Abstract
Posttranslational modifications play a key role in recruiting chromatin remodeling and modifying enzymes to specific regions of chromosomes to modulate chromatin structure. Alc1 (amplified in liver cancer 1), a member of the SNF2 ATPase superfamily with a carboxy-terminal macrodomain, is encoded by an oncogene implicated in the pathogenesis of hepatocellular carcinoma. Here we show that Alc1 interacts transiently with chromatin-associated proteins, including histones and the poly(ADP-ribose) polymerase Parp1. Alc1 ATPase and chromatin remodeling activities are strongly activated by Parp1 and its substrate NAD and require an intact macrodomain capable of binding poly(ADP-ribose). Alc1 is rapidly recruited to nucleosomes in vitro and to chromatin in cells when Parp1 catalyzes PAR synthesis. We propose that poly(ADP-ribosyl)ation of chromatin-associated Parp1 serves as a mechanism for targeting a SNF2 family remodeler to chromatin.
Keywords: Alc1, chromatin remodeling enzyme, macrodomain, poly-(ADP-ribose) polymerase, Snf2-like ATPase
In eukaryotic cells, chromosomal DNA is packaged into nucleosomes, which are in turn folded into higher order nucleosome arrays in chromatin fibers. This packaging allows the ≈2 m of DNA that make up the human genome to fit into nuclei with diameters on the order of 2–6 μm; however, it also blocks access to DNA of the machinery responsible for transcription, replication, and DNA repair. Eukaryotic organisms have evolved a set of chromatin modifying and remodeling enzymes that alter the structure of chromatin to control accessibility to the machineries responsible for DNA replication and repair and for transcription. These enzymes have been shown to be targeted to regions of modified chromatin by such domains as bromodomains, which can bind acetylated histones, or chromodomains, tudor domains, or MBT domains, which can interact with methylated histones (1–4).
ALC1 (amplified in liver cancer 1), alternatively known as CHD1L, is a member of the SNF2 superfamily of ATPases, some of which function as chromatin remodeling enzymes (5–7). Sequence alignments suggest that Alc1 is similar to chromatin remodeling ATPases Snf2, Iswi, and Chd1, which have been implicated in transcription, DNA repair, and replication (7). Alc1 lacks identifiable chromo-, bromo-, tudor-, MBT, or other domains known to have chromatin targeting functions. Instead, it contains a carboxy-terminal macrodomain. Macrodomains have been shown through biochemical and structural analyses to bind ADP-ribose (8).
Over 50% of human hepatocellular carcinoma (HCC) patients contain a chromosomal amplification at 1q21, which includes the ALC1 gene (9–11). Alc1-overexpressing cells exhibit increased colony formation in soft agar and increased tumorigenicity in nude mice (11), suggesting that ALC1 functions as an oncogene.
While mounting evidence points to a potential role for Alc1 in oncogenesis, the molecular function of the Alc1 ATPase has not been studied. Here, we show that Alc1 is a chromatin remodeling enzyme that is recruited to nucleosomes and activated in a manner dependent on poly(ADP-ribosylation) (PARylation), most likely via interactions with chromatin-associated poly(ADP-ribosyl)ated Parp1.
Results and Discussion
To investigate possible Alc1 interactors, we generated an HEK293/FRT cell line stably expressing ALC1 with an N-terminal FLAG tag (F-Alc1). Initial immunopurification of F-Alc1 from nuclear extracts with M2 agarose suggested that unlike many SNF2 superfamily members, Alc1 does not reside in a stable multisubunit complex (Fig. S1A); however, MudPIT mass spectrometry indicated that preparations of F-Alc1 contained small amounts of histones and Parp1 and several Parp1-interacting proteins (Table S1).
To gain further insight into the molecular function of Alc1, we expressed and purified recombinant wild-type F-Alc1; a DEAH box mutant F-Alc1(E175Q), which is mutated at a position expected to prevent ATP binding and hydrolysis; a macrodomain mutant F-Alc1(D723A), which is mutated at a position shown previously to decrease substantially the affinity of ADP-ribose binding by AF1521, a macrodomain-containing protein from Archaeoglobus fulgidus (8); and the Alc1 macrodomain (amino acids 666–897) (Fig. 1A and Fig. S1B). To determine if Alc1 can bind poly(ADP)-ribose (PAR), purified recombinant proteins were dot-blotted on nitrocellulose after incubation with 32P-labeled PAR (Figs. 1B and C). Alc1 and the DEAH box mutant Alc1(E175Q) bound PAR. PAR binding was abolished by heat treatment and was substantially reduced by high salt. Indicating that the Alc1 macrodomain is necessary and sufficient for PAR binding, the isolated Alc1 macrodomain bound PAR, while PAR binding by the macrodomain mutant Alc1(D723A) was greatly reduced.
