiltrates (Fig. S4). Finally, analyses of cultured blastocysts obtained at E3.5 from Bub1 mutant females crossed to WT males showed that a majority of blastocysts did not grow in culture (Fig. S5) suggesting that preimplantation development is also impaired in Bub1+/m females.
Table 1.
Comparison of implantation rates and embryo phenotypes in Bub1+/m and WT females between 6.5 and 9.5 dpc
| Parent genotype (total implantation sites analyzed) | Normal embryos, % | Arrested or abnormal embryos, % | Complete loss of embryo, % | Implantation sites (average) |
|---|---|---|---|---|
| Het ♀ × Het ♂ (n = 24) | 29 | 25 | 46 | 6.0 |
| Het ♀ × WT ♂ (n = 23) | 22 | 65 | 13 | 7.6 |
| WT ♀ × Het ♂ (n = 19) | 95 | 0 | 5 | 9.5 |
| WT ♀ × WT ♂ (n = 10) | 90 | 0 | 10 | 10 |
Bub1 Mutation Leads to Maternal Age-Dependent Germ Cell Aneuploidy.
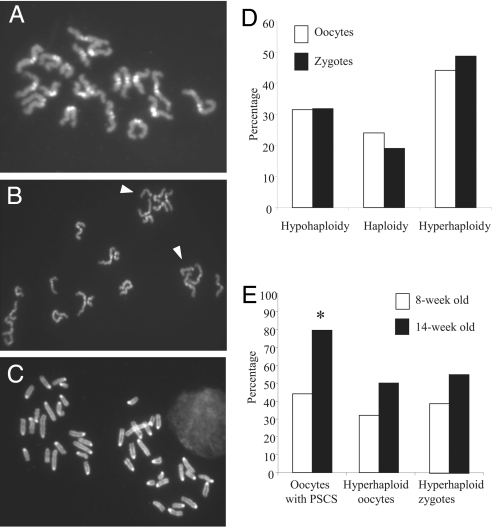
To determine whether the reduced litter size in crosses with Bub1+/m females was due to the generation of aneuploidy during oogenesis, we mated superovulated Bub1+/m females with B6C3F1 males and collected zygotes from the oviducts to analyze the chromosome complements of maternally and paternally derived pronuclei. Normal numbers of eggs were recovered from Bub1+/m females (28.1 average per Bub1+/m female versus 26.8 for WT females). As shown in Table 2, 38 of 47 zygotes from 8- to 14-week-old Bub1+/m females had aneuploid maternal chromosome complements (15 were hypohaploid, i.e., missing chromosomes; and 23 were hyperhaploid, i.e., had extra chromosomes) (Figs. 2B and C). These frequencies were significantly different (P < 0.001; χ2) from those observed in zygotes from WT females (Table 2). To determine whether these chromosomal segregation errors were occurring during meiosis I, we analyzed metaphase II (MII) oocytes from Bub1+/m and WT females. Fifty-nine of 78 MII oocytes from 8- to 14-week-old Bub1+/m females were aneuploid (25 were hypohaploid and 34 were hyperhaploid), compared with only 3 of 75 in WT females (Table 2). Comparisons of the results obtained in oocytes and zygotes showed good agreement between the incidences of hypohaploid and hyperhaploid maternal chromosome complements (Fig. 2D) suggesting that the majority of chromosomal segregation errors were occurring during meiosis I. These results show that Bub1 heterozygosity induced extremely high levels of aneuploidy in female germ cells and that inherited aneuploidy is most likely the cause of the embryonic lethality observed in matings with Bub1+/m females.
Table 2.
Aneuploidy frequencies in germ cells of Bub1 heterozygous female mice
| Female strain | Male strain | Total cells | Maternal chromosome number (%) |
|||
|---|---|---|---|---|---|---|
| <20 | 20 | >20† | 40 | |||
| Metaphase II oocytes | ||||||
| Bub1 WT | 75 | 2 (2.7) | 72 (96.0) | 1 (1.3) | 0 (0.0) | |
| Bub1+/m | 78 | 25 (32.1)* | 19 (24.3) | 34 (43.6)* | 0 (0.0) | |
| Zygotes | ||||||
| Bub1 WT | B6C3F1 | 70 | 3 (4.3) | 60 (85.7) | 4 (5.7) | 3 (4.3) |
| Bub1+/m | B6C3F1 | 47 | 15 (31.9)* | 9 (19.1) | 23 (48.9)* | 0 (0.0) |
*, P < 0.001 vs Bub1 WT females (χ2).
