Abstract
Substrates of the proteasome are recognized and unfolded by the regulatory particle (RP), then translocated into the core particle (CP) to be degraded1. A hetero-hexameric ATPase ring, containing subunits Rpt1-Rpt6, is situated within the base subassembly of the RP1. The ATPase ring sits atop the CP, with the Rpt C-termini inserted into pockets in the CP2–6. We have identified a novel function of the Rpt proteins in proteasome biogenesis through deleting the C-terminal residue from each Rpt. Our results indicate that assembly of the hexameric ATPase ring is templated on the CP. We have also identified an apparent intermediate in base assembly, BP1, which contains Rpn1, three Rpts, and Hsm3, a chaperone for base assembly. The Rpt proteins with the strongest assembly phenotypes, Rpt4 and Rpt6, were absent from BP1. We propose that Rpt4 and Rpt6 form a nucleating complex to initiate base assembly, and that this complex is subsequently joined by BP1 to complete the Rpt ring. Our studies show that assembly of the proteasome base is a rapid yet highly orchestrated process.
The core particle (20S CP) of the 26S proteasome is associated with the regulatory particle (19S RP), which in turn consists of two subassemblies, the base and lid1. The CP is composed of α-type and β-type subunits, with each family of subunits arranged into heteroheptameric rings, which are stacked to form a barrel-like α7-β7-β7-α7 complex1,7. The proteolytic sites are inside the CP barrel, and substrate access to these sites is achieved by opening a gate in the α ring. Binding of the RP opens the gate. Rpt1-6, six ATPases of the base, are thought to form a hetero-hexameric ring, and the C-termini of these Rpts insert into pockets in the CP α ring2–6. The extreme C-termini of four Rpts contain the motif Hb-Y-X (hydrophobic residue-Tyr-X), which mediates opening of the CP gate3,4,6 (Supplementary Table 1).
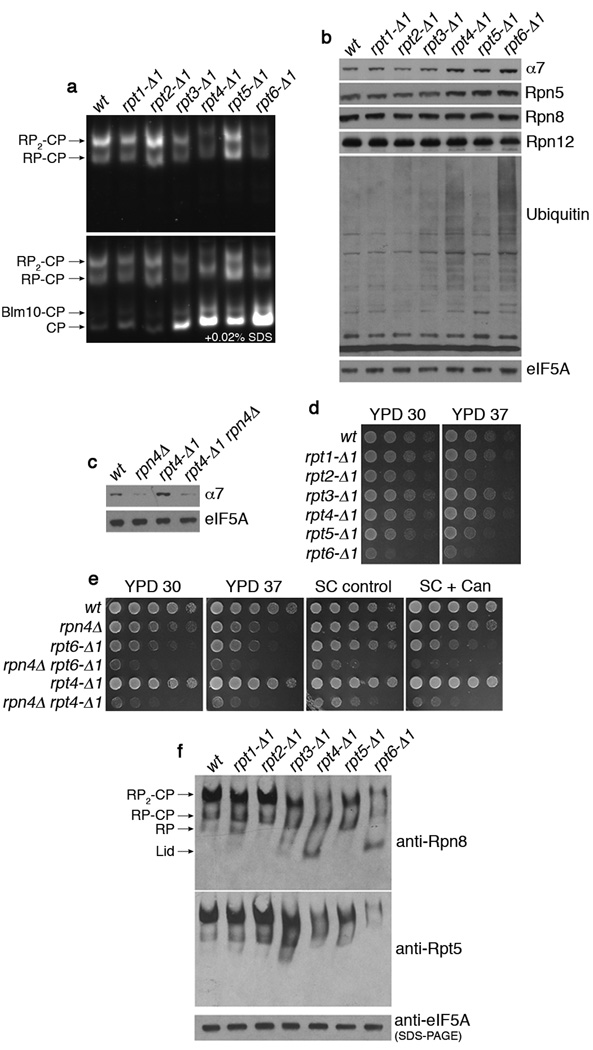
The C-terminal carboxylate of a given Rpt protein forms a salt bridge to a lysine residue at the bottom of the CP α ring pocket2,3. To eliminate these salt bridges, we deleted the C-terminal residue from each Rpt protein, generating six chromosomal mutations (rpt1-Δ1 to rpt6-Δ1). Proteasomes in whole cell extracts from each mutant were analyzed by native PAGE, combined with an in-gel peptidase assay using a fluorogenic substrate, LLVY-AMC (Fig. 1a). Free CP was strongly elevated in rpt3-Δ1, rpt4-Δ1, rpt5-Δ1, and rpt6-Δ1 mutants as revealed by the addition of 0.02% SDS (Fig. 1a). As previously shown, SDS opens the CP gate, which is closed in the free CP1,7. The CP, and a representative CP subunit, α7, were most strongly induced in rpt4-Δ1 and rpt6-Δ1 mutants (Fig. 1a and 1b). Interestingly, Rpt4 and Rpt6 are the two Rpts that lack the gating-directed HbYX motifs4 (Supplementary Table 1). These mutants also showed strong reductions in proteasome holoenzyme (Fig. 1a, RP-CP and RP2-CP).