Fig. 1.
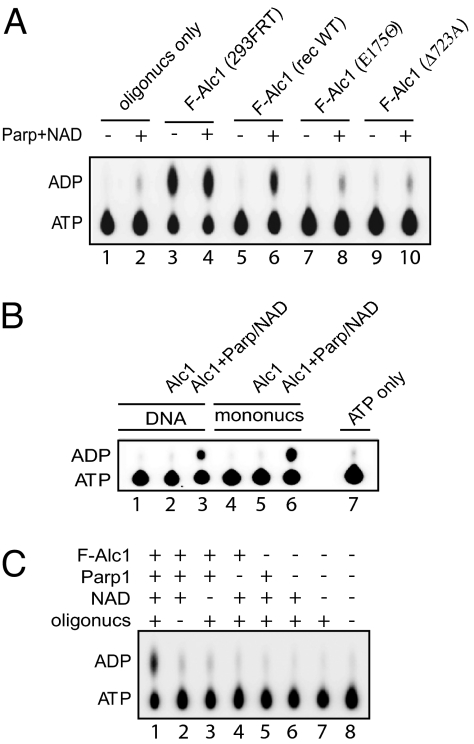
Alc1 binds poly(ADP-ribose). (A) Alc1 domain structure. Sequences below diagram show amino acid changes in catalytically inactive Alc1 mutant E175Q and macrodomain mutant D723A. The mutated macrodomain region is compared to the homologous sequence from the AF1521 macrodomain. Snf2N, SNF2 family N-terminal domain; HelicC, Helicase superfamily C-terminal domain, associated with DEXDc-, DEAD-, and DEAH-box proteins; macro, macrodomain. (B) Approximately 100 ng of each protein was incubated with 32P-labeled PAR in buffer with the indicated NaCl concentrations. PAR binding was detected with a nitrocellulose filter binding assay. (C) Approximately 100 ng (1×) or 200 ng (2×) wild-type or mutant Alc1 was incubated with PAR in buffer containing 0.15 M NaCl. PAR binding was measured as in panel B. F-Alc1, Flag epitope-tagged Alc1; rec, recombinant.
Many SNF2 superfamily members have both DNA- and nucleosome-activated ATPase activities (7). To determine whether Alc1 has similar activities, we assayed anti-Flag agarose eluates from F-Alc1 expressing HEK293/FRT cells and wild-type and mutant versions of recombinant F-Alc1, expressed in and purified from Sf21 cells, for ATPase activity. F-Alc1 from HEK293/FRT cells exhibited robust nucleosome-dependent ATPase. However, ATPase activity was lost after size exclusion chromatography (Fig. S1C), and recombinant F-Alc1 lacked activity (Fig. 2A, compare lanes 3 and 5), suggesting a requirement for an activating factor or cofactor.
Fig. 2.
Alc1 has Parp1- and NAD-dependent ATPase. (A) ATPase assays performed with ≈100 ng wild-type or mutant recombinant (rec) F-Alc1 or F-Alc1 from HEK 293/FRT (293FRT) cells and 150 ng HeLa cell oligonucleosomes, with or without Parp1 and NAD. (B) ATPase assays performed with recombinant F-Alc1, with or without Parp1 and NAD, in the presence of DNA or an equimolar amount of mononucleosomes assembled on the same DNA with HeLa cell histones. (C) ATPase assays performed as in panel B with the indicated combinations of recombinant F-Alc1, Parp1, NAD, and oligonucleosomes.
Parp1 catalyzes nicotinamide adenine dinucleotide (NAD)-dependent mono- and PARylation of protein residues in a reaction strongly activated by Parp1 binding to DNA or nucleosomes (12–14). The major PAR acceptor in cells appears to be Parp1 itself; however, many other nuclear proteins, including histones, can be ADP-ribosylated. Biochemical studies have revealed that Parp1 can be incorporated into nucleosomes in place of histone H1 (15). Parp1 has been implicated in both transcriptional regulation and DNA damage repair in vivo (12, 14–17). In addition, Parp1 is localized to a large fraction of active promoters (18), and Parp1 and PAR accumulate at sites of DNA damage (14) in cells.
