Abstract
Background
Phylogenetic analyses of the Annonaceae consistently identify four clades: a basal clade consisting of Anaxagorea, and a small 'ambavioid' clade that is sister to two main clades, the 'long branch clade' (LBC) and 'short branch clade' (SBC). Divergence times in the family have previously been estimated using non-parametric rate smoothing (NPRS) and penalized likelihood (PL). Here we use an uncorrelated lognormal (UCLD) relaxed molecular clock in BEAST to estimate diversification times of the main clades within the family with a focus on the Asian genus Pseuduvaria within the SBC. Two fossil calibration points are applied, including the first use of the recently discovered Annonaceae fossil Futabanthus. The taxonomy and morphology of Pseuduvaria have been well documented, although no previous dating or biogeographical studies have been undertaken. Ancestral areas at internal nodes within Pseuduvaria are determined using dispersal-vicariance analysis (DIVA) and weighted ancestral area analysis (WAAA).
Results
The divergence times of the main clades within the Annonaceae were found to deviate slightly from previous estimates that used different calibration points and dating methods. In particular, our estimate for the SBC crown (55.2-26.9 Mya) is much younger than previous estimates (62.5-53.1 ± 3.6 Mya and ca. 58.76 Mya). Early diversification of Pseuduvaria was estimated to have occurred 15-8 Mya, possibly associated with the 'mid-Miocene climatic optimum.' Pseuduvaria is inferred to have originated in Sundaland in the late Miocene, ca. 8 Mya; subsequent migration events were predominantly eastwards towards New Guinea and Australia, although several migratory reversals are also postulated. Speciation of Pseuduvaria within New Guinea may have occurred after ca. 6.5 Mya, possibly coinciding with the formation of the Central Range orogeny from ca. 8 Mya.
Conclusion
Our divergence time estimates within the Annonaceae are likely to be more precise as we used a UCLD clock model and calibrated the phylogeny using new fossil evidence. Pseuduvaria is shown to have dispersed from Sundaland after the late Miocene. The present-day paleotropical distribution of Pseuduvaria may have been achieved by long-distance dispersal, and speciation events might be explained by global climatic oscillations, sea level fluctuations, and tectonic activity.
Background
The Annonaceae are a large pantropical family of flowering plants, consisting of ca. 135 genera and ca. 2,500 species in predominantly tropical and subtropical lowland forests [1]. The phylogeny of the family has previously been reconstructed based on morphological [2-4] and molecular data [5-7]. Four main clades are consistently recognised in the molecular analyses: two of these clades (consisting of Anaxagorea and the small 'ambavioid' clade) form a heterogeneous basal grade, basal to two large clades known as the 'long branch clade' (LBC) and 'short branch clade' (SBC) to reflect differing rates of nucleotide substitutions [7,8].
Evolutionary divergence times based on molecular data have been estimated for the Annonaceae as a whole [5,7] as well as for several individual genera and clades, including Anaxagorea [9], 'Andean-centred' genera in the SBC [8], Guatteria [10], and an African clade including Isolona and Monodora [11]. These studies were based on between two and five commonly used chloroplast regions (matK, trnL-F, trnT-L, rbcL, and psbA-trnH spacer). Comparatively few fossil calibration points were used in these studies due to the scarcity of unequivocal Annonaceae fossils. The most widely applied calibration point is the stem age of the Magnoliaceae, based on the fossil Archaeanthus from North America, which either provides a minimum age of 98 Mya in the early Cenomanian [12], or 100 Mya in the late Albian [13]. Other possible calibration points include: 68 Mya for the split between the ambavioids and the combined LBC-SBC clade, derived using Anonaspermum seeds from Nigeria [14]; 112 Mya for the split between the Eupomatiaceae and Annonaceae, using the fossil Endressinia [15]; and 120 Mya for core Magnoliales, based on Aptian granular monosulcate pollen [16]. In addition, secondary calibration points have been derived from earlier studies of divergence times, including 82 Mya [17] and 90.93 Mya (unpublished estimate by M.D. Pirie) for the stem of the Annonaceae. Richardson et al. [7] have accordingly estimated the Annonaceae stem age at 90.6 ± 1.3 Mya, based on three calibration points [7]. The age estimates for the family have enabled significant palaeobiogeographical insight, although the most important work has focused on dispersal patterns in Africa and South America (western Gondwana) since the genera studied were largely neotropical.
The program r8s [18] has been widely adopted in studies of divergence times in the Annonaceae [7,9,10]. This program incorporates both non-parametric rate smoothing (NPRS) methods and penalized likelihood (PL) methods. NPRS methods estimate ages through a smoothing criterion [19], whereas PL is a semiparametric approach [20] that combines parametric methods with the robustness of non-parametric methods. Other dating methods utilized in studies of the Annonaceae [21] include PAML [22] and MULTIDIVTIME [23], based on Bayesian dating methods. BEAST (Bayesian Evolutionary Analysis Sampling Trees) [24] is the only software that simultaneously co-estimates phylogeny, node ages and substitution rates. BEAST has recently been used to estimate the origins of the East African lineages within the Annonaceae [11].