Figure 1. rpt4-Δ1 and rpt6-Δ1 mutants are proteasome hypomorphs with defective proteasome assembly.
a, Native PAGE (3.5%) and two consecutive LLVY-AMC assays with whole cell extracts (85 µg). Immediately after the first LLVY-AMC assay (top panel), the second assay was conducted in the presence of 0.02% SDS (bottom panel).
b, c, SDS-PAGE and immunoblotting with whole cell extracts. eIF5A is a loading control.
d, e, Growth phenotypes of rpt mutants. Cells were spotted in 4-fold dilutions on YPD, synthetic medium (control), or synthetic medium containing canavanine at 0.5 µg/ml. Plates were incubated for 2–4 days at 30°C or at 37°C (YPD 37).
f, 3.5% native PAGE and immunoblotting of whole cell extracts (85 µg) against Rpn8 (lid subunit) and Rpt5 (base subunit). Extracts (20 µg) were resolved by SDS-PAGE and immunoblotting for loading control.
CP induction was mediated principally by Rpn4, a transcription factor that upregulates the expression of proteasome subunits when proteasome activity is compromised8,9; RPN4 deletion in rpt4-Δ1 or rpt6-Δ1 mutants markedly decreased the level of α7 and free CP (Fig. 1c and Supplementary Fig. 1). Assembly defects in the rpt mutant proteasomes correlated with the accumulation of ubiquitin-conjugates and, in the case of the strongest mutant, rpt6-Δ1, a growth defect at 30°C (Fig. 1b and 1d). Phenotypes of rpt4-Δ1 and rpt6-Δ1 were exacerbated by RPN4 deletion, indicating that the proteasome stress response masks the severity of these mutations (Fig. 1e).
Proteolytically inactive subassemblies such as the RP and lid cannot be visualized by activity assay. Therefore, to further address the assembly defects in rpt4-Δ1 and rpt6-Δ1 mutants, proteasomes were analyzed by immunoblot following native PAGE. Given the apparent role of Rpt C-termini in CP binding2–6, the increase in free CP and decrease in RP2-CP and RP-CP might predict an increase in free RP in rpt4-Δ1 and rpt6-Δ1 mutants. On the contrary, free RP was not detected in these mutants; instead free lid was observed (Fig. 1f and Supplementary Fig. 2). The production of free lid was unexpected because the Rpt C-termini, if docking to the CP, cannot be in contact with or proximal to the lid. Base-CP complex, previously observed in a lid mutant10, was not apparent in rpt4-Δ1 or rpt6-Δ1 mutants (Fig. 1f and Supplementary Fig. 2). Thus, productive interaction between the CP and the Rpt C-termini of the base is apparently required for stable incorporation of the lid to complete proteasome assembly.