Our observation that the Alc1 macrodomain binds PAR, together with evidence from MudPIT mass spectrometry that anti-FLAG agarose eluates from F-Alc1 expressing HEK293/FRT cells contained substoichiometric amounts of Parp1, raised the possibility that addition of NAD and Parp1 to reactions might stimulate ATPase. Indeed, we observed that the ATPase activity of recombinant F-Alc1 was strongly stimulated by addition of Parp1 and NAD in the presence of either DNA or nucleosomes (Fig. 2B). ATPase was not activated in the absence of DNA or nucleosomes or when either NAD or Parp1 were omitted from reactions, suggesting Parp1-dependent PAR synthesis is required for the reaction (Fig. 2C). Consistent with this possibility, addition of poly(ADP-ribose) glycohydrolase (Parg), an enzyme known to catalyze the hydrolysis and breakdown of PAR (14), blocks activation of Alc1 ATPase by Parp1 and NAD (Fig. S2).
Suggesting a coupling of ATPase and PAR binding activities, we found that ATPase activity depends on an intact macrodomain. F-Alc1 (D723A), which does not bind PAR, lacks ATPase activity in either the presence or absence of Parp1 and NAD (Fig. 2A, compare lanes 6 and 10). PAR binding is not, however, sufficient to activate ATPase. Neither free PAR nor ADP-ribose activate Alc1 ATPase, even when present at concentrations (expressed in mole equivalents of adenosine) nearly 5 times higher than the maximal amount of poly(ADP-ribosyl)ated species that could be synthesized in reactions containing Parp1 and NAD (Table S2). Taken together, our data suggests that Alc1 ATPase activity depends on automodification of Parp1 and/or on PARylation of Alc1 itself. As discussed later, our data are most consistent with the former possibility.
Many Snf2 superfamily members, including Chd1, Iswi, and Ino80, can catalyze the ATP-dependent remodeling of nucleosomes in vitro (7, 19–21). To determine if Alc1 also has chromatin remodeling activity, we used a previously described assay (22–24) that takes advantage of the fact that DNA on the octamer surface is largely protected from cleavage by restriction enzymes, while DNA outside the nucleosome boundary is accessible.
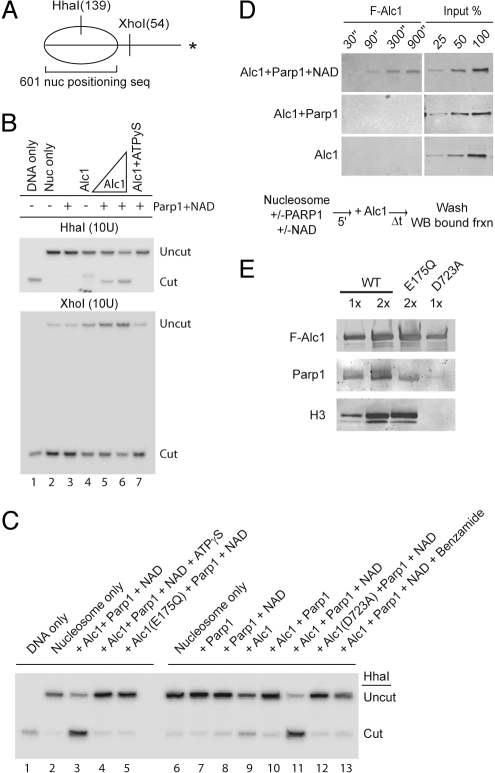
We assayed for nucleosome remodeling using mononucleosomes assembled from purified recombinant histones or HeLa oligonucleosomes on a 32P-end-labeled DNA probe containing a nucleosome positioning sequence (Fig. 3A) (25). The accessibility of a HhaI site, initially protected by the positioned nucleosome, is increased after incubation with recombinant F-Alc1, Parp1, and NAD. Arguing that Alc1 moves the nucleosome from its initial lateral position toward a more central position on the DNA, we observe a concomitant decrease in accessibility of an XhoI site outside the initial nucleosomal boundary (Fig. 3B). The DEAH box mutant F-Alc1 (E175Q) fails to remodel mononucleosomes (Fig. 3C). Additionally, nucleosome remodeling by F-Alc1 is inhibited by ATPγS (Fig. 3 B and C).
