Abstract
HDAC4 is a Class II histone deacetylase (HDAC) that is highly expressed in the brain but whose functional significance in the brain is not known. We show that forced expression of HDAC4 in cerebellar granule neurons protects them against low potassium-induced apoptosis. HDAC4 also partially protects cultured cortical neurons from 6-hydroxydopamine-induced neurotoxicity and HT22 neuroblastoma cells from death induced by oxidative stress. HDAC4-mediated neuroprotection does not require its HDAC catalytic domain and cannot be inhibited by chemical inhibitors of HDACs. Neuroprotection by HDAC4 also does not require the Raf-MEK-ERK or the PI-3 kinase - Akt signaling pathways, and occurs despite the activation of c-jun, an event that is generally believed to condemn neurons to die. The protective action of HDAC4 occurs in the nucleus and is mediated by a region that contains the nuclear localization signal. HDAC4 inhibits the activity of cyclin-dependent kinase-1 (CDK1) and the progression of proliferating HEK293T and HT22 cells through the cell cycle. Mice lacking HDAC4 have elevated CDK1 activity and display cerebellar abnormalities including a progressive loss of Purkinje neurons postnatally in posterior lobes. Surviving Purkinje neurons in these lobes have duplicated soma. Furthermore, large numbers of cells within these affected lobes incorporate BrdU, indicating cell cycle progression. These abnormalities along with the ability of HDAC4 to inhibit CDK1 and cell cycle progression in cultured cells suggest that neuroprotection by HDAC4 is mediated by preventing abortive cell cycle progression.
Keywords: HDAC4, apoptosis, neuronal survival, histone deacetylase, cell cycle
Histone deacetylases (HDACs) are the catalytic subunits of multiprotein complexes that deacetylate specific lysines in the tail residues of histones, resulting in the compactation of chromatin into a transcriptionally repressed state (for review, see de Ruijter et al., 2003; Verdin et al., 2003; Yang and Gregoire, 2005). Although best studied for their effects on histones and transcriptional activity, it is now known that HDACs regulate the acetylation status of a number of other non-histone proteins suggesting complex functions of HDACs (for review, see de Ruijter et al., 2003; Verdin et al., 2003). In fact, HDACs have been shown to participate, or have been implicated in a variety of cellular functions including cell transformation, proliferation, senescence, differentiation, survival, and death (for review, see Verdin et al., 2003; Yang and Gregoire, 2005).
Vertebrates express at least 18 distinct HDACs which have been grouped into four classes based on their similarity with yeast HDACs (for review, see de Ruijter et al., 2003; Verdin et al., 2003; Yang and Gregoire, 2005). Class I HDACs (HDACs 1, 2, 3 and 8) are composed primarily of a catalytic domain. These HDACs are ubiquitously expressed, localized to the nucleus, and serve as transcriptional repressors. Class II HDACs (HDAC 4, 5, 6, 7, 9 and 10) are larger proteins with an extended N-terminus extension involved in protein-protein interaction. These HDACs are further sub-classified into Class IIa (HDACs 4, 5, 7, and 9) and IIb (HDACs 6 and 10). In contrast to Class I HDACs, Class II HDACs display stage and tissue-specific expression. These HDACs are able to shuttle in and out of the nucleus in response to certain cellular signals. Phosphorylation of these HDACs at two N-terminal serine residues by the calcium-calmodulin-dependent kinase (CaMK) creates a docking site for 14-3-3 proteins which results in their export out of the nucleus (Lu et al., 2000; McKinsey et al., 2000; for review, see de Ruijter et al., 2003; Verdin et al., 2003; Chang et al., 2005). HDAC11 is the sole member of the Class III HDAC family. The fourth class of deacetylases, called sirtuins (SIRT1 - 7), have catalytic domains similar to the yeast NAD+-dependent deacetylase Sir2. While serving as HDACs in yeast, in mammalian cells SIRTs are involved in the deacetylation of proteins other than histones, and are therefore not considered to be “classical HDACs”.
Class IIa HDACs are abundantly expressed in the brain although their significance to brain function is just beginning to be explored. A recent study found that the downregulation of HDAC5 expression is an important aspect in the efficacy of certain antidepressants (Tsankova et al., 2006). These investigators found that overexpression of HDAC5 inhibited the effectiveness of antidepressants in a rodent model of depression (Tsankova et al., 2006). Overexpression of HDAC5 in cultured neurons induces apoptosis (Linseman et al., 2003).. In contrast to the pro-apoptotic action of HDAC5, a spliced variant of HDAC9 called HDAC-related protein (HDRP) promotes the survival of neurons (Morrison et al., 2006). Death of cerebellar granule neurons (CGNs) induced by low potassium (LK) treatment is preceded by a downregulation of HDRP (Morrison et al., 2006). Overexpression of HDRP protects against LK-induced neuronal apoptosis whereas forced downregulation of HDRP expression induces death in otherwise healthy neurons (Morrison et al., 2006). In this study we have focused on HDAC4, another Class IIa HDAC protein which is highly expressed in brain, heart and skeletal muscle (Grozinger et al., 1999; Wang et al., 1999). We report that HDAC4 is a neuroprotective protein. Although reminiscent of HDRP in this regard, we demonstrate that the mechanism by which HDAC4 mediates it neuroprotective effect is distinct from that utilized by HDRP. While HDRP protects neurons by inhibiting c-jun via direct interaction with c-jun N-terminal kinase (JNK), HDAC4 represses CDK1 activity and suppresses cell cycle progression.
MATERIALS AND METHODS
Materials
All cell culture media and reagents were from Invitrogen (Carlsbad, CA). Unless otherwise noted, all chemicals were purchased from Sigma (St. Louis, MO). Antibodies against HDAC4, HDRP, Flag (mouse origin) and 5-bromo-2-deoxyuridine (BrdU) were from Sigma (St. Louis, MO) (BrdU antibody purchased from sigma was used for immunocytochemistry experiments). The following antibodies were purchased from Santa Cruz Biotechonology (Santa Cruz, CA): Actin, HA, GFP, α-tubulin, c-Jun, Flag (rabbit origin), CDK5 and CDK1. Antibodies against p-Akt, Akt, p-ERK, ERK, p-MEK, MEK, p-GSK3 and p-JNK were purchased from Cell Signaling Technology (Beverly, MA). MEF2D antibody was from BD Transduction (San Jose, CA). Primary antibodies used on brain sections include Ki-67 (Lab Vision, Fremont, CA), Calbindin (Sigma, St. Louis, MO) and BrdU (Vision BioSystems Inc., Norwell, MA). ABC kit (Vector Inc, Burlingame, CA) and Texas Red conjugated secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA) were used for immunohistochemical experiments followed by either diaminobenzidine (DAB) (Vector Inc, Burlingame, CA) or 4′,6′-diamidino-2-phenylindole hydrochloride (DAPI) staining. All secondary antibodies for western blotting were from Santa Cruz Biotechnology (Santa Cruz, CA) and for immunocytochemistry were from Jackson Immunoresearch Laboratories (West Grove, PA). pTarget mammalian expression vector and restriction enzymes were purchased from Promega (Madison, WI). Primers were ordered through Sigma-Genosys (St. Louis, MO).
Plasmids and adenoviruses
A recombinant adenovirus expressing HDAC4 (Flag tag) was cloned in Eric N. Olson’s laboratory (University of Texas Southwestern Medical Center, Dallas, Texas). GFP Adenoviral vector was a kind gift from Kim A. Heidenreich (University of Colorado Health Sciences Center, Denver, CO). HDAC4 and its truncation mutants were Flag tagged by PCR and were cloned in pTarget vector (Promega, Madison, WI). CDK1-HA plasmid was purchased from Addgene, Inc. (Cambridge, MA).
Cell lines
Human embryonic kidney cells (HEK293T clone 17cells, referred in the manuscript as HEK293T) were purchased from ATCC (Manassas, VA) and cultured in DMEM (supplemented with L-glutamine, 110 mg/L sodium pyruvate and 4.5 g/L D-Glucose) plus 10% fetal bovine serum (FBS). Mouse neuroblastoma HT22 cells were kindly provided by Rajiv Ratan (Burke Medical Research Institute, New York, NY) and were cultured in DMEM (supplemented with L-glutamine and 4.5 g/L D-Glucose) plus 10% FBS.
Cerebellar granule neuron cell culture, transfection and treatments
CGNs were cultured from dissociated cerebella of 7-8 day old Wistar rats or C57BL/6 mice as previously described (D’Mello et al., 1993). In short, processed CGNs were plated in basal Eagle’s medium with Earle’s salts (BME) supplemented with 10% FBS, 25 mM KCl, 2 mM glutamine (Invitrogen, Carlsbad, CA), and 100 ug /ml gentamicin. Cells were plated on dishes coated with poly-L-lysine at a density of 106 cells/well (24-well dishes), 5×106cells/35 mm dishes and 107 cells/60 mm dishes. 10 uM of Cytosine arabinofuranoside (Ara-C) was added to the culture medium 18 to 22 hrs after plating to prevent proliferation of non-neuronal cells. Cells were then maintained for 4 to 5 days prior to transient transfection experiments. Transfection of CGNs was done as previously described (Koulich et al., 2001). Briefly, 1ug/well of DNA was precipitated by calcium phosphate method for 30 min at room temperature. Cell culture media of neurons were changed to DMEM without L-glutamine after one wash. Precipitated DNA was added to the cell culture media for 1 hr. Cells were then washed twice with DMEM without L-glutamine media and placed in their original medial. A day after transfection, the cells were rinsed once and then maintained in serum-free BME medium containing either 5 mM KCl (low potassium, LK) or 25 mM KCl (high potassium, HK).