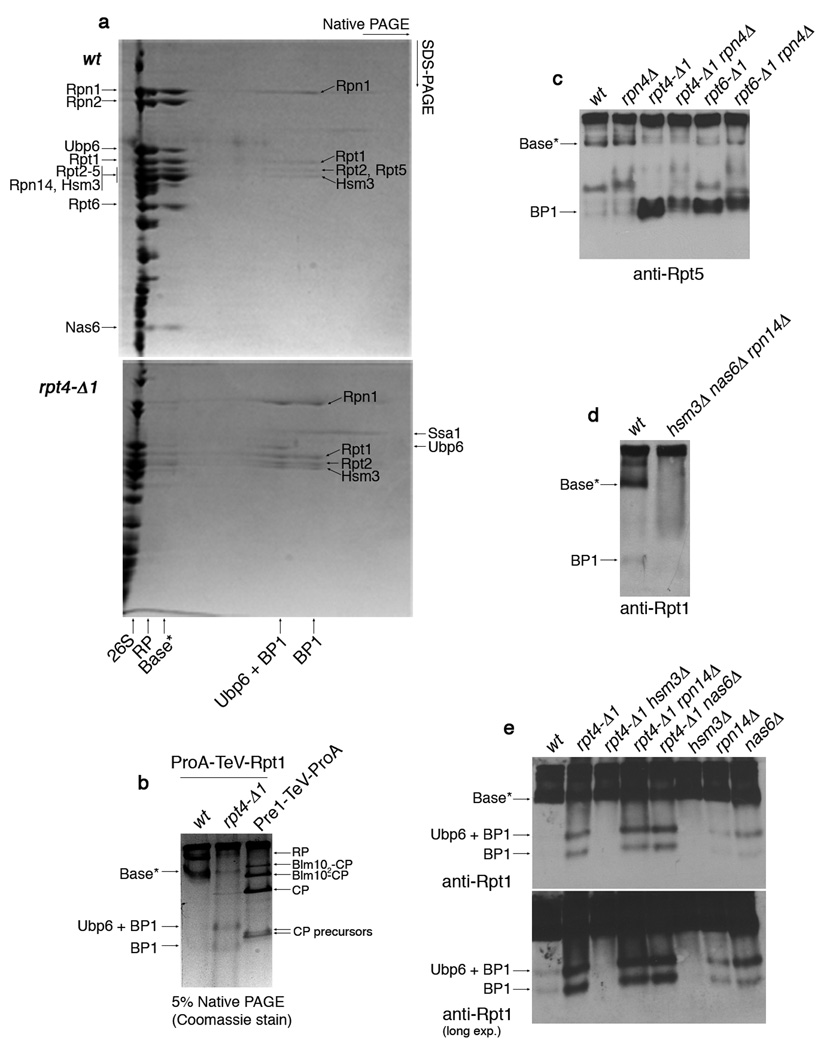
Because base joins the lid to the CP in proteasome holoenzymes, the accumulation of free lid and free CP in rpt4-Δ1 and rpt6-Δ1 mutants may reflect underlying defects in base assembly11,12. The base can be produced by exposing assembled proteasome to 1M NaCl13,14. The resulting particle represents the mature form of the base (Supplementary Fig. 3). If the base exists as a free endogenous complex, its isolation should be possible by affinity purification via Protein A-tagged Rpt1, using a physiological salt concentration. Such samples were analyzed by 2-D native/SDS-PAGE. A base variant, which we refer to as base*, was detected by this method (Fig. 2a, top). Interestingly, mass spectrometric analysis revealed that base* contains, in addition to Ubp6 and subunits of the base15, three proteins that are not proteasome components: Rpn14, Nas6, and Hsm3 (Fig. 2a). An accompanying paper16 shows that this set of proteins functions as molecular chaperones and more specifically as base assembly factors16. We show below that the function of these proteins, termed RP-chaperones, is tightly linked to the Rpt C-termini.
Figure 2. Identification of a base assembly intermediate.
a, 2-D native/SDS-PAGE of affinity-purifications with ProA-TeV-Rpt1 (Protein A tag appended to the N-terminus of Rpt1, a base subunit). Following a first-dimension native PAGE (5%), native gel lanes were individually excised and subjected to second-dimension SDS-PAGE (12.5%). Gels were stained with Coomassie blue. Individual spots of the base*, BP1, and Ubp6 + BP1 complexes were excised for mass spectrometry. Labels on the left indicate spots within base*. The presence of Rpn10 and Rpn13 in base* was not determined. Note that Rpt5 is absent from BP1 purified from rpt4-Δ1 mutants, but present in BP1 in untagged rpt4-Δ1 extracts (panel 4c). This appears to reflect a labile association of Rpt5 with BP1 in rpt4-Δ1 mutants. The presence of Ubp6 in BP1 is explained by its interaction with Rpn125. Note: Base* and BP1 species appear comparable between rpt6-Δ1 and rpt4-Δ1 mutants in their level and composition.
b, 5% Native PAGE following affinity purification via a ProA-TeV-Rpt1 as in (a) or a Pre1 (CP subunit)-TeV-ProA. Native gels were stained with Coomassie blue.
c, d, Immunoblotting following 5% native PAGE of whole cell extracts (100 µg).
e, Immunoblotting following 5% native PAGE of affinity-purified samples (2 µg) from indicated strains carrying ProA-TeV-Rpt1.