†Indicates oocytes with at least 20 chromosomes (i.e., 20 dyads plus a single chromatid or 19 dyads plus 3 single chromatids) and zygotes with at least 21 chromosomes (see Table S1 for a complete list of all aneuploid oocytes and zygotes).
Fig. 2.
Metaphase II oocytes and zygotes from Bub1+/m females have abnormal chromosome numbers. Bub1+/m or 8- to 16-week-old WT females were mated with B6C3F1 males and zygotes collected ≈30 h after the induction of superovulation. Oocytes were collected from females that did not mate ≈20 h after the induction of superovulation. (A) Normal metaphase II oocyte with 20 dyads from a WT female. (B) Metaphase II oocyte from a Bub1+/m female with a total of 21 chromosomes. Arrowheads indicate single chromatids. (C) Zygote from a Bub+/m female with 20 paternal chromosomes on the left and 23 maternal chromosomes on the right. (D) Comparisons of aneuploidy frequencies in metaphase II oocytes and zygotes from Bub+/m females showing similar levels of hypohaploid and hyperhaploid maternal complements before and after fertilization. (E) Age-dependent increase in the incidences of PSCS and hyperploid oocytes and zygotes in Bub1+/m female of different ages. ∗, P < 0.002 (χ2).
An important insight into the event(s) that underlie the generation of aneuploidy during meiosis I in Bub1+/m females was provided by the observation that ≈70% of the oocytes from mutant females presented with single chromatids (or half-dyads) (Fig. 2B) compared with <7% in WT females. This strongly suggests that the Bub1 mutation affected chromatid cohesion and resulted in premature sister chromatid separation (PSCS). Of the 38 aneuploid oocytes with unpaired chromatids found in Bub1+/m females (Table S1), 30 had an odd number of single chromatids, suggesting that the majority of PSCS occurred during meiosis I. However, PSCS could not explain all of the aneuploid events observed, but was likely involved in the genesis of ≈85% of aneuploidy. Specifically, of the 34 hyperhaploid MII oocytes observed in Bub1+/m females, 19 (56%) originated from PSCS, 5 (15%) by nondisjunction, that is, had more than 20 dyads and no single chromatids, and 10 (29%) by a combination of PSCS and nondisjunction.
The effects of the Bub1 mutation seem to aggravate with maternal age. As shown in Fig. 2E, older Bub1+/m females (14-week-old) had significantly higher (P < 0.002; χ2) frequencies of oocytes with PSCS than younger Bub1+/m females (8-week-old). Older females also had higher frequencies of hyperhaploid oocytes and zygotes, although these differences were not statistically significant (P > 0.1; χ2). In addition, no litters were obtained from Bub1+/m heterozygous females that were at least 24 weeks old, further supporting an effect of maternal age on aneuploidy induction.
The dramatic effects of heretozygosity for the Bub1 mutation on meiotic chromosome segregation are restricted to female germ cells. Only 3 out 109 zygotes recovered from WT females mated to Bub1+/m males were aneuploid (2 hypohaploid and 1 hyperhaploid). These frequencies were not different from those observed in control matings (Table S2). To exclude the possibility that the lack of aneuploidy in zygotes fathered by Bub1+/m males was due to selective elimination of aneuploid spermatids during postmeiosis, we analyzed MII spermatocytes isolated from testes from the same males used for the zygote studies. Analysis of 108 MII spermatocytes (Table S2) showed no hyperhaploidy and no PSCS.