Pharmacological inhibitors were added in LK condition at the time of treating. Times of treatments are described in the figure legends. Transfected neurons were detected by GFP fluorescence or by immuno staining using Flag antibody as described previously (Koulich et al., 2001). Neuronal viability was quantified by staining cell nuclei with DAPI or Syto 13 as previously described (Koulich et al., 2001; Yalcin et al., 2003). If the nucleus of a cell was condensed or fragmented, the cell was counted as dead and if the nucleus was rounded with a diffuse fluorescence color, it was counted as living. For immunocytochemistry, antibodies purchased from Sigma (St. Louis, MO) were used at a 1:200 dilution and antibodies from Santa Cruz Biotechnology (Santa Cruz, CA) at a 1:50 dilution.
Cortical neuron cell culture
Cortical cultures were obtained from the cerebral cortex of Wistar rats (day 19 of gestation) as described previously (Murphy et al., 1990) and were plated in dishes covered with poly-L-lysine at the density of 7 × 105 cells/well (24 well plates) in MEM plus GlutaMAX-1 media supplemented with 10% FBS. 24 hrs after plating 20 uM 5-fluoro-2′-deoxyuridine and 20 uM uridine was added to prevent proliferation of non-neuronal cells. Infection experiments were initiated one day after plating. 6-hydroxy dopamine (6-OHDA) was used to induce apoptosis 24 hrs after infection.
Adenovirus-mediated overexpression
For CGNs, infection with HDAC4 and GFP adenoviral vectors was performed at a multiplication of infection (MOI) of 50, 4 - 5 days after plating. The original culture medium was removed, saved, and replaced with serum-free HK medium containing the adenovirus. After 1-1.5 hrs the infection media was removed and the original serum-containing medium was added back. Cortical neurons were infected similarly and at the same MOI except that infection was performed on 1-day old cultures in serum-free medium. Treatments were performed 24 hrs after infection. Times of treatments are described in the figure legends. The cultures were then either subjected to western blotting or were fixed and subjected to immunocytochemistry as described previously (Koulich et al., 2001). Viability of cells positive for GFP or Flag was assessed by Syto 13 or DAPI staining. Nuclei that were condensed or fragmented were scored as dead and rounded nuclei with a diffuse florescence color were counted as living. For immunocytochemistry, antibodies purchased from Sigma (St. Louis, MO) were used at a 1:200 dilution and antiobodies from Santa Cruz Biotechnology (Santa Cruz, CA) at a 1:50 dilution.
Infection of HT22 cells was performed as follows. The cells were split at the density of 1 × 104/well (24 well dishes). The day after splitting, the cells were infected in serum free media for 2 hrs. Fresh serum was added and infection was allowed to persist for 24 hrs before treatments with Homocysteic acid (HCA).
Analysis of RNA expression by RT-PCR
RNAs were extracted from cells using Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. 5 ug of RNA was used for cDNA synthesis using the Thermoscript reverse transcriptase PCR system (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. PCR was performed using PCR master mix (Promega, Madison, WI). The primers used for PCR amplification were as follows:
actin forward: 5′-AGGACTCCTATGTGGGTGACGA-3′,
actin reverse: 5′-CGTTGCCAATAGTGATGACCTG-3′,
HDAC4 forward: 5′- ACAGTTCCCTTGACCAGAGTTC-3′
HDAC4 reverse: 5′- GTCTGCACCAACCAAGGACT-3′
HDRP forward: 5′- AACTTGAAGGTGCGGTCCA-3′
HDRP reverse: 5′- TTACAAATCCTGGAGCTAAAT-3′
GAPDH forward: 5′- -CCATCACCATCTTCCAGGAG-3′
GAPDH reverse: 5′- CCTGCTCACCACCTTCTTG-3′
MnSOD forward:
MnSOD reverse:
BCL-XL forward:
BCL-XL reverse:
BCL2A1 forward:
BCL2A1 reverse:
BAD forward:
BAD reverse:
BCL-XL forward:
BCL-XL reverse:
Western blotting
Upon treatments, the culture medium was removed, and the cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed in lysis buffer (Cell Signaling Technology, Beverly, MA) with an addition of one protease inhibitor cocktail tablet (Roche, Basel, Switzerland) in 10 ml of lysis buffer. The lysates were centrifuged at 12000g for 10 min at 4° C. Supernatant were taken and protein concentrations were normalized using the Bradford protein assay reagent (Bio-Rad, Hercules, CA). 40-80 ug of protein was subjected to Western blotting as previously described (Koulich et al., 2001; Zhao et al., 2002). Immunoreactivity was examined using enhanced chemiluminescence (Amersham Bioscience, Piscataway, NJ). MEF2D primary antibody and Flag were used at a 1:2500 dilutions. Other primary antibodies were used at a 1:1,000 dilution followed by secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) at a 1:10,000 dilution.
In vitro kinase assay
CDK1 kinase activity of transfected HEK293T cells was performed 24 hrs after transient transfection and CDK1 or CDK5 kinase activity of whole brain lysates were performed on mice lacking HDAC4 and wild-type littermates at P1. Briefly, samples were lysed in lysis buffer (Cell Signaling Technology, Beverly, MA) containing protease inhibitor cocktail tablet (Roche, Basel, Switzerland). Protein concentration was measured using the Bradford protein assay reagent (Bio-Rad, Hercules, CA). 1ug of CDK1 antibody was added to 500 ug of protein followed by agitation overnight at 4° C. The next day, 30 ul of protein A/G plus agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) was added and the tubes were rocked at 4° C for an additional 1.5 hr. The bead/immune complexes were pelleted and washed three times with lysis buffer without a protease inhibitor cocktail tablet, followed by three washes with kinase buffer (Cell Signaling Technology, Beverly, MA). The immune complex was then resuspended in 30 ul of kinase buffer, 1 mM cold ATP, 1 uCi [γ-32P] ATP (MP Biomedicals, Solon, OH) and 5 ug of histone H1 (Sigma, St Louis, MO) and incubated for 30 min at 30° C. The reaction was stopped by addition of 2X SDS buffer (187.5 mM Tris-HCl, pH 6.8 at 25° C, 6% SDS, 30% glycerol, 150 mM DTT, 0.03% bromphenol blue). The samples were ran on a 10% SDS gel and transferred to a polyvinylidene difluoride membrane (Bio-Rad Laboratories, Hercules, CA). Following Ponceau S staining, membranes were visualized by autoradiography on a Storm 840 (Amersham Biosciences, Pittsburgh, PA). The membrane was then blocked using 5% non-fat dry milk with 1% Tween 20 and probed with CDK1 or CDK5 antibodies to ensure equal protein pull down.
Cell proliferation assay
HEK293T cells were plated in 35 mm dishes in DMEM media supplemented with 10% FBS. The next day, cells were transfected using Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA) according to manufacture’s protocol. Four hours after transfection, media was changed to DMEM plus 2% serum. Cells were counted using a hematocytometer 5 days later.
BrdU incorporation assay of cell lines
HEK293T or HT22 cells were split in 24 well dishes one day prior to transfection experiment. Cells were then transfected using calcium phosphate procedure. 1 ug/well of DNA was precipitated by calcium phosphate method for 30 min at room temperature. Cell culture media of HEK293T was changed to DMEM plus 2% serum and that of HT22 to DMEM plus 10% serum followed by drop wise addition of precipitated DNA to culture media. After 22 hrs, 20 uM of BrdU reagent was added to the cultures. After 2 hrs of BrdU incorporation, cells were fixed and stained for BrdU followed by immunocytochemistry to detect the exogenous proteins. BrdU staining protocol was as follows. Cells were fixed in 4% paraformaldehyde for 30 min followed by 3, 5 min washes in PBS containing 0.5% Triton on ice. Cells were then incubated in 1 N HCl for 10 min on ice to break open the DNA structure of labeled cells. This was followed by 30 min incubation at 37°C in presence of 2 N HCl. Immediately after the acid washed, Borate buffer (0.1 M, pH 8.5) was added for 12 min at room temperature. Three 5 min washes of PBS were followed at room temperature. Cells were then blocked in diluent (1% BSA, 0.5% Tween 20, 0.1% Sodium Azide; in PBS) plus 5% goat serum for 15 min at 37°C. BrdU primary antibody dilution of 1:200 along with antibody against protein of interest were made in diluent and used overnight at room temperature. The remaining steps were done as in a regular immunocytochemistry (Koulich et al., 2001). For immunocytochemistry, antibodies purchased from Sigma (St. Louis, MO) were used at a 1:200 dilution and antiobodies from Santa Cruz Biotechnology (Santa Cruz, CA) at a 1:50 dilution.
Histology and immunohistochemistry
The HDAC4-/- mice was obtained from Eric N. Olson’s laboratory (The University of Texas Southwestern Health Science Center, Dallas, Texas). Whole brains from HDAC4-/- and wild-type littermate mice were examined at the following ages: Postnatal day 1 (P1), P3, P7, P9 and P11. The sectioning procedures were performed as previously described (Mitchell et al., 2001). Briefly, brains were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer. Samples were allowed to sink in 30% sucrose solution in PBS overnight following fixation. Whole brains were then frozen in cryopreservative solution (Cryo-STAT™, StatLab Medial Products, Inc, Lewisville, TX) on dry ice. Frozen brains were then sectioned at 20 μm on a cryostat in either the coronal or sagittal planes. Sections were either stained with DAPI or cresyl violet for structural studies or subjected to immunohistochemistry. Primary antibodies used on sections were as follows: Ki-67 (1:200), Calbindin (1:1000) or BrdU (1:200). Sections were incubated with primary antibody at 4°C overnight followed by secondary antibody incubation as in manufacture’s protocol (ABC kit, vector Inc, Burlingame, CA). This was followed by DAB (Vector Inc, Burlingame, CA) staining of the sections. For fluorescence imaging, Texas Red secondary antibody (Jackson Immunoresearch Laboratories, West Grove, PA) (1:200) was used for 2 hrs at room temperature and DAPI was used to stain cell nuclei.