In rpt4-Δ1 mutants, base* levels are severely reduced, suggesting a defect in base assembly (Fig. 2a and 2b). Base* can also be visualized in unfractionated extract from untagged wild-type cells (Fig. 2c and 2d). Base* was deficient in rpt6-Δ1 cells (Fig. 2c) and undetectable in the RP-chaperone triple mutant, hsm3Δ nas6Δ rpn14Δ (Fig. 2d).
The disappearance of base* in rpt4-Δ1 mutants suggested an upstream assembly defect that would be manifest in the accumulation of smaller assembly intermediates of the base. Accordingly, affinity purification from ProA-TeV-Rpt1 rpt4-Δ1 mutants revealed a novel species that we termed Base Precursor 1, or BP1 (Fig. 2a and 2b). BP1 has two forms; the lower-mobility form contains deubiquitinating enzyme Ubp6, and is referred to as Ubp6 + BP1 (Fig. 2a and 2b). BP1 was also observed in wild-type, though at lower levels (Fig. 2a and 2b). The proportion of BP1 loaded with Ubp6 is higher in the mutant; most likely base assembly, which consumes BP1, is faster than Ubp6 loading in wild-type, while slower than Ubp6 loading in the mutant (see below). By mass spectrometry following 2-D native/SDS-PAGE, BP1 was found to contain Rpn1, Rpt1, Rpt2, Rpt5, and the RP-chaperone Hsm3 (Fig. 2a). The presence of Hsm3 in BP1 suggests that BP1 is an assembly intermediate. BP1 was detected in untagged whole cell extracts, and thus, like base*, it is not an artifact of tagging or purification (Fig. 2c and 2d). BP1 levels in extracts are elevated in rpt4-Δ1 and rpt6-Δ1 mutants (Fig. 2c). In contrast, HSM3 deletion resulted in an apparently complete elimination of BP1 (Fig. 2e). Thus, Hsm3 is not a bystander in BP1, but strongly regulates its abundance.
The absence of RP-chaperones Rpn14 and Nas6 from BP1 can be explained by their binding specificities: whereas Hsm3 binds to the BP1 component Rpt112,16, the other RP-chaperones bind to Rpt proteins that are absent from BP1: Nas6 to Rpt316,17, and Rpn14 to Rpt4 and Rpt616. RP-chaperones bind to the C-domains of Rpt proteins, but not to their extreme C-termini16. In contrast to HSM3, deletion of RPN14 or NAS6 had little effect on the level of BP1 (Fig. 2e), indicating that their roles in base assembly are distinct from the regulation of BP1.