Discussion
Despite the prevalence of aneuploidy in human female germ cells, its importance to female fertility and the well-established maternal age effect, we know little about the causes and factors that predispose to chromosome nondisjunction during oogenesis (2). In recent years, the use of mouse models with mutations in genes involved in meiotic recombination and chromosome segregation have shed light on factors that assure proper chromosome segregation. It is becoming clear that disturbances in meiotic recombination are important contributors to meiotic nondisjunction. Increased female germ cell aneuploidy rates have been observed in mice lacking the synaptonemal complex protein 3 (Scp3) (30, 31), the meiosis-specific cohesin Smc1β (32), and the homologous recombination protein Rad51c (33). An important role for histone acetylation in chromosome alignment and meiotic spindle organization has also been demonstrated (34, 35). It has so far been difficult to assess the role of SAC proteins during gametogenesis, because their deletion has invariably resulted in embryonic lethality (22, 27–29). However, a recent study that used a floxed Bub1 allele and a Zp3-Cre recombinase transgene approach to specifically delete Bub1 in mouse oocytes (26) demonstrated the important role of Bub1 in assuring proper chromosome segregation in mammalian oocytes. We show here that heterozygosity for a Bub1 mutation predisposes female germ cells, but not male germ cells, to chromosome missegregation and produces a phenotype that is indistinguishable from that generated by the complete deletion of Bub1 protein (26). Furthermore, we demonstrate that the Bub1 mutation affects sister chromatid cohesion resulting in the induction of aneuploidy primarily during meiosis I, that aneuploidy increases with advancing maternal age, and that such aneuploidy is inherited in zygotes. More importantly, the incidence of aneuploidy in zygotes is sufficient to explain the reduced litter size observed in matings with Bub1 heterozygous females providing a direct link between heterozygosity for Bub1 in females and aneuploid frequencies in their offspring. Our results suggest that a functional deficiency in the Bub1 protein, either through age-related decline in mRNA levels or age-related accumulation of mutations that inactivate the protein, may represent a mechanism that contributes to the known maternal age-related increase in aneuploidy (2) and fertility defects in women with multiple abortions.
How does a mutation for Bub1 result in chromosome missegregation and why are the effects limited to female germ cells? Unlike a recent Bub1 mouse model with a hypomorphic allele that expressed ≈20% of the WT protein and did not exhibit fertility defects (22), our mutational strategy generated a mutant Bub1 protein that lacks the kinase domain but that still contains the kinetochore binding domain. The mutant protein is able to bind to kinetochores (Fig. 1E) and may compete with the WT protein resulting in a dominant negative mutation. Support for this hypothesis comes from another study that showed that a truncated mutant for Bub1 (amino acids 1–331) disrupted the SAC in mitotic cells by competing with the endogenous Bub1 kinase for kinetochore localization (24). Alternatively, the mutated protein may be unable to recruit other SAC proteins to the kinetochore or to properly interact with the cohesin protein complex resulting in premature centromeric separation of sister chromatids (19, 20). It was recently demonstrated that deletion of the Bub1 gene in mouse oocytes causes premature activation of the APC/C and separase and that the precocious activation of these 2 SAC components was responsible for many of the phenotypes caused by Bub1 depletion (26). Interestingly, these authors also demonstrated that a mutated Bub1 protein that lacked the kinase domain, as the mutated version of Bub1 in our heterozygous mouse model, prevented the precocious activation of the APC/C (26). Earlier studies have shown that Bub1 is necessary for the centromeric localization of the shugoshin proteins (Sgol in meiosis and SgoII in mitosis of fission yeast) (36), as well as Rec8 in fission yeast (37). Furthermore, recruitment of PP2A that regulates Rec8 cleavage also depends on SgoI in fission and budding yeast (38). Rec8 localization to centromeres in mouse oocytes depends on SgoII and deletion of SgoII leads to fertility defects in mice (23, 39). Together with these findings, our data suggest that the Bub1 function affected in our heterozygous model is its role in protecting centromere cohesion through recruitment of shugoshin, PP2A, and Rec8 proteins and prevention of separase-mediated removal of cohesin (19, 20). Further evidence for the role of Bub1 in sister chromatid cohesion is strengthened by earlier observations that have shown a dual role for Bub1 in chromosomal congression (involving the regulation of kinetochore-microtubule attachments) in addition to a role in SAC signaling (18). Our data show that abnormal Bub1 function results in the premature separation of chromosomes into separated sister chromatids, a mechanism that has been proposed to be involved in the genesis of the majority of human germ cell aneuploidy (40).