BrdU injections and staining
7 days old HDAC4-/- and wild-type littermates were injected with 200 mg/kg of BrdU dissolved in PBS twice a day for 2 days. Brains were harvested on day 9. Fixation and section preparation were performed as described in the histology section. BrdU detection protocol was as follows. Sections were incubated at 65° C for 10 min and then cooled to room temperature and incubated in 0.1 mg/ml pepsin in 0.1 N HCl. After being rinsed in PBS, sections were incubated in 2 N HCl for 10 min at 37° C. Washes with PBS were followed and then the sections were blocked in 5% horse serum, 1% BSA, 0.5% Tween 20 and 0.1% Sodium Azide; in PBS. BrdU primary antibody (Vision BioSystems Inc., Norwell, MA) incubation was followed for 1 hr in PBS plus 1% horse serum and 0.2% Triton X-100. Secondary antibody incubation was followed as in manufacture’s protocol (ABC kit, vector Inc, Burlingame, CA). This was followed by DAB (Vector Inc, Burlingame, CA) staining of the sections.
Statistical analysis
All graphs with standard deviations present mean values for data obtained from at least three independent experiments. Statistical significance for all data sets was determined by using the two tailed unpaired t- test as is shown by an asterisk.
RESULTS
HDAC4 protects against neuronal apoptosis
We recently reported that HDRP, a splice variant of HDAC9 lacking the entire C-terminus deacetylase domain, is a neuroprotective protein that can completely prevent LK-induced death of CGNs when overexpressed (Morrison et al., 2006). In contrast, HDAC5, which shares structural similarity with HDAC9, has pro-apoptotic activity in neurons (Linseman et al. 2003b). In this study we investigated whether a third member of this subclass of HDAC proteins, HDAC4, was capable of regulating neuronal apoptosis. As shown in Fig. 1, HDAC4 expression is not downregulated in CGNs primed to die by LK treatment. This is in contrast to HDRP the expression of which is reduced under these conditions (Fig. 1) (Morrison et al., 2006). However, the overexpression of HDAC4 in CGNs efficiently prevents apoptosis caused by LK treatment (Fig. 2B and supplemental Fig. 1). HDAC4 also partially protects cortical neurons against 6-OHDA-induced neurotoxicity (supplemental Fig. 2) and HT22 cells against HCA-induced oxidative stress (Fig. 2C). HDAC4 normally localizes to the cytoplasm in neurons but is capable of translocating to the nucleus in response to specific stimuli (Chawla et al., 2003; Bolger and Yao, 2005). Consistent with these previous findings, HDAC4 is present in the cytoplasm of CGNs maintained in HK medium but translocates to the nucleus when the cultures are switched to LK medium (Fig. 2A and supplemental Fig. 1). As observed for other Class II HDAC proteins such as HDAC5 and HDRP (Potter et al., 2001; Zhang et al., 2001), overexpressed HDAC4 displays a punctate staining pattern in the nucleus. Taken together, neurons expressing HDAC4 are completely protected against LK induced apoptosis even after extended treatments of cells in LK condition (72 hrs) (Fig. 2B and supplemental Figure 1).
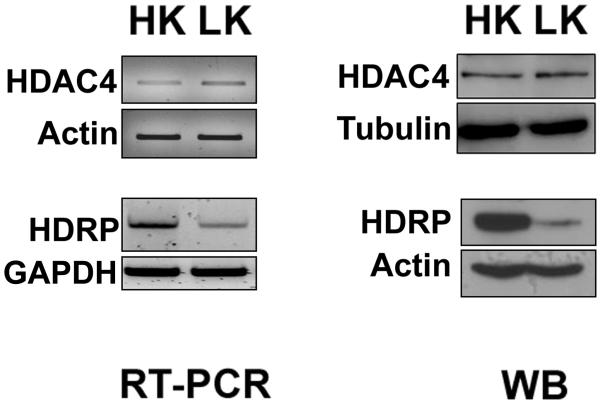
Figure 1. Expression of HDAC4 and HDRP in neurons.
6-7 days old CGNs were treated with HK and LK for 6 hrs. RT-PCR panel; RNA was extracted from cells followed by cDNA synthesis. PCR was performed using primers specific to HDAC4, actin, HDRP and glyceraldehyde-3- phosphate dehydrogenase (GAPDH). Actin and GAPDH were used to demonstrate that similar quantities of cDNA were used. Western blot (WB) panel; cells were lysed and subjected to western blotting using HDAC4 and HDRP antibodies. The same membranes were reprobed with α-tubulin and actin antibodies respectively to demonstrate equal loading.
Figure 2. Effect of HDAC4 overexpression on neuronal apoptosis.
(A and B) 5 day old culture of CGNs were infected with Ad-GFP or Ad-HDAC4 (Flag tag) and switched to HK or LK medium for 24 hrs. Infected neurons were identified by GFP fluorescence or Flag immunoreactivity and the status of the nuclei were assessed by Syto 13 staining. (A) Subcellular localization of HDAC4 in HK and LK medium. HDAC4 (red) is primarily in the cytoplasm in HK but translocates to the nucleus (green) in LK. Panel B quantifies survival of HDAC4 and GFP infected cells in HK and LK medium as percent of control (viability of GFP-infected neurons in HK). P < 0.006 for comparison with HK-GFP. (C) HT22 cells were infected with HDAC4 and GFP adenovirus and then treated with 2 mM HCA. Because most cells in the culture were infected viability was measured using the MTT assay. Cultures overexpressing HDAC4 proliferate slower than control cultures precluding a direct comparison with GFP infected cultures. Hence, viability (after 24 hrs of treatment) in the presence of HCA is compared with the viability of cultures infected with the same construct (either GFP or HDAC4) in the absence of HCA. P < 0.02 for comparison with control--GFP.
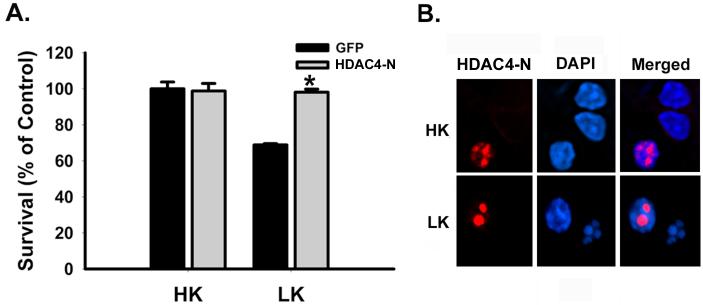
To examine whether deacetylase activity of HDAC4 was necessary for neuroprotection, a truncated form of the protein (spanning amino acids 1-653) lacking the catalytic domain was expressed in CGNs. This protein, HDAC4-N, was as effective as the full form of HDAC4 demonstrating that the deacetylase activity of HDAC4 is not required for neuroprotection (Fig. 3A). Since HDAC4-N also lacks the nuclear export signal situated in the C-terminus of HDAC4, it is trapped within the nucleus in HK or LK medium (Fig. 3B). Thus the protective action of HDAC4 is mediated within the nucleus. To map the region within HDAC4 that mediates protection, we generated and expressed additional deletion mutants. Two such deletion mutants spanning regions 1 - 288 and 1 - 435 of HDAC4 were both capable of preventing LK-induced neuronal death whereas a construct spanning amino acids 1 - 215 failed to protect (Fig. 4A). HDAC4435 resides in the nucleus, however, HDAC4215 lacks the NLS and is therefore localized to the cytoplasm (Fig. 4B). It was unclear whether HDAC4215 construct lacked the neuroprotective domain or whether its failure to inhibit cell death was due to its inability to localize to the nucleus. Another construct spanning the region between 216 - 653 was capable of neuroprotection, indicating that the neuroprotection domain was not located within the first 215 amino acids of HDAC4. Taken together, our analyses maps the neuroprotective domain to the 72 amino acid segment spanning residues 216 - 288 and containing the nuclear localization signal (NLS).
Figure 3. Effect of overexpression of HDAC4 lacking the deacetylase domain (HDAC4-N) on neuronal apoptosis.
(A and B) 5 day old cultures of CGNs were transfected with HDAC4 lacking the deacetylase domain (HDAC4-N, Flag tag) or GFP plasmids and switched to HK or LK medium for 24 hrs. Transfected neurons were identified by GFP fluorescence or Flag immunoreactivity and the status of the nuclei assessed by DAPI staining. Panel A quantifies survival of HDAC4-N and GFP transfected cells in HK and LK medium as percent of control (viability of GFP-transfected neurons in HK). P < 0.0003 for comparison with HK-GFP. (B) Subcellular localization of HDAC4-N in HK and LK medium. HDAC4-N (red) is trapped in the nucleus (blue) in both HK and LK conditions.
Figure 4. Analysis of neuroprotection by HDAC4 truncation mutants.
(A and B) 5 day old cultures of CGNs were transfected with HDAC4 truncation mutants or GFP plasmids and switched to HK or LK medium for 24 hrs. Transfected neurons were identified by GFP fluorescence or Flag immunoreactivity and the status of the nuclei assessed by DAPI staining. Panel A demonstrates survival of cells overexpressing HDAC4 truncation mutants and GFP in LK medium as percent of control (Survival of GFP-transfected neurons in HK were set to 100%). The asterisks correspond to constructs that demonstrate statistically significant protection in LK when compared with control (survival of GFP transfected neurons in LK) (P<0.05). (B) Subcellular localization of HDAC4435 and HDAC4215 in HK and LK medium. Shown in red is Texas Red conjugated secondary antibody that binds the Flag tag present on the c-terminus of HDAC4435 and HDAC4215. Cell nuclei are stained using DAPI (blue).