The absence of Rpt4 and Rpt6 from BP1 is interesting because these are the Rpt proteins whose C-termini are most critical for base assembly. The accumulation of BP1 in rpt4-Δ1 and rpt6-Δ1 mutants likely reflects perturbation of an assembly event upstream of BP1 incorporation into nascent base. Also absent from BP1 were CP subunits (Supplementary Fig. 4); the failure to observe a stable BP1-CP complex suggests that such complexes are not involved in assembly. Rather, BP1 may load onto a separate, complementary assembly intermediate that includes Rpt4 and Rpt6 and is capable of direct interaction with CP.
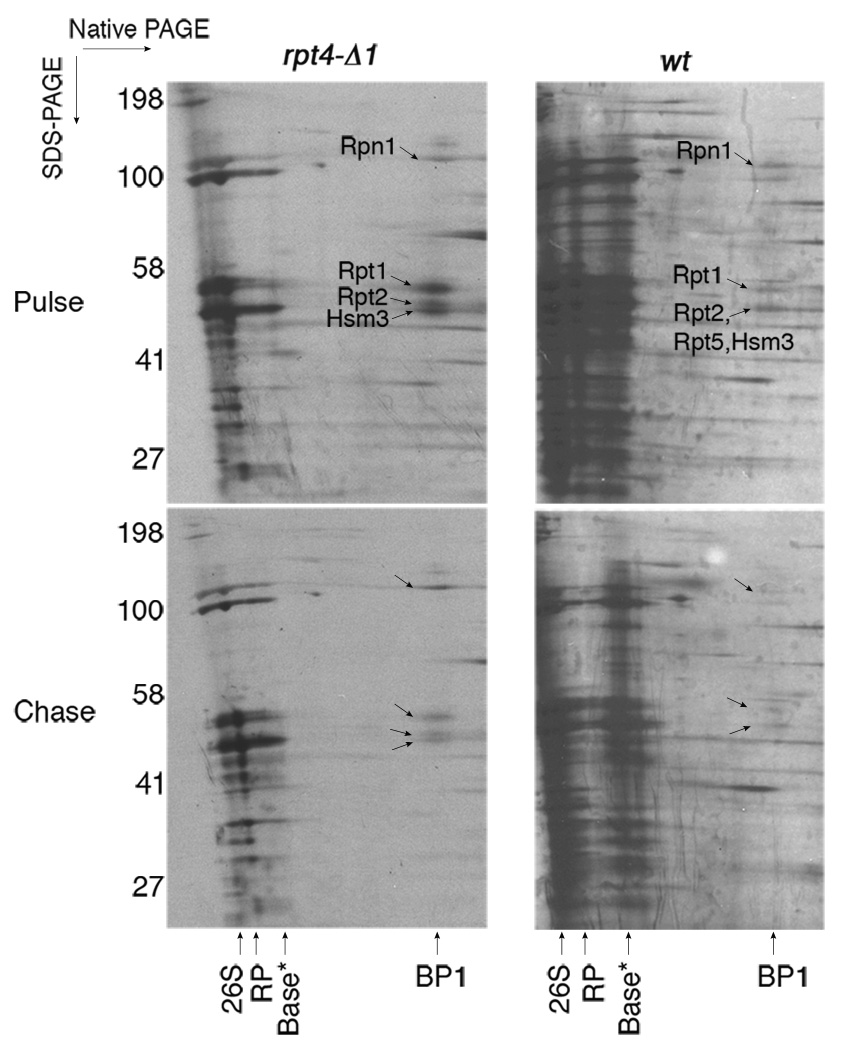
To verify that BP1 is a nascent complex utilized for proteasome assembly, we performed a pulse-chase experiment using in vivo S35 metabolic labeling (Fig. 3). BP1 was visualized by pulse-labeling, and after a 30-minute chase with excess unlabeled methionine, S35-labeled BP1 was significantly decreased (Fig. 3). Thus, BP1 is a transient complex in vivo. Consistent with steady-state experiments, S35-labeled BP1 was more prominent in rpt4-Δ1 than in wild-type, suggesting that its incorporation into nascent base depends on proper interaction between Rpt4 C-termini and CP.
Figure 3. Pulse-chase analysis of the BP1 base assembly intermediate.
Strains were metabolically labeled with S35-methionine in vivo for 4–5 min (pulse) and chased with excess methionine for 30 min (chase). Samples were then subjected to affinity-purification with ProA-TeV-Rpt1 and 2-D native/SDS-PAGE (5% and 4–12%, respectively) followed by autoradiography. BP1, but not Ubp6 + BP1, was labeled with S35-methionine during the pulse in both wild-type and the rpt4-Δ1 mutant.
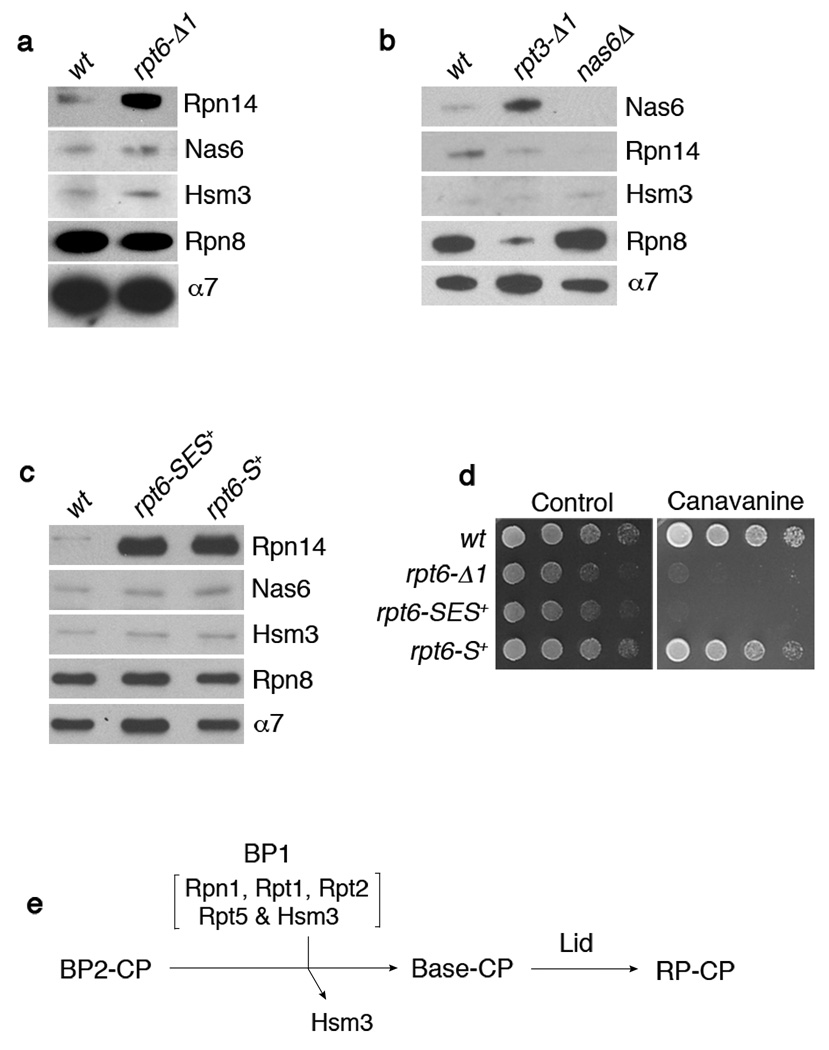
The interaction of CP with Rpt4 and Rpt6 is regulated by Rpn14, which specifically interacts with the C-domains of these two Rpts16. Rpn14 associates with RP or base, but little or no Rpn14 is found on CP-containing complexes such as proteasome holoenzyme16. Strikingly, Rpn14 was readily detected on Rpt6-Δ1 proteasome holoenzyme that had been affinity-purified via a CP tag (Fig. 4a and Supplementary Fig. 5). In contrast, Hsm3 and Nas6 did not associate with Rpt6-Δ1 proteasomes (Fig. 4a), showing that the effect of the rpt6-Δ1 mutation