Fig. 3.
Alc1 has Parp1- and NAD-dependent nucleosome remodeling and binding activities. (A) Schematic showing location of positioned nucleosome (nuc) and length of HhaI and XhoI cleavage products. Asterisk, 32P-labeled DNA end. (B) DNA or nucleosomes reconstituted with HeLa cell histones were monitored for restriction enzyme accessibility after incubation with ATP (lanes 3–6) or ATPγS (lane 7) and Alc1, Parp1, and NAD as indicated. (C) DNA or nucleosomes reconstituted with recombinant histones were monitored for restriction enzyme accessibility after incubation with ATP (lanes 3, 5, 7–13) or ATPγS (lane 4) and wild-type or mutant Alc1, Parp1, NAD, or 2 mM benzamide. (D) Mononucleosomes reconstituted with HeLa cell histones on biotinylated DNA and immobilized on streptavidin beads were incubated for the indicated times with recombinant F-Alc1, with or without Parp1 and NAD. Bound fractions were analyzed by anti-Flag western blotting. (E) Whole cell lysates from HEK 293/FRT cells expressing wild-type or mutant F-Alc1 were immunoprecipitated with anti-FLAG (M2) agarose. Precipitated proteins were analyzed by western blotting.
Nucleosome remodeling activity depends strongly on Parp1 and NAD (Fig. 3 B and C) and is inhibited by benzamide, a potent inhibitor of Parp1 (Fig. 3C, lane 13). In addition, the macrodomain mutant F-Alc1(D723A), which exhibits reduced PAR binding, is inactive in our nucleosome remodeling assays (Fig. 3C, lane 12). To confirm further the association of ATPase and nucleosome remodeling activities with Alc1, we subjected anti-FLAG agarose eluates from F-Alc1 expressing HEK293/FRT cells to size exclusion chromatography. F-Alc1 and Parp1- and NAD-dependent ATPase and nucleosome remodeling activities co-eluted from the column as a mono-disperse peak (Fig. S1C). Taken together, our findings argue that Alc1 posseses ATP-dependent nucleosome remodeling activity and that nucleosome remodeling, like ATPase, is closely coupled to PAR binding.
Alc1, unlike other chromatin remodeling and modifying enzymes or complexes, lacks targeting domains, such as bromo- or chromodomains, that contribute to targeted recruitment to regions of specifically marked chromatin. However, our observation that Alc1 ATPase and chromatin remodeling activities require Parp1 and NAD raises the possibility that Alc1 could be targeted to chromatin by PARylation via its macrodomain. We tested this hypothesis using biochemical and in vivo assays.
First, we tested Alc1's ability to bind mononucleosomes formed on biotinylated DNA and immobilized on streptavidin beads (Fig. 3D). In the presence, but not in the absence, of Parp1 and NAD, Alc1 was rapidly recruited to nucleosomes and remained bound after extensive washing. In addition, we observed that F-Alc1 and the ATPase mutant F-Alc1(E175Q), but not the macrodomain mutant Alc1(D723A), could be co-purified from cell extracts with Parp1 and histones (Fig. 3E and Table S2). Thus, PARylation and an intact Alc1 macrodomain regulate binding of the Alc1 ATPase to nucleosomes.