Effect of HDAC4 on signaling molecules regulating neuronal survival
Two of the best studied survival-promoting signaling pathways are the PI-3 kinase - Akt pathway and the Raf - MEK - ERK pathway (for review, see D’Mello and Chin, 2005). As shown in Fig. 5A, there is no difference in the profile of MEK or ERK phosphorylation in LK-treated neurons overexpressing HDAC4 compared with those overexpressing GFP. Similarly, Akt phosphorylation in LK-treated neurons occurs to a similar extent in neurons overexpressing HDAC4 or GFP (Fig. 5A). These results suggest that neuroprotection by HDAC4 is independent of both the MEK-ERK and the PI-3K-Akt signaling pathways. This conclusion is supported by pharmacological studies, the results of which are shown in Fig. 5B. Blocking Raf-MEK-ERK signaling using the highly selective MEK inhibitor, U0126, and PI-3k-Akt signaling using the PI-3 kinase inhibitors, LY294002, or the Akt inhibitor, ML-9, did not reduce the ability of HDAC4 to neuroprotect (Fig. 5B).
Figure 5. Effect of HDAD4 overexpression on key players of Raf-MEK-ERK and PI3K-Akt signal transduction pathways.
(A) 5 days old CGNs were infected with Ad-HDAC4, Ad-GFP or left uninfected. 24 hrs after infection media was switched to HK or LK medium. Cell lysates were prepared 3 hrs after the treatments and subjected to Western blotting using antibodies against HDAC4, c-jun, p-JNK, pMEK, total MEK, pERK, total EREK, pGSK3β, MEF2D, pAkt, total Akt and α-tubulin (as loading control). “Input” shows an abundant protein in the Ponceau-S stained membrane. (B) 5 days old CGNs were transfected with HDAC4-N (Flag tag) or GFP plasmids 24 hrs before treatments with HK, LK or LK plus inhibitors. After 24 hrs of treatment, cells were subjected to immunocytochemistry using Flag antibody followed by DAPI staining. Following concentrations were used for each inhibitor: Gleevec (50 uM); LY294002 (10 uM); TSA (1 uM); U0126 (10 uM); KN62 (50 uM) and ML9 (20 uM). The graph demonstrates cell survival as percent of control (viability of GFP-transfected neurons in HK).
Moreover, HDAC4-mediated neuroprotection is also not affected by KN-62, a highly selective inhibitor of calcium calmodulin-dependent kinase II (CamKII). Phosphorylation by CamKII regulates the export of Class II HDACs such as HDAC4 out of the nucleus and the inhibition of this kinase traps HDACs within the nucleus (McKinsey et al., 2000; Morrison et al., 2006).
Activation of GSK3β is a common feature of neuronal apoptosis and can be detected by dephosphorylation of this kinase at an inhibitory site, Ser-9 (Chin et al., 2005; Kaytor et al., 2002). While GSK3β is activated in GFP-expressing neurons switched to LK, this activation is not affected by HDAC4 (Fig. 5A). In LK-treated CGNs and in some other paradigms of neuronal apoptosis, cell death is accompanied by the phosphorylation of MEF2D and the inhibition of MEF2D phosphorylation prevents neuronal death (Okamoto et al., 2000; Butts et al., 2003; Linseman et al., 2003). While increased phosphorylation of MEF2D is observed following LK treatment, this is also not affected by HDAC4 expression (Fig. 5A).
HDRP-mediated neuroprotection involves the downregulation of c-jun expression as well as its phosphorylation by JNK (Morrison et al., 2006). C-jun phosphorylation is inhibited by the direct interaction of HDRP with JNK. As shown in Fig. 5A, HDAC4 overexpression has no effect on JNK phosphorylation (Fig. 5A). The inhibition of c-jun expression by HDRP is mediated via its recruitment of HDAC1 to the c-jun promoter resulting in the inhibition of histone deacetylation and hence transcriptional repression of the c-jun gene (Morrison et al., 2006). Treatment with HDAC inhibitors blocks histone deacetylation at the c-jun promoter and the ability of HDRP to neuroprotect (Morrison et al., 2006). HDAC4 does not interact directly with HDAC1 (data not shown) and pharmacological inhibition of HDACs using Trichostatin A (TSA) does not reduce protection by HDAC4 (Fig. 5B). Finally, treatment with Gleevec, a pharmacological inhibitor of c-Abl that inhibits HDRP-mediated protection, has no effect on the protective action of HDAC4 (Fig. 5B). Taken together, our results demonstrate that the mechanism by which HDAC4 confers neuroprotection is fundamentally different from that of HDRP. Arguing against the possibility that inefficient infection underlies the lack of responsiveness of these molecules to HDAC4 overexpression is our finding that the expression of several other genes encoding apoptosis-regulatory molecules is altered by HDAC4 (supplemental Figure 3). More studies need to be done to determine the extent of the relevance of this alteration in gene expression in regulation of HDAC4 mediated neuroprotection.
Mice lacking HDAC4 display brain abnormalities
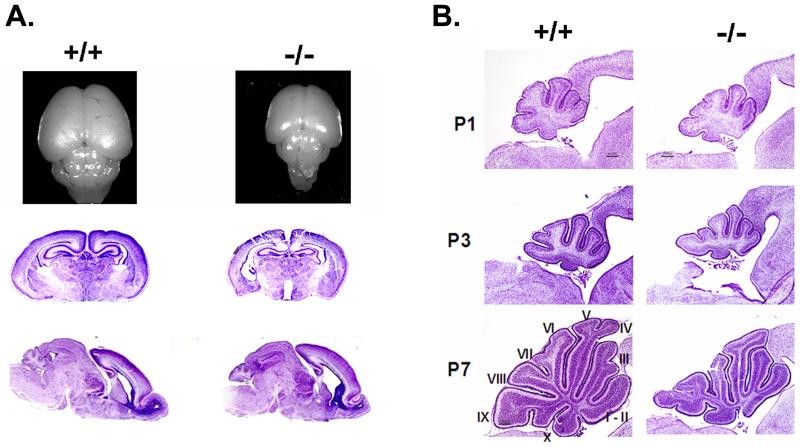
As reported previously, HDAC4-deficient mice are severely runted, display skeletal abnormalities, and die within 2 weeks of birth (Vega et al., 2004). The mutant brain is ∼40% smaller than wild-type consistent with the smaller body size (Fig. 6A, top panel). Relative to other brain structures, the cerebral hemispheres and cerebellum are reduced in size while the lateral ventricles and third ventricle are slightly larger than normal (Fig. 6A, lower panels).
Figure 6. Abnormalities in the HDAC4-/- brain.
(A) Outward appearance of the HDAC4-/- brain at P7 along with the brain of a wild-type littermate (top panel). The lower two panels show cresyl violet-stained coronal and sagittal sections of wild-type and mutant brain at P0. The HDAC4-/- has larger ventricles. The cortex, cerebellum, and olfactory bulb are slightly reduced in size. (B) Appearance of the HDAC4-/- cerebellum. Sagittal sections of the cerebellum of HDAC4-/- and wild-type littermates at P1, P3 and P7. The fissures separating the lobes are not as defined in the mutant. The shapes of the posterior lobes are also abnormal.
Examination of the cerebellum of HDAC4-/- mice and wild-type littermates at P1, P3 and P7 revealed a delay in the formation of folia with less developed fissures in the mutant (Fig. 6B). Moreover, the pattern of foliation and shape of the lobes, particularly the posterior lobes, is altered. Cellular lamination of the cerebellar cortex is normal in perinatal animals (at P3) suggesting normal neuronal migration (Fig. 7A). By P7 however, disruption in the lamination pattern of lobe IX can be clearly observed (Fig. 7A and B). We examined the status of Purkinje neurons whose axons extend into the molecular layer using a calbindin antibody. Although appearing normal at P3, a considerable number of Purkinje neurons in lobe IX had degenerated by P7 (Fig. 7C). Degeneration of Purkinje cells is more extensive at P11 when it is evident even in lobe VIII (Fig. 7D). The processes of the surviving Purkinje neurons in lobe X as well as those in other lobes of the HDAC4-/- cerebellum were stunted and dystrophic (Fig. 7E). Interestingly, most of the Purkinje neurons within lobe X and adjacent lobes appear to exist with duplicated cell bodies (Fig. 8).
Figure 7. Purkinje cell degeneration in the HDAC4-/- cerebellum.
(A and B) Disrupted lamination pattern in the HDAC4-/- cerebellum. In (A), cell lamination of lobe IX was visualized by DAPI-staining. While the cell sparse molecular layer is clearly delineated in the mutant cerebellum at P3, disruption of the molecular layer is discernible at P4 and extensive at P7. In (B), lamination is visualized using cresyl violet staining of wild-type and HDAC4-/- cerebellum at P7. Circle shows area with disrupted lamination in lobe IX of the HDAC4-/- cerebellum. (SR) Purkinje cell degeneration in the HDAC4-/- cerebellum. In (C), cerebella from HDAC4-/- mice and control littermates were dissected at P3, P4, and P7 and subjected to immunohistochemical analysis with a calbindin antibody. The figure shows the appearance of region containing lobe IX. A normal layer of Purkinje neurons can be observed in lobe IX of the mutant cerebellum at P3. By P4 however, patches lacking Purkinje neurons are visible. Neurodegeneration is more prominent at P7. In (D), calbindin immunohistochemistry was performed on P11 cerebella from HDAC4-/- and mutant cerebellum. Purkinje cell degeneration is also evident in lobe VIII at this stage. (E) Stunted dendritic arborization of Purkinje neurons in the HDAC4-/- cerebellum. Calbindin immunstaining was performed at P7. Figure shows appearance of lobe X.