Maternal age is a well-established etiological factor in the genesis of human aneuploidy (2). Therefore, we investigated whether the effects of heterozygosity for the Bub1 mutation increased with maternal age. The results showed that 14-week-old Bub1+/m females had significantly (P < 0.05) higher levels of PSCS than 8-week-old Bub1+/m females (Fig. 2E). The frequencies of hyperhaploid oocytes and zygotes, although higher in older females, did not show significant age-related increases. However, this was more likely due to the already high frequencies of these events in the 8-week-old females, than to a lack of an age effect. To further substantiate age-dependent increase in aneuploidy, there was complete loss of fertility in Bub1+/m females that were older than 24 weeks (6 months). Interestingly, decline of Bub1 mRNA levels has been reported in human oocytes of older women, particularly during meiosis I (41). This suggests an additive, or possibly synergistic, effect between the presence of the Bub1 mutation and the age-dependent reduction of Bub1 mRNA levels that results in the progressive reduction in the amount of the WT Bub1 protein with augmented loss of chromatid cohesion and possibly SAC dysfunction. Indeed, weakening of sister chromatid cohesion has been also suggested as an important component of the maternal age effect (42).
Sexually dimorphic phenotypes in mice have been reported for several mutations involving genes with roles in meiotic chromosome segregation (31, 33, 43). However, the lack of an effect in male germ cells was unexpected given the recently demonstrated importance of Bub1 in spermatogenesis (21). It is important to note that the spermatogenesis defects reported in the conditional deletion of Bub1 in males resulted from mitotic defects that lead to the loss of proliferation of spermatogonial cells (21), whereas our data implicate a specific role for Bub1 in meiosis that affects female fertility. Bub1 is expressed in testis (44), and a recent conditional Bub1 nullizygous model clearly demonstrated impaired chromosome segregation and spermatogenic progression in the absence of Bub1 (21). Our results strongly suggest that Bub1 has different functions during oogenesis and spermatogenesis or that a yet unidentified testis-specific protein compensates for the reduced level of normal Bub1 protein in heterozygote males. Genomic studies have shown the presence of a testis-specific mRNA species, in addition to the full-length Bub1 mRNA found in testis and other tissues, that encodes for a protein that lacks the first 421 aa of the Bub1 protein (44). This testis-specific transcript derived from an alternative start site or alternative splicing event would generate a testis-specific protein that would not be affected by our mutational scheme and may be sufficient to prevent chromosome missegregation during spermatogenesis. Furthermore, it is interesting to note that heterozygosity for another SAC component, Mad2, affects chromosome segregation in oocytes only (8), and differences in spindle association of Mad2 and BubR1 have been reported between spermatogenesis and oogenesis (45, 46). Therefore, there may be qualitative and quantitative differences in the SAC machinery between oogenesis and spermatogenesis.
The generation of various mutant mouse models have led to major advances in dissecting the molecular mechanisms of germ cell aneuploidy and to a better understanding of the causes of infertility during the last decade (47). The mouse model described here should be valuable for elucidating the mechanisms responsible for the known sex differences in the susceptibility to chromosome missegregation during meiosis and understanding the basis of the maternal age-dependent increase in germ cell aneuploidy. Our results suggest that heterozygous mutations in the human BUB1 gene that lead to truncated versions of the protein and mutations interfering with recruitment/binding to other SAC components could be etiological causes for fertility defects in women with multiple abortions and be involved in the known maternal-age effect for germ cell aneuploidy.
Materials and Methods
Generation of Bub1 Mutant Mice.
We generated Bub1-deficient mice using the Baygenomics gene-trap ES cell resource (48). The Baygenomics insertional mutagenesis strategy utilizes a gene-trap cassette consisting of a splice-acceptor-βgeo cassette (β-galactosidase-neomycin fusion gene). The Bub1 trapped ES cells were obtained and characterized further. Bub1 disruption was confirmed by using primers specific for Bub1 exon 8 (5′-AAGAAGCATGAGCAGTGGGTT-3′) and the gene-trap sequences (5′-ACCTGGCTCCTATGGGATAG-3′). A schematic of the Bub1 gene-trap insertion, its effect on the Bub1 gene product and genotype analyses of the mutant and WT alleles are shown in Fig. S1. Sequencing of the PCR product indicated that the gene trap was integrated within intron 8 of the Bub1 gene. Bub1-targeted ES cells were used for blastocyst injections to generate founder mice and F1 progeny. The insertion of the gene trap leads to the loss of expression of the 17 downstream exons. At the protein level, the gene-trap insertion was determined to be downstream of the kinetochore localization domain and upstream of the C-terminal kinase domain. The gene-trap strategy used to generate the Bub1 mutant ES cells also leads to the generation of a putative truncated Bub1-βgeo fusion protein containing the first 269 aa (lacking the kinase domain) of the 1,057-aa-long WT protein (16). The use of animals in this study was approved by the Institutional Animal Care and Use Committee at both the University of Tennessee and Lawrence Berkeley National Laboratory.