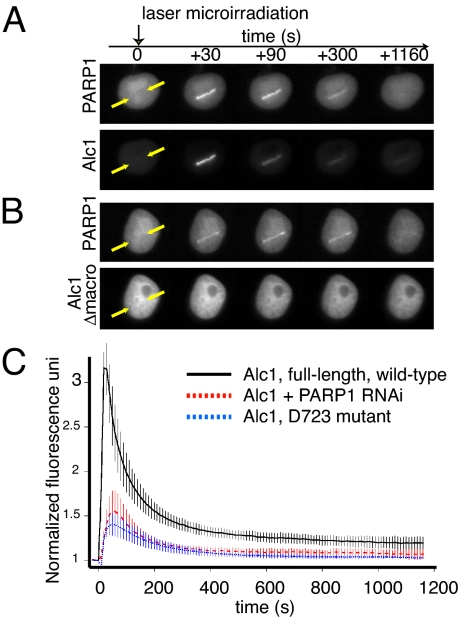
Second, we tested whether Alc1 is recruited to locally induced PARylation sites in living cells. We fused the full-length Alc1 cDNA to EYFP and used a pulsed-laser to microirradiate a small section of DNA in a human cell nucleus. The laser rapidly induces a highly localized region of DNA damage that recruits and enzymatically activates cellular PARP1 (26, 27). Parp1 and Alc1 are recruited rapidly to the microirradiated region. Alc1 and Parp1 fluorescence appears within seconds, and most is lost from the irradiated site within 10 min (Fig. 4A and C and Movie S1). Deletion of the macrodomain results in a complete loss of recruitment to the microirradiated region (Fig. 4B and Movie S2), while the macrodomain point mutant Alc1(D723A), which exhibits greatly reduced PAR binding in vitro, also exhibits reduced recruitment to the microirradiated region (Fig. 4C and Movie S3). Arguing that Alc1 recruitment requires the presence of Parp1 protein and PAR synthesis, we observed a substantial reduction in Alc1 recruitment when endogenous Parp1 was knocked down using short hairpin-mediated RNAi (Fig. 4C and Fig. S3) or in the presence of the Parp inhibitor PJ34 (Fig. S4). Thus, Parp1 and Alc1 are co-recruited to irradiation-induced sites of localized PAR synthesis in living cells, and Alc1 association with chromatin in vivo depends on an intact macrodomain.
Fig. 4.
Alc1 recruitment after microirradiation depends on its macrodomain and on PARP1 activity. Microirradiated HeLa cells were imaged for recruitment of EYFP-Alc1 wild-type or EYFP-Alc1Δmacrodomain (Δmacro) and PARP1-mCherry. (A) Recruitment of EYFP-Alc1 and PARP1-mCherry to site of microirradiation (between arrows). (B) Loss of Alc1's macrodomain abrogates PARylation-induced recruitment of Alc1 to chromatin. The background in Alc1 images is lower because the integration time of the CCD camera was lower to allow accurate quantitation of the recruitment kinetics. (C) Kinetics of recruitment (n ≥ 6) to microirradiated sites of wild-type (black) and D723A macrodomain mutant (blue) Alc1, or recruitment of wild-type Alc1 after Parp1 knockdown (red).
In summary, our in vivo results indicate that a Parp1-dependent PARylation event directs the recruitment of Alc1 to chromatin in cells. Further, our biochemical assays reveal that Parp1-dependent PARylation promotes the recruitment of Alc1 to nucleosomes and activates its associated ATPase and chromatin remodeling activites. Upon binding to DNA or chromatin, Parp1 can catalyze local PARylation of chromatin associated proteins, including histones; however the primary PAR acceptor in vitro and in cells is Parp1 itself (12, 15, 28, 29). While we cannot rule out the possibility that modification of histones or other proteins contributes to recruitment of Alc1, our biochemical data are most consistent with the model that automodification of Parp1 represents the key PARylation event for Alc1 activation. Indicating that poly(ADPribosyl)ation of histones is not required, Parp1, NAD, and DNA are sufficient to activate Alc1 ATPase activity (Fig. 2B). To address the alternative possibility that modification of Alc1 leads to its activation, we performed order of addition experiments using the Parp1 inhibitor benzamide (30). When added at the beginning of the reaction, benzamide blocked nucleosome remodeling (Fig. 3C, compare lane 13 to lanes 3 and 11); however, when Parp1 was preincubated with nucleosomes and NAD before addition of benzamide and Alc1, robust chromatin remodeling activity was detected (Fig. S2B), suggesting that the essential PARylation events occur before Alc1 addition.
It remains to be determined whether the apparent PARylation-dependent increase in the affinity of Alc1 for nucleosomes is sufficient to explain the activation of its ATPase and nucleosome remodeling activities in the presence of Parp1 and NAD. It will be of interest to determine whether binding of a PARylated species, most likely Parp1 itself, to the Alc1 macrodomain results in allosteric activation of the enzyme.