Figure 8. Aberrant proliferation in the HDAC4-/- cerebellum.
Duplication of Purkinje cell soma in the HDAC4-/- cerebellum. Cerebella of wild-type and HDAC4-/- mice were dissected at P11 and sagittal sections subjected to calbindin immunostaining. The figure shows a portion of lobe X that has not degenerated. Several of the surviving Purkinje neurons appear as doublets.
In addition to the effect on Purkinje neurons, we find increased proliferation in the external granule layer of the HDAC4-/- cerebellum as judged by Ki-67 immunostaining (Supplemental Fig. 4A, upper panel). The quantification of Ki-67 positive cells are shown in supplemental Fig. 4A, lower panel. This result was further confirmed by BrdU incorporation assay (supplemental Fig. 4B).
HDAC4 inhibits cell cycle progression
Aberrant activation of the cell cycle machinery has been implicated as a mechanism underlying neuronal cell death in a variety of experimental paradigms as well as human neurodegenerative diseases (for review, see Copani et al., 2001; Becker and Bonni, 2004; Greene et al., 2004). The duplication of Purkinje neurons in the HDAC4-deficient cerebellum raised the possibility that HDAC4 protected neurons by preventing aberrant cell cycle reentry. To directly examine whether HDAC4 was capable of inhibiting cell cycle progression we used the HEK293T cell line. HEK293T cells are highly proliferative and are hence ideally suited to evaluate regulatory influences on cell cycle progression. As shown in Fig. 9A, HEK293T cultures transfected with HDAC4 and allowed to grow for 5 days contain fewer cells than cultures transfected with GFP. The phase contrast image of representative cultures are shown in Fig. 9B to demonstrate that the decrease in cell numbers by HDAC4 overexpression is not a result of toxicity caused by HDAC4. To further confirm that this was due to an inhibition of cell cycle progression rather than increased cell death, we performed BrdU-incorporation assays. The reduced incorporation of BrdU in HEK293T cultures transfected with HDAC4 (Fig. 10A) confirms that it inhibits the rate of cell cycle progression. A similar reduction in cell cycle progression was observed in HT22 cells (supplemental Fig. 5)
Figure 9. Effect of HDAC4 overexpression on cell cycle progression.
(A) HEK293T cells were transfected using GFP and HDAC4 plasmids. Total cell numbers were counted 5 days after transfection using a hematocytometer (P < 0.05).
(B) HEK293T cells were transfected with GFP or HDAC4 plasmids. Protein expression was allowed to persist for 22 hrs after which BrdU was added for an additional 2 hrs. Cells were then harvested and stained for HDAC4, GFP and BrdU. The graph demonstrates percentage of cells expressing protein of interest that are BrdU positive (the data has been normalized to % of GFP expressing BrdU positive cells) (P < 0.002).
(C) Images of HEK293T cells expressing GFP or HDAC4. Shown in blue is DAPI staining of these cells. Shown in red are BrdU positive cells (Texas Red). GFP and HDAC4 overexpressing cells are shown in green (FITC).
Figure 10. Effect of HDAC4 overexpression on CDK1 activity.
(A) HEK293T cells were transfected with GFP, CDK1 and/or HDAC4 plasmids. Protein expression was allowed to persist for 22 hrs after which BrdU was added for an additional 2 hrs. Cells were then harvested and stained for HDAC4, GFP and BrdU. The graph demonstrates percentage of cells expressing protein of interest that are BrdU positive (the data has been normalized to % of GFP expressing BrdU positive cells) (P < 0.003). (B) CDK1 was immunoprecipitated from HEK293T cultures transfected with HDAC4, HDAC4-N or GFP plasmids and an in vitro kinase assay was performed in the presences of [γ-32P] ATP and Histone H1 as substrate. The extent of Histone H1 phosphorylation was detected by autoradiography. The panel showing Histone H1 is taken from staining of the membrane with Ponceau-S to demonstrate the similarity in the amount of substrate used. The membrane was also probed with CDK1antibody to show that similar amounts of kinase were pulled down. (C) CDK1 and CDK5 were immunoprecipitated from whole brain lysates obtained from mice lacking HDAC4 and wild-type littermates at P1. Kinase assays were performed as described in part B.
Cell cycle progression is dependent on the activation of cyclin-dependent kinases (CDKs) and an inappropriate activation of specific CDKs has been reported to occur in several different in vitro and in vivo paradigms of neurodegeneration (for review, see Krantic et al., 2005; Aulia and Tang, 2006). Among these is CDK1. Activation of CDK1 has been shown to play an essential role in the death of CGNs caused by LK treatment (Konishi et al., 2002; Konishi and Bonni, 2003). We therefore examined whether the anti-proliferative effect of HDAC4 was mediated via the inhibition of CDK1. As shown in Fig. 10B, CDK1 activity was a little lower in HEK293T cells overexpressing HDAC4. Moreover, in BrdU incorporation assays HDAC4 inhibited cell cycle progression when co-expressed with CDK1 in HEK293T cells (Fig. 10A, top panel). Consistent with experiments performed using HEK293T cells; HDAC4 overexpression had the same effect in HT22 cells as well (supplemental Fig. 5).
Although HDAC4 overexpression inhibits CDK1 in cell lines, it was unclear if it had the same effect in the brain. To investigate this issue we examined the activity of CDK1 in brain lysates prepared from newborn HDAC4-/- mice and control littermates. As shown in Fig. 10C, mice lacking HDAC4 have elevated CDK1 activity. In contrast, kinase activity of the non-mitotic CDK, CDK5, was not changed in HDAC4-/- brains.
DISCUSSION
The administration of HDAC inhibitors can protect against neurodegeneration in a variety of cell culture and in vivo models of neurodegeneration (for review, see Langley et al., 2005; Butler and Bates, 2006). This observation has led to the growing belief that HDACs play an important role in promoting neuronal death and that inhibiting the catalytic activity of these proteins might therefore serve as a potential therapeutic strategy for human neurodegenerative diseases. We have been examining the role of individual HDAC proteins in the regulation of neuronal survival and death. In this report we demonstrate that contrary to expectations one may derive from the use of HDAC inhibitors, the overexpression of the Class IIa HDAC, HDAC4, can protect cultured neurons. Although much of our analyses have been performed using LK-treated CGNs, HDAC4 overexpression also protects HT22 cells from oxidative stress induced cell death and cortical neurons from 6-OHDA neurotoxicity. Furthermore, the absence of HDAC4 has a detrimental effect on the survival of Purkinje neurons in vivo. The effectiveness of HDAC4 to act as a neuroprotective protein thus extends to different neuronal types and against different death-inducing stimuli.
HDAC4 localizes to the nucleus under apoptotic conditions indicating that neuroprotection is mediated in the nucleus. Supporting this conclusion is the finding that protection is also observed with HDAC4-N, a truncation mutant which is trapped in the nucleus because it lacks a nuclear export signal. Using deletion constructs we have localized the neuroprotective domain to a 72 amino acid segment of HDAC4 which spans the NLS. Our findings contradict those of Bolger and Yao (Bolger and Yao, 2005) who reported that HDAC4 has a modest pro-apoptotic effect when overexpressed in cultured CGNs. One difference in the experimental conditions between the two studies is the age of the CGN cultures. In the Bolger and Yao study cultures that were relatively immature (2 days after plating) were used compared with our cultures which were typically 6 - 7 days old. We have used less mature cultures but have not observed neurotoxicity with HDAC4 overexpression. On the contrary, we find that HDAC4 is neuroprotective even in younger neurons although the extent of cell death induced by LK in these immature neuronal cultures is somewhat less than that observed in more mature neurons (data not shown).
We have previously reported that HDRP, a spliced variant of HDAC9 that completely lacks the catalytic domain, prevents LK-induced apoptosis. Neuroprotection by HDRP involves the inhibition of c-jun phosphorylation through direct interaction with JNK. Furthermore, HDRP interacts with HDAC1 recruiting it to the c-jun promoter resulting in deacetylation of histones and a repression of c-jun gene transcription. Not unexpectedly, the inhibition of HDAC activity using pharmacological inhibitors attenuates the neuroprotective effects of HDRP. HDRP-mediated neuroprotection can also be inhibited by Gleevec, an inhibitor of c-Abl. HDAC4 does not interact with HDAC1 and its effectiveness to protect is not reduced by pharmacological inhibition of HDACs or by c-Abl inhibition. HDAC4 has also no effect on the activation of c-jun or JNK phosphorylation triggered by LK-treatment. C-jun activation is an invariable feature of neuronal apoptosis and is seen both in tissue culture models and in vivo paradigms of neurodegeneration (Estus et al., 1994; Ham et al., 1995; Watson et al., 1998; Schenkel, 2004). The normal and robust activation of c-jun in HDAC4-overexpressing neuronal cultures indicates that HDAC4 acts at a step downstream of c-jun induction, or that HDAC4 activates survival mechanisms that are able to neutralize the pro-apoptotic effects of c-jun.