was local, and restricted to Rpt6’s binding partner. We conclude that Rpn14 is released as a specific consequence of the engagement of its cognate Rpt’s C-terminus with the CP. Proteasome holoenzyme or base-CP assembly per se is not sufficient for displacement. The RP-chaperone competes with CP for occupancy of the Rpt C-domain, providing a simple mechanism by which RP-chaperones can regulate base assembly and subsequently exit the complex16. Could the failure to release Rpn14 account for the phenotypic defects of rpt4-Δ1 and rpt6-Δ1 mutants? This is unlikely, because rpn14Δ is not a suppressor of either allele (data not shown).
Figure 4. Interactions between Rpt C-termini and the CP control RP-chaperone release.
a-c, SDS-PAGE and immunoblotting followed by affinity purification using a CP tag (Pre1-TeV-ProA). Rpn8 (lid subunit), and α7 (CP subunit) are loading controls. Nas6Δ proteasome (b) is a negative control. The rpt6-SES+ or rpt6-S+ mutants in (c) contain either a three (Ser-Glu-Ser) or one (Ser) residue insertion prior to the fourth residue from the C-terminus.
d, Phenotypic analysis of RPT6 insertion mutants. 4-fold serial dilutions were spotted on synthetic medium (control) or canavanine (1 µg/ml) plates. Cells were grown for 2–4 days at 30°C.
e, Model for late-stage base assembly. It is proposed that the base is formed by the addition of the BP1 complex, containing Rpn1, Rpt1, Rpt2, Rpt5, and Hsm3, to the putative BP2 complex. Initial contact with the CP is provided by BP2, whereas BP1 exists independently of the CP until it joins the BP2-CP complex. BP2 has not been identified and its exact composition is unknown. However, likely components of BP2 are Rpt4, Rpt6, Rpn2, and perhaps Rpt3. Rpn14 and Nas6 (not shown) may function prior to the formation of BP2-CP.
Analysis of Rpt3-Δ1 proteasomes also revealed a defect in chaperone expulsion during assembly. Rather than Rpn14, Rpt3-Δ1 proteasomes retained Nas6, the binding partner of Rpt316,18 (Fig. 4b). The rpt3-Δ1 mutation had no effect on the association of Hsm3 or Rpn14 with proteasomes. The association of Rpn14 or Nas6 with the holoenzyme was not due to an increase in their protein expression level in rpt6-Δ1 or rpt3-Δ1 mutants, respectively (data not shown).