Our in vivo assays take advantage of the ability to induce DNA breaks by pulsed-laser microirradiation, resulting in Parp1-dependent PAR synthesis and consequent Alc1 recruitment at a discrete nuclear location. In DNA damage repair, Parp1 is thought to bind and be allosterically activated by DNA ends. Parp1 can also be activated by other mechanisms, including interaction with the signaling kinase ERK2 (31) or binding to DNA hairpin and other unbroken DNA structures (32, 33), transcription regulatory proteins (17), and nucleosomes (15). In addition, Parp1 has been recently shown to be localized to many promoters and to contribute to transcriptional regulation (18). Thus, our data opens the possibility that Parp1-activated nucleosome remodeling by Alc1 could contribute to the control of chromatin structure during DNA repair, transcription, or other processes requiring Parp1. Future experiments will be necessary to illuminate the precise role of Alc1 in these processes.
Materials and Methods
Purification of Flag-Alc1.
For expression in human cells, Alc1 cDNA (accesssion no. BC001171) was cloned into pcDNA5 with an N-terminal FLAG tag and introduced into HEK293/FRT cells as described (34). Cells were grown to 70–80% confluence. Nuclear extracts were prepared according to the method of Dignam et al. (35), and FLAG-Alc1 and associated proteins were purfied on anti-FLAG (M2) agarose beads (Sigma) as described (34). Alternatively, whole cell lysates were prepared as described in SI Methods, and Flag-Alc1 and associated proteins were immunopurified as described (34), except beads were washed with 0.2 M KCl. For expression in Sf21 insect cells, Flag-Alc1 was cloned into a pBacPAK8 (Clontech) derivative, and purified from lysates of infected cells as described (36).
Poly(ADP)ribose Binding Assays.
Recombinant proteins (1 pmol) were incubated for 30 min at 32 °C in 15 μL 40 mM HEPES-NaOH, pH 7.9, 0.1 M NaCl, 0.1 mM EDTA, 10% glycerol, and 32P-labeled PAR purified as described (8). Reaction mixtures were applied to nitrocellulose and washed overnight with TBS-T containing 100 mM NaCl. Bound 32P-labeled PAR was detected using a Typhoon phosphorimager.
ATPase Assays.
ATPase assays were performed as described (34). Where indicated, reaction mixtures contained ≈100 ng (1 pmol) Flag-Alc1 (wild-type, E175Q, or D723A) from HEK293/FRT cells or SF9 cells, ≈115 ng (1 pmol) Parp1 (Trevigen), recombinant Parg (Trevigen), 34 μM nicotine adenine dinucleotide, and 150 ng mono- or oligonucleosomes from HeLa cells (37).
Nucleosome Remodeling Assays.
Mononucleosomes were reconstituted by dilution transfer from HeLa oligonucleosomes on a 32P-end-labeled 216-bp DNA fragment (601-lat Gal4) generated by PCR from pGEM3Z-601-Gal4 (37, 38). F-Alc1 (1 pmol) from HEK293/FRT or SF9 cells was incubated at 32 °C for 30 min with mononucleosomes (≈0.01 pmol labeled mononucleosome, ≈0.25 pmol unlabeled oligonucleosomes) in 20 mM HEPES-NaOH, pH 7.9, 50 mM NaCl, 4.5 mM MgCl2, 2 mM DTT, 0.5 mM PMSF, 45 μg/mL BSA, 10% glycerol, 0.02% Triton X-100, 0.02% Nonidet P-40, and 2 mM ATP. Where indicated, reactions contained 2 mM ATPγS, 1 pmol Parp1, 34 μM NAD, or 2 mM benzamide. Reaction products were incubated for a further 30 min with 10 U of either HhaI or XhoI and resolved on gels containing 7% polyacrylamide (19:1 acrylamide:bis), 7 M urea, and 45 mM Tris-borate/1 mM EDTA, pH 8.3 (39).
Nucleosome Binding Assay.
Mononucleosomes (40 pmol) were assembled on a 5′-biotinylated 601-lat Gal4 fragment, bound to 400 μL streptavidin dynabeads, washed, and resuspended in a final volume of 400 μL (100 fmol mononucleosome/μL beads). Recombinant F-Alc1 (1 pmol) was incubated with 100 fmol immobilized nucleosomes in 45 μL 20 mM HEPES, pH 7.9, 50 mM NaCl, 0.5% Nonidet P-40, 10% glycerol, 5 mM MgCl2, 1 mM DTT, 0.5 mM PMSF, 1 mM ATP, and 100 μg/mL BSA. Where indicated 1 pmol Parp1 and NAD (34 μM) were included in reaction mixtures. Beads were washed 3 times with 200 μL 40 mM HEPES-NaOH, pH 7.9, 0.2 M NaCl, 0.2% Triton X-100, and 10% glycerol, transferred to a fresh microcentrifuge tube, and bound proteins were eluted with 3× SDS sample buffer and analyzed by western blot.