Mice lacking HDAC4 display developmental abnormalities including slightly larger ventricles, a reduction in the thickness of the cortex, and a smaller cerebellum. Within the cerebellum, the fissures defining the lobes are somewhat underdeveloped and the shapes of the posterior lobes are severely deformed. Since the skull of the HDAC4-/- is deformed in shape it is conceivable that some of these abnormalities occur secondary to the established skeletal defects (Vega et al., 2005). A closer examination of the cerebellar cortex reveals that the lamination pattern is normal at birth in HDAC4-deficient mice. By P11 however, disruption of cellular lamination is evident within lobes VIII and IX along with a significant degeneration of Purkinje neurons. Although degeneration is not obvious in the other cerebellar lobes, dendritic arborization of Purkinje neurons in all lobes is stunted. It is possible that axons and dendrites of the Purkinje cells are affected before cell death occurs and that the degeneration begins in lobe IX and subsequently in the other cerebellar lobes. The premature death of the mutant mice by P14 precludes the investigation of this possibility.
A notable abnormality in the cerebellum is the duplication of Purkinje neuron soma in posterior lobes. This occurrence along with the degeneration of these neurons is consistent with the possibility that the absence of HDAC4 activates cell cycle progression resulting in neuronal apoptosis. In support of this possibility is the finding that a substantial number of cells in the HDAC4-/- cerebellum incorporate BrdU, a commonly used indicator of cell cycle progression (Majdzadeh and D’Mello; unpublished observation). To test whether HDAC4 was a negative regulator of cell cycle progression we used the proliferating HEK293T cell line. We find that the overexpression of HDAC4 in HEK293T cells, which normally does not express detectable amounts of HDAC4, reduces their rate of cell cycle progression. A similar inhibition of cell cycle progression was observed in HT22 cells upon HDAC4 overexpression. While the mechanism by which HDAC4 inhibits cell cycle progression remains to be completely elucidated, we have found that one target of HDAC4 in the cell cycle machinery is CDK1. HDAC4 inhibits CDK1 activity in HEK293T cells and the ability of CDK1 to stimulate cell proliferation.
Although HDAC4 blocks cell proliferation in cell lines, more work is needed to confirm that its protective effect in neurons is also mediated by inhibiting aberrant cell cycle progression. Several lines of evidence support this possibility, however. In addition to the effect on Purkinje neurons, we find increased proliferation in the external granule layer of the HDAC4-/- cerebellum as judged by Ki-67 immunostaining and BrdU incorporation assays.At P10, Ki-67-positive cells can also be observed in the internal granule layer which is composed of postmitotic granule neurons (data not shown). Moreover, CDK1 activity is higher in the brains of mice lacking HDAC4.
The expression of HDRP mRNA and protein is reduced in cultured CGNs primed to die by LK treatment (Morrison et al., 2006). In contrast, the expression of HDAC4 is not altered at the onset of apoptosis although at late stages of degeneration decreased HDAC4 expression can be observed (Majdzadeh and D’Mello, unpublished observation). Moreover, CGNs cultured from HDAC4-deficient mice do not display increased vulnerability to apoptosis (data not shown), a feature that is displayed by HDRP-deficient neurons (Morrison et al., 2006). These results suggest that HDAC4 is not required for CGN survival although its overexpression is sufficient to prevent the death of these neurons in response to LK treatment. It is possible that HDRP and possibly other Class II HDACs are more important players in ensuring the survival of CGNs whereas HDAC4 is more critical for Purkinje neurons. A lack of compensation by other HDAC proteins in Purkinje neurons might also contribute to their sensitivity to the absence of HDAC4.
The effectiveness of pharmacological HDAC inhibitors to reduce neurodegeneration is striking. Indeed, these inhibitors reduce degeneration in cultured neurons, Caenorhabditis elegans, Drosophila, and mouse models of polyglutamine toxicity (Steffan et al., 2001; Ferrante et al., 2003; Hockly et al., 2003; Jeong et al., 2003; Ryu et al., 2003; Kanai et al., 2004; Gardian et al., 2005; Bates et al., 2006; Chen et al., 2006). Administration of histone deacetylase inhibitors also protects against neurotoxicity in transgenic fly models of Parkinson’s disease and ALS, and reduces ischemic stroke-induced neuronal death in rats (Gardian et al., 2004; Ren et al., 2004; Faraco et al., 2006; Petri et al., 2006). It remains unclear which specific HDAC proteins are targeted by the inhibitors in these various experimental models. In cultured CGNs, however, the same inhibitors actively promote cell death (Salminen et al., 1998; Boutillier et al., 2003; Morrison et al., 2006). One possibility to explain these seemingly contradictory findings is that individual members of the HDACs family might act differently to regulate neuronal survival, with some proteins functioning to promote survival whereas other family members having pro-apoptotic actions. Since the commonly used HDAC inhibitors inhibit all HDAC proteins efficiently whether they promote or reduce neuronal death may be context dependent and influenced substantially by the relative levels of pro and anti-apoptotic HDAC proteins in the specific cell type. The finding that HDRP and HDAC4 are protective while HDAC5 promotes death of CGNs demonstrates the existence of individual family members with opposing actions on cell survival. HDAC proteins with opposing contributions to neuronal survival have also been described in Caenorhabditis elegans (Bates et al., 2006). In the nematode, HDA-1 (homolog of mammalian HDAC1 and HDAC2) suppresses neurotoxicity whereas HDA-3 (homolog of mammalian HDAC3) promotes it. It is also conceivable that HDAC inhibitors protect neurons by activating global gene expression through increased histone acetylation and chromatin remodeling. The general activation of gene expression could have beneficial or detrimental outcomes depending on the extent of expression of pro-survival versus pro-apoptotic genes. Knockout mice lacking several different HDAC proteins, including HDACs 4, 7, 5, and 9 have been generated (Chang et al., 2004; Vega et al., 2004; Chang et al., 2006). None of these mutant mouse lines display signs of defective developmentally-regulated neuronal death such as ectopic lumps in the brain or an enlargement of the brain. Such abnormalities are observed in knockouts of other pro-apoptotic molecules such as caspase-3, caspase-9, or Apaf-1 (Kuida et al., 1996; Cecconi et al., 1998; Hakem et al., 1998; Kuida et al., 1998; Yoshida et al., 1998). The effect of HDAC inhibitors on developmentally-regulated neuronal death has also not been systematically studied. Whether HDACs play a role during brain development in the context of neuronal survival and death is therefore not known.
In summary, we show that HDAC4 can protect neurons from death when overexpressed. Along with HDRP, this is the second member of the HDAC family of proteins that has been found to have neuroprotective efficacy. Overexpression of HDRP or HDAC4 or increasing the endogenous expression of these proteins in vulnerable neuronal populations through pharmacological means may serve as a potential therapeutic strategy for human neurodegenerative diseases. Although, further research is needed to determine exactly how HDAC4 exerts its neuroprotective effect, our studies points to CDK1 inhibition and a blockade of cell cycle progression as the likely mechanism.
Supplementary Material
Supplemental figure 1. HDAC4 neuroprotection after extended LK treatments. 5 day old culture of CGNs were infected with Ad-GFP or Ad-HDAC4 (Flag tag) and switched to HK or LK medium for 24, 48 and 72 hrs. Infected neurons were identified by GFP fluorescence or Flag immunoreactivity and the status of the nuclei were assessed by DAPI staining. As shown in the upper panel HDAC4 (red) is primarily in the cytoplasm in HK but translocates to the nucleus (blue) in LK. Lower panel quantifies survival of HDAC4 and GFP infected cells in HK and LK medium as percent of control (viability of GFP-infected neurons in HK).
Supplemental figure 2. Effect of HDAC4 overexpression on cortical neuron survival. (A) Cortical neurons were infected using HDAC4 (Flag tag) adenoviral construct one day after plating or left uninfected. The next day, cells were treated with 100 uM of 6- OHDA for 24 hrs after which immunocytochemistry was performed using Flag antibody followed by DAPI staining. Panel A. quantifies the survival of HDAC4 and uninfected cells in control and 6-OHDA treated cells as a percent of control (cell viability in control uninfected cells). (B) Images of cells infected with HDAC4 in control and 6-OHDA treatment cells. HDAC4 is shown in red and cell nuclei are stained with DAPI (blue).
Supplemental figure 3. HDAC4 overexpression modulates transcription of a number of apoptosis regulatory genes. 6-7 days old CGNs were treated with HK and LK for 6 hrs. RT-PCR panel; RNA was extracted from cells followed by cDNA synthesis. PCR was performed using primers specific to the listed genes. Actin was used to demonstrate that similar quantities of cDNA were used.
Supplemental figure 4. Increased cell proliferation in the EGL of the HDAC4-/- cerebellum. (A) Sections of HDAC4-/- and wild-type cerebellum was examined at P3 following Ki-67 immunostaining. Increased Ki-67 staining in the external granule layer of lobe IX is evident. Lower panel; Quantification of increased Ki-67 staining is shown in the graph. Ki-67 positive cells were counted in the EGL of 6 randomly chosen fields from cerebellum of two HDAC4-/- mice along with their wild-type littermates at P3 (P < 0.0009). Although increased proliferation is observed at P3, a similar analysis at P10 does not show higher proliferation in the EGL. At this age however, more Ki-67 immunorective cells were observed in the inner layers of the cerebellar cortex. (B) Increased BrdU-staining in the HDAC4-/- cerebellum. 7-day old HDAC4-/- mice and wild-type littermates were injected intraperitoneally with BrdU as described in Methods and sacrificed 2 days later. The brains were sectioned and subjected to immunohistochemical analysis using a BrdU antibody. The figure shows a higher number of BrdU-positive cells in the HDAC4-/- cerebellum. The positions of the external granule layer (EGL), molecular layer (ML), Purkinje cell layer (PCL) and internal granule layer (Chin et al.) is marked.