Cytogenetic Analyses of Oocytes, Spermatocytes, and Zygotes.
Bub1+/m and WT littermates were generated by mating Bub1+/m males and WT females at the University of Tennessee and then shipped to Lawrence Berkeley National Laboratory. B6C3F1 mice were purchased from Harlan Sprague–Dawley. Mice 8–14 weeks of age were maintained under a 12-h light/dark photoperiod at room temperature of 21–23 °C and relative humidity of 50% ± 5%. Female mice were superovulated by an i.p. injection of 7.5 I.U. of pregnant mare's serum (PMS; Sigma) followed 48 h later by an i.p. injection of 5.0 I.U. of human chorionic gonadotrophin (hCG; Sigma). Females were caged with males (1:1) ≈4 h after HCG injection and checked for vaginal plugs the next morning. Unmated females were used for isolation of MII oocytes (≈20 h after HCG). Mated females received an i.p. injection of 0.08 mg/kg colchicine (CAS No. 64-86-8; Sigma), dissolved in 0.2 mL distilled water 24 h after HCG to prevent the union of the 2 parental pronuclei and arrest zygotic development at the metaphase stage of the first cleavage division (49), and were euthanized by CO2 inhalation 6 h after colchicine injection. Zygotes were flushed out from isolated oviducts and processed according to the mass harvest procedure (50). The prepared slides were air-dried for at least 24 h at room temperature and stored in nitrogen atmosphere at −20 °C until use. 4,6-Diamidino-2-phenylindole (DAPI) at 0.25 μg/mL diluted in Vectashield mounting medium (Vector Laboratories) was used as counterstaining. An oocyte was considered aneuploid if it contained a chromosomal number different from 20 when both dyads and single chromatids (or half-dyads) were counted. For example, oocytes with 19 dyads plus 2 single chromatids (or 18 dyads plus 4 monads) were considered euploid whereas oocytes with 19 dyads plus 3 single chromatids were considered hyperhaploid.
Testis preparations were made according to a standard method (51). Slides from 4 Bub1+/m animals were scored. They were stained with 0.25 μg/mL DAPI diluted in Vectashield mounting medium (Vector Laboratories). Each slide was examined using a 40× objective for localizing MII spermatocytes and then with a 100× objective for identifying chromosomal abnormalities. At least 25 MII spermatocytes per mouse were analyzed.
Immunofluorescence Analysis of Bub1 Mutant MEFs.
Bub1+/m mouse embryonic fibroblasts were grown on glass cover slides and fixed with 4% formaldehyde at 25 °C (for 10 min) and blocked (10% donkey serum, 0.1% Triton X-100 in PBS) for 1 h. Slides were incubated with antibodies against β-galactosidase (1:200 dilution in blocking solution overnight at 4 °C) followed with Texas-red conjugated secondary antibodies (1:100 dilution in blocking solution at 25 °C for 1 h). Slides were then incubated with FITC-conjugated anti-α-tubulin antibodies (1:200 dilution) overnight and counterstained with 0.1 μg/mL DAPI to visualize mitotic cells during immunofluorescence analysis.
For additional information, see SI Text.
Supplementary Material
Acknowledgments.
We thank Drs. Bruce McKee, MaryAnn Handel, Andrew Wyrobek, and John Mailhes for comments. Work performed in part under the auspices of the U.S. Department of Energy by the University of California, Lawrence Berkeley National Laboratory under contract DE-AC02–05CH11231. S.V. was supported by the University of Tennessee start-up funds.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.M.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903075106/DCSupplemental.