Transient Transfections.
HeLa-Kyoto and AGS cells were grown in HEPES-buffered DMEM-Glutamax-I (Invitrogen) containing 4.5 g/L glucose and 10% FCS US/certified (Invitrogen) and supplemented with 50 U/mL penicillin, 50 μg/mL streptomycin (Invitrogen) and MEM-nonessential amino acids (MEM NEAA; Invitrogen). AGS cells stably expressing scrambled or 2 different short hairpin RNAs targeting PARP1 were generated at the Institute of Veterinary Biochemistry and Molecular Biology (IVBMB) using a shRNA SIN-lentivirus approach. Wild-type and mutant ALC1 cDNAs were amplified by PCR and cloned into the BglII and EcoRI sites of pEYFP-C1 (Clontech) for expression of EYFP-Alc1. PARP1 cDNA was amplified by PCR and introduced into the NheI and SmaI sites of pmCherry-N1 for expression of Parp1-mCherry. For pulsed-laser microirradiation experiments, AGS cells were grown without puromycin. Where indicated, 1 μM PARP inhibitor PJ-34 (Alexis) was added 30 min before laser microirradiation.
Pulsed Laser Microirradiation, Live Imaging, and Image Analysis.
Pulsed laser microirradiaton was performed through a Zeiss C-Apo 63×/1.2 water immersion objective lens on a Zeiss Axiovert 200M epifluorescence microscope equipped with a frequency tripled 355 nm Nd:YAG pulsed laser (JDS Uniphase), scanned with galvo mirrors (40) and an ORCA CCD camera (Hamamatsu Photonics KK). DNA damage was induced by focusing in the nucleus an ≈6–8 μm line target including 40–42 points with a pulse energy of 200–300 nJ for 3 times. Cells were imaged every 10 s for 20 min. Cells were kept at 37 °C in a CO2 independent HEPES-based imaging medium (Invitrogen) supplemented with 20% FBS (Invitrogen), 1 mM sodium pyruvate (Sigma), 2 mM L-glutamine (Sigma), 50 U/mL penicillin, 50 μg/mL streptomycin (Sigma) in MatTek glass bottom dishes. Live images were registered and analyzed using ImageJ. Igor Pro (WaveMetrics) was used for analyzing and plotting the data. Cell motions were corrected using ImageJ plug-in MultiStackReg (41). To quantify protein recruitment following laser microirradiation, data were background-subtracted, normalized to premicroirradiation, and corrected for fluorescence loss: R(t) = [(I(t) − Iback(t))/(I(t0) − Iback(t0))]*[(T(to) − Iback(t0))/(T(t) − Iback(t))], where R is recruitment, I is the intensity acquired along the laser path region, Iback is the background region outside the cell of interest, and T is the total fluorescence within the nucleus.
Supplementary Material
Acknowledgments.
We thank Kym Delventhal and Paul Hassa for molecular biology assistance, Tingting Yao for helpful discussions, and Julien Colombelli, Ernst Stelzer, and Jan Ellenberg for advice with live cell imaging. This work was supported by National institutes of Health General Medical Sciences Grant R37 GM41628 (to R.C.C.) and the Stowers Institute and by the European Molecular Biology Laboratory (EMBL) and the Human Frontier Science Program (A.L. and G.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906920106/DCSupplemental.