Supplemental figure 5. Effect of HDAC4 overexpression on cell cycle progression in HT22 cells. (A and B) HT22 cells were transfected with GFP, HDAC4 and/or CDK1 plasmids. Protein expression was allowed to persist for 22 hrs after which BrdU was added for an additional 2 hrs Cells were then harvested and stained for HDAC4, GFP and BrdU. The graph demonstrates percentage of cells expressing protein of interest that are BrdU positive (the data has been normalized to % of GFP expressing BrdU positive cells).
Acknowledgements
The research described in this report was funded by NIH - NINDS grants and NS40408 and NS047201 to SRD. We thank Li Liu for her thoughtful and constructive suggestions in the course of our study.
REFERENCES
- Aulia S, Tang BL. Cdh1-APC/C, cyclin B-Cdc2, and Alzheimer’s disease pathology. Biochem Biophys Res Commun. 2006;339:1–6. doi: 10.1016/j.bbrc.2005.10.059. [DOI] [PubMed] [Google Scholar]
- Bates EA, Victor M, Jones AK, Shi Y, Hart AC. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J Neurosci. 2006;26:2830–2838. doi: 10.1523/JNEUROSCI.3344-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker EB, Bonni A. Cell cycle regulation of neuronal apoptosis in development and disease. Prog Neurobiol. 2004;72:1–25. doi: 10.1016/j.pneurobio.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Bolger TA, Yao TP. Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J Neurosci. 2005;25:9544–9553. doi: 10.1523/JNEUROSCI.1826-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutillier AL, Trinh E, Loeffler JP. Selective E2F-dependent gene transcription is controlled by histone deacetylase activity during neuronal apoptosis. J Neurochem. 2003;84:814–828. doi: 10.1046/j.1471-4159.2003.01581.x. [DOI] [PubMed] [Google Scholar]
- Butler R, Bates GP. Histone deacetylase inhibitors as therapeutics for polyglutamine disorders. Nat Rev Neurosci. 2006;7:784–796. doi: 10.1038/nrn1989. [DOI] [PubMed] [Google Scholar]
- Butts BD, Linseman DA, Le SS, Laessig TA, Heidenreich KA. Insulin-like growth factor-I suppresses degradation of the pro-survival transcription factor myocyte enhancer factor 2D (MEF2D) during neuronal apoptosis. Horm Metab Res. 2003;35:763–770. doi: 10.1055/s-2004-814148. [DOI] [PubMed] [Google Scholar]
- Cecconi F, Alvarez-Bolado G, Meyer BI, Roth KA, Gruss P. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell. 1998;94:727–737. doi: 10.1016/s0092-8674(00)81732-8. [DOI] [PubMed] [Google Scholar]
- Chang S, Bezprozvannaya S, Li S, Olson EN. An expression screen reveals modulators of class II histone deacetylase phosphorylation. Proc Natl Acad Sci U S A. 2005;102:8120–8125. doi: 10.1073/pnas.0503275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Young BD, Li S, Qi X, Richardson JA, Olson EN. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126:321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Chawla S, Vanhoutte P, Arnold FJ, Huang CL, Bading H. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J Neurochem. 2003;85:151–159. doi: 10.1046/j.1471-4159.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- Chen PS, Peng GS, Li G, Yang S, Wu X, Wang CC, Wilson B, Lu RB, Gean PW, Chuang DM, Hong JS. Valproate protects dopaminergic neurons in midbrain neuron/glia cultures by stimulating the release of neurotrophic factors from astrocytes. Mol Psychiatry. 2006;11:1116–1125. doi: 10.1038/sj.mp.4001893. [DOI] [PubMed] [Google Scholar]
- Chin PC, Liu L, Morrison BE, Siddiq A, Ratan RR, Bottiglieri T, D’Mello SR. The c-Raf inhibitor GW5074 provides neuroprotection in vitro and in an animal model of neurodegeneration through a MEK-ERK and Akt-independent mechanism. J Neurochem. 2004;90:595–608. doi: 10.1111/j.1471-4159.2004.02530.x. [DOI] [PubMed] [Google Scholar]
- Copani A, Uberti D, Sortino MA, Bruno V, Nicoletti F, Memo M. Activation of cell-cycle-associated proteins in neuronal death: a mandatory or dispensable path? Trends Neurosci. 2001;24:25–31. doi: 10.1016/s0166-2236(00)01663-5. [DOI] [PubMed] [Google Scholar]
- D’Mello SR, Chin PC. Treating neurodegenerative conditions through the understanding of neuronal apoptosis. Curr Drug Targets CNS Neurol Disord. 2005;4:3–23. doi: 10.2174/1568007053005118. [DOI] [PubMed] [Google Scholar]
- D’Mello SR, Galli C, Ciotti T, Calissano P. Induction of apoptosis in cerebellar granule neurons by low potassium: inhibition of death by insulin-like growth factor I and cAMP. Proc Natl Acad Sci U S A. 1993;90:10989–10993. doi: 10.1073/pnas.90.23.10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estus S, Zaks WJ, Freeman RS, Gruda M, Bravo R, Johnson EM., Jr. Altered gene expression in neurons during programmed cell death: identification of c-jun as necessary for neuronal apoptosis. J Cell Biol. 1994;127:1717–1727. doi: 10.1083/jcb.127.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraco G, Pancani T, Formentini L, Mascagni P, Fossati G, Leoni F, Moroni F, Chiarugi A. Pharmacological Inhibition of Histone Deacetylases by Suberoylanilide Hydroxamic Acid Specifically Alters Gene Expression and Reduces Ischemic Injury in the Mouse Brain. Mol Pharmacol. 2006;70:1876–1884. doi: 10.1124/mol.106.027912. [DOI] [PubMed] [Google Scholar]
- Ferrante RJ, Kubilus JK, Lee J, Ryu H, Beesen A, Zucker B, Smith K, Kowall NW, Ratan RR, Luthi-Carter R, Hersch SM. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington’s disease mice. J Neurosci. 2003;23:9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardian G, Browne SE, Choi DK, Klivenyi P, Gregorio J, Kubilus JK, Ryu H, Langley B, Ratan RR, Ferrante RJ, Beal MF. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington’s disease. J Biol Chem. 2005;280:556–563. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- Gardian G, Yang L, Cleren C, Calingasan NY, Klivenyi P, Beal MF. Neuroprotective effects of phenylbutyrate against MPTP neurotoxicity. Neuromolecular Med. 2004;5:235–241. doi: 10.1385/NMM:5:3:235. [DOI] [PubMed] [Google Scholar]
- Greene LA, Biswas SC, Liu DX. Cell cycle molecules and vertebrate neuron death: E2F at the hub. Cell Death Differ. 2004;11:49–60. doi: 10.1038/sj.cdd.4401341. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci U S A. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakem R, Hakem A, Duncan GS, Henderson JT, Woo M, Soengas MS, Elia A, de la Pompa JL, Kagi D, Khoo W, Potter J, Yoshida R, Kaufman SA, Lowe SW, Penninger JM, Mak TW. Differential requirement for caspase 9 in apoptotic pathways in vivo. Cell. 1998;94:339–352. doi: 10.1016/s0092-8674(00)81477-4. [DOI] [PubMed] [Google Scholar]
- Ham J, Babij C, Whitfield J, Pfarr CM, Lallemand D, Yaniv M, Rubin LL. A c-Jun dominant negative mutant protects sympathetic neurons against programmed cell death. Neuron. 1995;14:927–939. doi: 10.1016/0896-6273(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, Sathasivam K, Ghazi-Noori S, Mahal A, Lowden PA, Steffan JS, Marsh JL, Thompson LM, Lewis CM, Marks PA, Bates GP. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc Natl Acad Sci U S A. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong MR, Hashimoto R, Senatorov VV, Fujimaki K, Ren M, Lee MS, Chuang DM. Valproic acid, a mood stabilizer and anticonvulsant, protects rat cerebral cortical neurons from spontaneous cell death: a role of histone deacetylase inhibition. FEBS Lett. 2003;542:74–78. doi: 10.1016/s0014-5793(03)00350-8. [DOI] [PubMed] [Google Scholar]
- Kanai H, Sawa A, Chen RW, Leeds P, Chuang DM. Valproic acid inhibits histone deacetylase activity and suppresses excitotoxicity-induced GAPDH nuclear accumulation and apoptotic death in neurons. Pharmacogenomics J. 2004;4:336–344. doi: 10.1038/sj.tpj.6500269. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Bonni A. The E2F-Cdc2 cell-cycle pathway specifically mediates activity deprivation-induced apoptosis of postmitotic neurons. J Neurosci. 2003;23:1649–1658. doi: 10.1523/JNEUROSCI.23-05-01649.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y, Lehtinen M, Donovan N, Bonni A. Cdc2 phosphorylation of BAD links the cell cycle to the cell death machinery. Mol Cell. 2002;9:1005–1016. doi: 10.1016/s1097-2765(02)00524-5. [DOI] [PubMed] [Google Scholar]
- Koulich E, Nguyen T, Johnson K, Giardina C, D’Mello S. NF-kappaB is involved in the survival of cerebellar granule neurons: association of IkappaBbeta [correction of Ikappabeta] phosphorylation with cell survival. J Neurochem. 2001;76:1188–1198. doi: 10.1046/j.1471-4159.2001.00134.x. [DOI] [PubMed] [Google Scholar]
- Krantic S, Mechawar N, Reix S, Quirion R. Molecular basis of programmed cell death involved in neurodegeneration. Trends Neurosci. 2005;28:670–676. doi: 10.1016/j.tins.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, Su MS, Rakic P, Flavell RA. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell. 1998;94:325–337. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- Kuida K, Zheng TS, Na S, Kuan C, Yang D, Karasuyama H, Rakic P, Flavell RA. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature. 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- Langley B, Gensert JM, Beal MF, Ratan RR. Remodeling chromatin and stress resistance in the central nervous system: histone deacetylase inhibitors as novel and broadly effective neuroprotective agents. Curr Drug Targets CNS Neurol Disord. 2005;4:41–50. doi: 10.2174/1568007053005091. [DOI] [PubMed] [Google Scholar]