Our structural modeling suggested that competition between CP and RP-chaperones for Rpt binding reflects the proximity of their binding sites16. The model predicts that separating the two binding sites would abrogate competition, and thus allow retention of RP-chaperones on the proteasome. Remarkably, insertion of a single residue into Rpt6 (rpt6-S+), four residues from the C-terminus, prevented expulsion of Rpn14 (Fig. 4c). Thus, the expulsion mechanism is very precise. Importantly, the failure to expel Rpn14 was not a result of failure to dock the mutant Rpt6 C-terminus at the CP, because rpt6-S+ mutants did not phenocopy rpt6-Δ1 mutants (Fig. 4d). The release of non-cognate RP-chaperones was unaffected by C-terminal insertions (Fig. 4c).
In bacterial AAA proteins, formation of the ATPase ring is thought to be spontaneous and unassisted19. In contrast, the proteasome’s ATPase ring is assembled through a programmatic and remarkably intricate pathway. This and the accompanying study16 suggest that the pathway of base assembly involves four distinct classes of factors, each found in multiple forms: the seven-membered subunit ring of the CP, the six ATPases of the Rpt ring, two scaffolding factors (Rpn1 and Rpn2), and three RP-chaperones.
The importance for assembly of the extreme C-termini of the Rpts indicates a prominent role of CP templating in Rpt ring formation. We propose that one subset of Rpts assembles directly on the CP, including Rpt4 and Rpt6, whose mutants confer the strongest assembly phenotypes. In contrast, another group of Rpts, including Rpt1, Rpt2, and Rpt5, is proposed to initially assemble free of the CP in the BP1 complex (Fig. 4e). Because BP1 disappears rapidly in pulse-chase experiments, it behaves as a true in vivo assembly intermediate. The interaction of the Rpt binding proteins with the α pockets of the CP appears to be negatively regulated by the RP-chaperones16. These chaperones may suppress α pocket-Rpt interactions during certain stages of assembly, and, during other stages, disengage from the Rpt to permit assembly to proceed forward (Fig. 4). In this way the RP-chaperones may temporally sequence the assembly process.
A simple though hypothetical scenario is that the base is formed by addition of BP1 to a preformed complex containing some or all of the base components absent from BP1. This proposed assembly intermediate is termed BP2 in our three-template model (Fig. 4e). Rpt4 and Rpt6 are proposed to mediate BP2 formation by CP templating. Slow maturation of BP2 may explain the accumulation of BP1 observed in rpt4-Δ1 and rpt6-Δ1 mutants, consistent with this model. We also propose that Rpn2 functions as a scaffold to organize BP2, based on evidence found in this and previous studies. For example, Rpt420 and Rpt621 have been shown to specifically interact with Rpn2 (whereas Rpt1 and Rpt2 interact with the Rpn122 scaffold of BP1). Moreover, Rpn2 binds CP directly23. Thus, a complex containing Rpn2, Rpt4, and Rpt6 is well suited to nucleate base assembly. Rpn2 and the CP may jointly template early ring assembly as the CP-Rpt binding events are most likely not strong enough to single-handedly specify Rpt ring assembly. A base subassembly containing Rpn1 and Rpn2 was isolated by in vitro dissociation of proteasomes23, but according to our findings it might not be formed during proteasome biogenesis. However, the Rpn1-Rpn2 interaction could be important for driving association of the BP1 complex with a complementary Rpn2-containing complex, such as the putative BP2.
Methods summary
Purification of BP1
Strains carrying ProA-TeV-Rpt1 were grown in 6 L of SD medium to OD600 0.8 to 1.2. Cells were harvested, washed once with ice-cold water and drop-frozen in liquid nitrogen. Frozen yeast samples were then ground using an MM301 grinding mill (Restch) under liquid nitrogen following manufacturer’s instructions, or using a mortar and a pestle as previously described24. Ground powder was hydrated in proteasome buffer (50 mM Tris-HCl [pH7.5], 5 mM MgCl2, 1 mM EDTA, and 10% glycerol) supplemented with 2 mM ATP, protease inhibitor tablets (Complete, Roche), 2 mM PMSF, 1 mM benzamidine, 10 µg/ml pepstatin A, and 1 µg/ml antipain. Cell extracts were cleared at 30,000 × g for 30 min at 4°C, and the supernatants were mixed with rabbit IgG resin (Cappel, MP Biomedicals) for 90 min at 4°C. Resins were collected at 900 × g for 2 min at 4°C and washed with proteasome buffer containing 50 mM NaCl three times, followed by a final wash with proteasome buffer alone. BP1 was then released from the resin by incubating with AcTEV protease (Invitrogen) at 2.5 unit/L culture in proteasome buffer containing 2 mM ATP and protease inhibitors for 1 hr at 30°C. Eluates were concentrated using Ultrafree-0.5 centrifugal filter device with 30 kDa NMWL (Millipore).