References
- 1.Hassold T, Hall H, Hunt P. The origin of human aneuploidy: Where we have been, where we are going. Hum Mol Genet. 2007;16(Spec No 2):R203–R208. doi: 10.1093/hmg/ddm243. [DOI] [PubMed] [Google Scholar]
- 2.Hassold T, Hunt P. To err (meiotically) is human: The genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 3.Vogt E, Kirsch-Volders M, Parry J, Eichenlaub-Ritter U. Spindle formation, chromosome segregation and the spindle checkpoint in mammalian oocytes and susceptibility to meiotic error. Mutat Res. 200;651:14–29. doi: 10.1016/j.mrgentox.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 5.Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 6.Hoyt MA, Geiser JR. Genetic analysis of the mitotic spindle. Annu Rev Genet. 1996;30:7–33. doi: 10.1146/annurev.genet.30.1.7. [DOI] [PubMed] [Google Scholar]
- 7.Yu H. Regulation of APC-Cdc20 by the spindle checkpoint. Curr Opin Cell Biol. 2002;14:706–714. doi: 10.1016/s0955-0674(02)00382-4. [DOI] [PubMed] [Google Scholar]
- 8.Niault T, et al. Changing Mad2 levels affects chromosome segregation and spindle assembly checkpoint control in female mouse meiosis I. PLoS ONE. 2007;2:e1165. doi: 10.1371/journal.pone.0001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homer HA, et al. Mad2 prevents aneuploidy and premature proteolysis of cyclin B and securin during meiosis I in mouse oocytes. Genes Dev. 2005;19:202–207. doi: 10.1101/gad.328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunet S, Pahlavan G, Taylor S, Maro B. Functionality of the spindle checkpoint during the first meiotic division of mammalian oocytes. Reproduction. 2003;126:443–450. doi: 10.1530/rep.0.1260443. [DOI] [PubMed] [Google Scholar]
- 11.Herbert M, et al. Homologue disjunction in mouse oocytes requires proteolysis of securin and cyclin B1. Nat Cell Biol. 2003;5:1023–1025. doi: 10.1038/ncb1062. [DOI] [PubMed] [Google Scholar]
- 12.Kudo NR, et al. Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell. 2006;126:135–146. doi: 10.1016/j.cell.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 13.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 14.Mailhes JB. Faulty spindle checkpoint and cohesion protein activities predispose oocytes to premature chromosome separation and aneuploidy. Environ Mol Mutagen. 2008;49:642–658. doi: 10.1002/em.20412. [DOI] [PubMed] [Google Scholar]
- 15.Johnson VL, Scott MI, Holt SV, Hussein D, Taylor SS. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci. 2004;117:1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- 16.Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: Model for spindle checkpoint regulation. Mol Cell Biol. 2001;21:5190–5199. doi: 10.1128/MCB.21.15.5190-5199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meraldi P, Sorger PK. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 2005;24:1621–1633. doi: 10.1038/sj.emboj.7600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Z, Sun Y, Harley SE, Zou H, Yu H. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci USA. 2004;101:18012–18017. doi: 10.1073/pnas.0408600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr Biol. 2005;15:353–359. doi: 10.1016/j.cub.2004.12.044. [DOI] [PubMed] [Google Scholar]
- 21.Perera D, et al. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev Cell. 2007;13:566–579. doi: 10.1016/j.devcel.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J, et al. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol. 2008;10:42–52. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
- 24.Tsurumi C, Hoffmann S, Geley S, Graeser R, Polanski Z. The spindle assembly checkpoint is not essential for CSF arrest of mouse oocytes. J Cell Biol. 2004;167:1037–1050. doi: 10.1083/jcb.200405165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wassmann K, Niault T, Maro B. Metaphase I arrest upon activation of the Mad2-dependent spindle checkpoint in mouse oocytes. Curr Biol. 2003;13:1596–1608. doi: 10.1016/j.cub.2003.08.052. [DOI] [PubMed] [Google Scholar]
- 26.McGuinness BE, et al. Regulation of APC/C activity in oocytes by a Bub1-dependent spindle assembly checkpoint. Curr Biol. 2009;19:369–380. doi: 10.1016/j.cub.2009.01.064. [DOI] [PubMed] [Google Scholar]