- Linseman DA, Bartley CM, Le SS, Laessig TA, Bouchard RJ, Meintzer MK, Li M, Heidenreich KA. Inactivation of the myocyte enhancer factor-2 repressor histone deacetylase-5 by endogenous Ca(2+) //calmodulin-dependent kinase II promotes depolarization-mediated cerebellar granule neuron survival. J Biol Chem. 2003;278:41472–41481. doi: 10.1074/jbc.M307245200. [DOI] [PubMed] [Google Scholar]
- Linseman DA, Cornejo BJ, Le SS, Meintzer MK, Laessig TA, Bouchard RJ, Heidenreich KA. A myocyte enhancer factor 2D (MEF2D) kinase activated during neuronal apoptosis is a novel target inhibited by lithium. J Neurochem. 2003;85:1488–1499. doi: 10.1046/j.1471-4159.2003.09799.x. [DOI] [PubMed] [Google Scholar]
- Lu J, McKinsey TA, Nicol RL, Olson EN. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci U S A. 2000;97:4070–4075. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci U S A. 2000;97:14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BD, Gibbons B, Allen LR, Stella J, D’Mello SR. Aberrant apoptosis in the neurological mutant Flathead is associated with defective cytokinesis of neural progenitor cells. Brain Res Dev Brain Res. 2001;130:53–63. doi: 10.1016/s0165-3806(01)00206-1. [DOI] [PubMed] [Google Scholar]
- Morrison BE, Majdzadeh N, Zhang X, Lyles A, Bassel-Duby R, Olson EN, D’Mello SR. Neuroprotection by histone deacetylase-related protein. Mol Cell Biol. 2006;26:3550–3564. doi: 10.1128/MCB.26.9.3550-3564.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TH, Schnaar RL, Coyle JT. Immature cortical neurons are uniquely sensitive to glutamate toxicity by inhibition of cystine uptake. Faseb J. 1990;4:1624–1633. [PubMed] [Google Scholar]
- Okamoto S, Krainc D, Sherman K, Lipton SA. Antiapoptotic role of the p38 mitogen-activated protein kinase-myocyte enhancer factor 2 transcription factor pathway during neuronal differentiation. Proc Natl Acad Sci U S A. 2000;97:7561–7566. doi: 10.1073/pnas.130502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri S, Kiaei M, Kipiani K, Chen J, Calingasan NY, Crow JP, Beal MF. Additive neuroprotective effects of a histone deacetylase inhibitor and a catalytic antioxidant in a transgenic mouse model of amyotrophic lateral sclerosis. Neurobiol Dis. 2006;22:40–49. doi: 10.1016/j.nbd.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Potter GB, Beaudoin GM, 3rd, DeRenzo CL, Zarach JM, Chen SH, Thompson CC. The hairless gene mutated in congenital hair loss disorders encodes a novel nuclear receptor corepressor. Genes Dev. 2001;15:2687–2701. doi: 10.1101/gad.916701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Leng Y, Jeong M, Leeds PR, Chuang DM. Valproic acid reduces brain damage induced by transient focal cerebral ischemia in rats: potential roles of histone deacetylase inhibition and heat shock protein induction. J Neurochem. 2004;89:1358–1367. doi: 10.1111/j.1471-4159.2004.02406.x. [DOI] [PubMed] [Google Scholar]
- Ryu H, Lee J, Olofsson BA, Mwidau A, Dedeoglu A, Escudero M, Flemington E, Azizkhan-Clifford J, Ferrante RJ, Ratan RR. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc Natl Acad Sci U S A. 2003;100:4281–4286. doi: 10.1073/pnas.0737363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Tapiola T, Korhonen P, Suuronen T. Neuronal apoptosis induced by histone deacetylase inhibitors. Brain Res Mol Brain Res. 1998;61:203–206. doi: 10.1016/s0169-328x(98)00210-1. [DOI] [PubMed] [Google Scholar]
- Schenkel J. Activation of the c-Jun transcription factor following neurodegeneration in vivo. Neurosci Lett. 2004;361:36–39. doi: 10.1016/j.neulet.2003.12.011. [DOI] [PubMed] [Google Scholar]
- SR CPaDM. 2004.
- Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, Kurokawa R, Housman DE, Jackson GR, Marsh JL, Thompson LM. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, Karsenty G, Olson EN. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- Wang AH, Bertos NR, Vezmar M, Pelletier N, Crosato M, Heng HH, Th’ng J, Han J, Yang XJ. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A, Eilers A, Lallemand D, Kyriakis J, Rubin LL, Ham J. Phosphorylation of c-Jun is necessary for apoptosis induced by survival signal withdrawal in cerebellar granule neurons. J Neurosci. 1998;18:751–762. doi: 10.1523/JNEUROSCI.18-02-00751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin A, Koulich E, Mohamed S, Liu L, D’Mello SR. Apoptosis in cerebellar granule neurons is associated with reduced interaction between CREB-binding protein and NF-kappaB. J Neurochem. 2003;84:397–408. doi: 10.1046/j.1471-4159.2003.01540.x. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Gregoire S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol Cell Biol. 2005;25:2873–2884. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Kong YY, Yoshida R, Elia AJ, Hakem A, Hakem R, Penninger JM, Mak TW. Apaf1 is required for mitochondrial pathways of apoptosis and brain development. Cell. 1998;94:739–750. doi: 10.1016/s0092-8674(00)81733-x. [DOI] [PubMed] [Google Scholar]
- Zhang CL, McKinsey TA, Olson EN. The transcriptional corepressor MITR is a signal-responsive inhibitor of myogenesis. Proc Natl Acad Sci U S A. 2001;98:7354–7359. doi: 10.1073/pnas.131198498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LJ, Zhang S, Chinnadurai G. Sox9 transactivation and testicular expression of a novel human gene, KIAA0800. J Cell Biochem. 2002;86:277–289. doi: 10.1002/jcb.10214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure 1. HDAC4 neuroprotection after extended LK treatments. 5 day old culture of CGNs were infected with Ad-GFP or Ad-HDAC4 (Flag tag) and switched to HK or LK medium for 24, 48 and 72 hrs. Infected neurons were identified by GFP fluorescence or Flag immunoreactivity and the status of the nuclei were assessed by DAPI staining. As shown in the upper panel HDAC4 (red) is primarily in the cytoplasm in HK but translocates to the nucleus (blue) in LK. Lower panel quantifies survival of HDAC4 and GFP infected cells in HK and LK medium as percent of control (viability of GFP-infected neurons in HK).
Supplemental figure 2. Effect of HDAC4 overexpression on cortical neuron survival. (A) Cortical neurons were infected using HDAC4 (Flag tag) adenoviral construct one day after plating or left uninfected. The next day, cells were treated with 100 uM of 6- OHDA for 24 hrs after which immunocytochemistry was performed using Flag antibody followed by DAPI staining. Panel A. quantifies the survival of HDAC4 and uninfected cells in control and 6-OHDA treated cells as a percent of control (cell viability in control uninfected cells). (B) Images of cells infected with HDAC4 in control and 6-OHDA treatment cells. HDAC4 is shown in red and cell nuclei are stained with DAPI (blue).
Supplemental figure 3. HDAC4 overexpression modulates transcription of a number of apoptosis regulatory genes. 6-7 days old CGNs were treated with HK and LK for 6 hrs. RT-PCR panel; RNA was extracted from cells followed by cDNA synthesis. PCR was performed using primers specific to the listed genes. Actin was used to demonstrate that similar quantities of cDNA were used.
Supplemental figure 4. Increased cell proliferation in the EGL of the HDAC4-/- cerebellum. (A) Sections of HDAC4-/- and wild-type cerebellum was examined at P3 following Ki-67 immunostaining. Increased Ki-67 staining in the external granule layer of lobe IX is evident. Lower panel; Quantification of increased Ki-67 staining is shown in the graph. Ki-67 positive cells were counted in the EGL of 6 randomly chosen fields from cerebellum of two HDAC4-/- mice along with their wild-type littermates at P3 (P < 0.0009). Although increased proliferation is observed at P3, a similar analysis at P10 does not show higher proliferation in the EGL. At this age however, more Ki-67 immunorective cells were observed in the inner layers of the cerebellar cortex. (B) Increased BrdU-staining in the HDAC4-/- cerebellum. 7-day old HDAC4-/- mice and wild-type littermates were injected intraperitoneally with BrdU as described in Methods and sacrificed 2 days later. The brains were sectioned and subjected to immunohistochemical analysis using a BrdU antibody. The figure shows a higher number of BrdU-positive cells in the HDAC4-/- cerebellum. The positions of the external granule layer (EGL), molecular layer (ML), Purkinje cell layer (PCL) and internal granule layer (Chin et al.) is marked.
Supplemental figure 5. Effect of HDAC4 overexpression on cell cycle progression in HT22 cells. (A and B) HT22 cells were transfected with GFP, HDAC4 and/or CDK1 plasmids. Protein expression was allowed to persist for 22 hrs after which BrdU was added for an additional 2 hrs Cells were then harvested and stained for HDAC4, GFP and BrdU. The graph demonstrates percentage of cells expressing protein of interest that are BrdU positive (the data has been normalized to % of GFP expressing BrdU positive cells).