References
- 1.Kim J, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horn PJ, Peterson CL. The bromodomain: A regulator of ATP-dependent chromatin remodeling? Front Biosci. 2001;6:D1019–D1023. doi: 10.2741/horn. [DOI] [PubMed] [Google Scholar]
- 3.Brehm A, Tufteland KR, Aasland R, Becker PB. The many colours of chromodomains. Bioessays. 2004;26:133–140. doi: 10.1002/bies.10392. [DOI] [PubMed] [Google Scholar]
- 4.Daniel JA, Pray-Grant MG, Grant PA. Effector proteins for methylated histones: an expanding family. Cell Cycle. 2005;4:919–926. doi: 10.4161/cc.4.7.1824. [DOI] [PubMed] [Google Scholar]
- 5.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Two related superfamilies of putative helicases involved in replication, recombination, repair, and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karras GI, et al. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchio A, et al. Recurrent chromosomal abnormalities in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer. 1997;18:59–65. [PubMed] [Google Scholar]
- 10.Wong N, et al. Positional mapping for amplified DNA sequences on 1q21–q22 in hepatocellular carcinoma indicates candidate genes over-expression. J Hepatol. 2003;38:298–306. doi: 10.1016/s0168-8278(02)00412-9. [DOI] [PubMed] [Google Scholar]
- 11.Ma NF, et al. Isolation and characterization of a novel oncogene, amplified in liver cancer 1, within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology. 2008;47:503–510. doi: 10.1002/hep.22072. [DOI] [PubMed] [Google Scholar]
- 12.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 13.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 14.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: Where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 17.Kraus WL. Transcriptional control by PARP-1: Chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishnakumar, et al. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 19.Tran HG, Steger DJ, Iyer VR, Johnson AD. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 2000;19:2323–2331. doi: 10.1093/emboj/19.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corona DF, et al. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. doi: 10.1016/s1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- 21.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 22.Polach KJ, Widom J. Restriction enzymes as probes of nucleosome stability and dynamics. Methods Enzymol. 1999;304:278–298. doi: 10.1016/s0076-6879(99)04017-3. [DOI] [PubMed] [Google Scholar]
- 23.Anderson JD, Thastrom A, Widom J. Spontaneous access of proteins to buried nucleosomal DNA target sites occurs via a mechanism that is distinct from nucleosome translocation. Mol Cell Biol. 2002;22:7147–7157. doi: 10.1128/MCB.22.20.7147-7157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 25.Anderson JD, Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol. 2000;296:979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- 26.Dantzer F, et al. Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006;409:493–510. doi: 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- 27.Kirsten E, Kun E, Mendeleyev J, Ordahl CP. Activity assays for poly-ADP ribose polymerase. Methods Mol Biol. 2004;287:137–149. doi: 10.1385/1-59259-828-5:137. [DOI] [PubMed] [Google Scholar]
- 28.Huletsky A, et al. The effect of poly(ADP-ribosyl)ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure. J Biol Chem. 1989;264:8878–8886. [PubMed] [Google Scholar]
- 29.Ogata N, Ueda K, Kawaichi M, Hayaishi O. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J Biol Chem. 1981;256:4135–4137. [PubMed] [Google Scholar]
- 30.Purnell MR, Whish WJ. Novel inhibitors of poly(ADP-ribose) synthetase. Biochem J. 1980;185:775–777. doi: 10.1042/bj1850775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen-Armon, et al. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: A link to histone acetylation. Mol Cell. 2007;25:297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Lonskaya I, et al. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J Biol Chem. 2005;280:17076–17083. doi: 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- 33.Soldatenkov VA, et al. Transcriptional repression by binding of poly(ADP-ribose) polymerase to promoter sequences. J Biol Chem. 2002;277:665–670. doi: 10.1074/jbc.M108551200. [DOI] [PubMed] [Google Scholar]
- 34.Cai Y, Jin J, Gottschalk AJ, Yao T, Conaway JW, Conaway RC. Purification and assay of the human INO80 and SRCAP chromatin remodeling complexes. Methods. 2006;40:312–317. doi: 10.1016/j.ymeth.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian cell nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong SE, Banks CAS, Shilatifard A, Conaway JW, Conaway R. ELL-associated factors 1 and 2 are positive regulators of RNA polymerase II elongation factor ELL. Proc Natl Acad Sci USA. 2005;102:10094–10098. doi: 10.1073/pnas.0503017102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owen-Hughes T, et al. Analysis of nucleosome disruption by ATP-driven chromatin remodeling complexes. Methods Mol Biol. 1999;119:319–331. doi: 10.1385/1-59259-681-9:319. [DOI] [PubMed] [Google Scholar]
- 38.Gutierrez JL, Chandy M, Carrozza MJ, Workman JL. Activation domains drive nucleosome eviction by SWI/SNF. EMBO J. 2007;26:730–740. doi: 10.1038/sj.emboj.7601524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Colombelli J, Grill SW, Stelzer EHK. UV diffraction limited nanosurgery of live biological tissues. Rev Sci Instr. 2004;75:472–478. [Google Scholar]
- 41.Thévenaz P, Ruttiman UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.