Pulse-chase analysis of BP1, native PAGE and immunoblot
Detailed methods and strain list are described in Supplementary information.
Supplementary Material
Acknowledgements
We thank J.D. Orth for discussion and technical assistance, and C. Hill for helpful discussions. We also thank J.W. Hanna, S. Elsasser and all members of Finley lab for comments on the manuscript. The work was supported by a grant from US National Institutes of Health (NIH) to D.F. (GM043601), an EMBO long-term fellowship to J.R. and a NIH NRSA postdoctoral fellowship (5F32GM75737-2) to S.P.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. doi: 10.1146/annurev.biochem.78.081507.101607. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol. Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 3.Rabl J, et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith DM, et al. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol. Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitby FG, et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 6.Gillette TG, Kumar B, Thompson D, Slaughter CA, Demartino GN. Differential roles of the C-termini of AAA subunits of PA700 (19S regulator) in asymmetric assembly and activation of the 26s proteasome. J. Biol. Chem. 2008;283:31823–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borissenko L, Groll M. 20S proteasome and its inhibitors: crystallographic knowledge for drug development. Chem. Rev. 2007;107:687–717. doi: 10.1021/cr0502504. [DOI] [PubMed] [Google Scholar]
- 8.Mannhaupt G, Schnall R, Karpov V, Vetter I, Feldmann H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999;450:27–34. doi: 10.1016/s0014-5793(99)00467-6. [DOI] [PubMed] [Google Scholar]
- 9.Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl Acad. Sci. USA. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isono E, et al. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol. Biol. Cell. 2007;18:569–580. doi: 10.1091/mbc.E06-07-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat. Struct. Mol. Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- 12.Le Tallec B, Barrault MB, Guerois R, Carre T, Peyroche A. Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol. Cell. 2009;33:389–399. doi: 10.1016/j.molcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Saeki Y, Toh-e A, Yokosawa H. Rapid isolation and characterization of the yeast proteasome regulatory complex. Biochem. Biophys. Res. Commun. 2000;273:509–515. doi: 10.1006/bbrc.2000.2980. [DOI] [PubMed] [Google Scholar]
- 14.Leggett DS, Glickman MH, Finley D. Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 2005;301:57–70. doi: 10.1385/1-59259-895-1:057. [DOI] [PubMed] [Google Scholar]
- 15.Glickman MH, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 16.Roelofs J, et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. doi: 10.1038/nature08063. (Submitted to Nature; Accompanying paper). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura Y, et al. Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26S proteasome. Biochem. Biophys. Res. Commun. 2007;359:503–509. doi: 10.1016/j.bbrc.2007.05.138. [DOI] [PubMed] [Google Scholar]
- 18.Dawson S, et al. Gankyrin is an ankyrin-repeat oncoprotein that interacts with CDK4 kinase and the S6 ATPase of the 26 S proteasome. J. Biol. Chem. 2002;277:10893–10902. doi: 10.1074/jbc.M107313200. [DOI] [PubMed] [Google Scholar]
- 19.Kress W, Mutschler H, Weber-Ban E. Assembly pathway of an AAA+ protein: tracking ClpA and ClpAP complex formation in real time. Biochemistry. 2007;46:6183–6193. doi: 10.1021/bi602616t. [DOI] [PubMed] [Google Scholar]
- 20.Richmond C, Gorbea C, Rechsteiner M. Specific interactions between ATPase subunits of the 26 S protease. J. Biol. Chem. 1997;272:13403–13411. doi: 10.1074/jbc.272.20.13403. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann-Petersen R, Tanaka K, Hendil KB. Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch. Biochem. Biophys. 2001;386:89–94. doi: 10.1006/abbi.2000.2178. [DOI] [PubMed] [Google Scholar]
- 22.Gorbea C, Taillandier D, Rechsteiner M. Mapping subunit contacts in the regulatory complex of the 26 S proteasome. S2 and S5b form a tetramer with ATPase subunits S4 and S7. J. Biol. Chem. 2000;275:875–882. doi: 10.1074/jbc.275.2.875. [DOI] [PubMed] [Google Scholar]
- 23.Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. The central unit within the 19S regulatory particle of the proteasome. Nat. Struct. Mol. Biol. 2008;15:573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verma R, et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leggett DS, et al. Multiple associated proteins regulate proteasome structure and function. Mol. Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.