- 27.Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 28.Kalitsis P, Earle E, Fowler KJ, Choo KH. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 2000;14:2277–2282. doi: 10.1101/gad.827500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker DJ, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 30.Kouznetsova A, Lister L, Nordenskjold M, Herbert M, Hoog C. Bi-orientation of achiasmatic chromosomes in meiosis I oocytes contributes to aneuploidy in mice. Nat Genet. 2007;39:966–968. doi: 10.1038/ng2065. [DOI] [PubMed] [Google Scholar]
- 31.Yuan L, et al. Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science. 2002;296:1115–1118. doi: 10.1126/science.1070594. [DOI] [PubMed] [Google Scholar]
- 32.Revenkova E, et al. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- 33.Kuznetsov S, et al. RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J Cell Biol. 2007;176:581–592. doi: 10.1083/jcb.200608130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De La Fuente R, Viveiros MM, Wigglesworth K, Eppig JJ. ATRX, a member of the SNF2 family of helicase/ATPases, is required for chromosome alignment and meiotic spindle organization in metaphase II stage mouse oocytes. Dev Biol. 2004;272:1–14. doi: 10.1016/j.ydbio.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Akiyama T, Nagata M, Aoki F. Inadequate histone deacetylation during oocyte meiosis causes aneuploidy and embryo death in mice. Proc Natl Acad Sci USA. 2006;103:7339–7344. doi: 10.1073/pnas.0510946103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 37.Bernard P, Maure JF, Javerzat JP. Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nat Cell Biol. 2001;3:522–526. doi: 10.1038/35074598. [DOI] [PubMed] [Google Scholar]
- 38.Riedel CG, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 39.Llano E, et al. Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev. 2008;22:2400–2413. doi: 10.1101/gad.475308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vialard F, et al. Evidence of a high proportion of premature unbalanced separation of sister chromatids in the first polar bodies of women of advanced age. Hum Reprod. 2006;21:1172–1178. doi: 10.1093/humrep/dei484. [DOI] [PubMed] [Google Scholar]
- 41.Steuerwald N, Cohen J, Herrera RJ, Sandalinas M, Brenner CA. Association between spindle assembly checkpoint expression and maternal age in human oocytes. Mol Hum Reprod. 2001;7:49–55. doi: 10.1093/molehr/7.1.49. [DOI] [PubMed] [Google Scholar]
- 42.Hodges CA, Revenkova E, Jessberger R, Hassold TJ, Hunt PA. SMC1beta-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nat Genet. 2005;37:1351–1355. doi: 10.1038/ng1672. [DOI] [PubMed] [Google Scholar]
- 43.Hunt PA, Hassold TJ. Sex matters in meiosis. Science. 2002;296:2181–2183. doi: 10.1126/science.1071907. [DOI] [PubMed] [Google Scholar]
- 44.Pangilinan F, et al. Mammalian BUB1 protein kinases: Map positions and in vivo expression. Genomics. 1997;46:379–388. doi: 10.1006/geno.1997.5068. [DOI] [PubMed] [Google Scholar]
- 45.Kallio M, Eriksson JE, Gorbsky GJ. Differences in spindle association of the mitotic checkpoint protein Mad2 in mammalian spermatogenesis and oogenesis. Dev Biol. 2000;225:112–123. doi: 10.1006/dbio.2000.9818. [DOI] [PubMed] [Google Scholar]
- 46.Jeganathan KB, van Deursen JM. Differential mitotic checkpoint protein requirements in somatic and germ cells. Biochem Soc Trans. 2006;34:583–586. doi: 10.1042/BST0340583. [DOI] [PubMed] [Google Scholar]
- 47.Matzuk MM, Lamb DJ. The biology of infertility: Research advances and clinical challenges. Nat Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stryke D, et al. BayGenomics: A resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 2003;31:278–281. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donahue RP. Cytogenetic analysis of the first cleavage division in mouse embryos (fertilization-pronuclei-T163H translocation) Proc Natl Acad Sci USA. 1972;69:74–77. doi: 10.1073/pnas.69.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mailhes JB, Yuan ZP. Cytogenetic technique for mouse metaphase II oocytes. Gamete Res. 1987;18:77–83. doi: 10.1002/mrd.1120180109. [DOI] [PubMed] [Google Scholar]
- 51.Evans EP, Breckon G, Ford CE. An air-drying method for meiotic preparations from mammalian testes. Cytogenetics. 1964;15:289–294. doi: 10.1159/000129818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.