The present study focuses on the palaeotropical genus Pseuduvaria, which is nested within the SBC. The genus consists of 56 species of trees and treelets (inclusive of Craibella phuyensis, which has recently been shown to be congeneric with Pseuduvaria [25,26], and three newly described species [26]). A comprehensive monograph of the genus has been completed [27] and the phylogeny of Pseuduvaria has been inferred using five chloroplast regions [25]. The genus was previously estimated to have evolved at least ca. 16 Mya ago based upon a published chronogram [7], although this may not represent the true age of the genus since the study included only five Pseuduvaria species (inclusive of C. phuyensis).
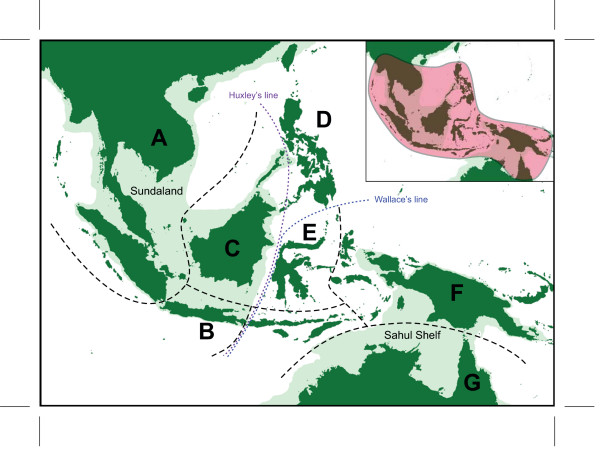
The Malesian phytogeographical region is separated into two main subregions by Huxley's line and Wallace's line (Figure 1: [28]): western Malesia, which includes Peninsular Malaysia, Sumatra, Java, Borneo and Palawan; and eastern Malesia, which includes Sulawesi, the Lesser Sunda Islands, the Moluccas, and New Guinea. Borneo and New Guinea represent the two most important centres of plant species richness and endemism within Malesia [29]. Pseuduvaria is widely distributed from Indochina to New Guinea and NE Australia (Figure 1, inset), with the main centre of diversity in New Guinea (with 20 species) and a secondary centre in Peninsular Malaysia (with 16 species, including three recently described species) [26]. In an earlier molecular phylogenetic study of Pseuduvaria, five clades were recognized, with the three basal clades occurring in the western Malesia [25]. A continuous land mass connecting Indochina, Thailand, Peninsular Malaysia, Borneo, Sumatra, and Java (collectively known as Sundaland) was formed during the Early Mesozoic [30], separated from the Australian-New Guinea plate by a major ocean barrier. The collision between the Asian and New Guinea land masses during the Cenozoic may have promoted the dispersal of Pseuduvaria species from Sundaland to New Guinea. The present paper aims to use molecular dating techniques to test whether this biogeographical hypothesis is supported by evidence of past geological events.
Figure 1.
Main biogeographical areas used in analyses, and (inset) distribution of Pseuduvaria in SE Asia. Eight areas are outlined and labelled: A, China, Indochina, Burma, Thailand, Peninsular Malaysia and Sumatra; B, Java and Bali; C, Borneo and Palawan; D, Philippines (excluding Palawan); E, Sulawesi; F, New Guinea; and G, Australia. The shaded light green area represents shallow continental shelves, as modified from [28].
We use Bayesian molecular dating techniques in BEAST to estimate the divergence times of major Annonaceae clades (including the date of origin of Pseuduvaria as well as clades within Pseuduvaria). Five chloroplast DNA regions, psbA-trnH spacer, trnL-F, matK, rbcL and atpB-rbcL spacer, were used in all analyses. The divergence times of major Annonaceae clades were estimated using a dataset ('matrix A') with a broad taxonomic sampling across the basal grade, LBC and SBC of the Annonaceae, together with representatives of related families in the Magnoliales. Two fossil calibration points were used to date the divergence times within the Annonaceae: the fossil Archaeanthus [12], providing a minimum age of 98 Mya for the stem of the Magnoliaceae (J.A. Doyle, pers. comm.); and the Late Cretaceous fossil Futabanthus, providing a minimum age of 89 Mya for the split between Anaxagorea and the combined ambavioid-LBC-SBC clade [31]. The ages of clades within Pseuduvaria were subsequently estimated using a smaller dataset ('matrix B') that included all Pseuduvaria species available (54 species) and selected members of the SBC. The age estimates inferred from BEAST analyses using matrix A were used as prior information in the subsequent analyses using matrix B. An additional objective was to identify the most likely biogeographical origin of Pseuduvaria, and to infer subsequent dispersal patterns using dispersal-vicariance analysis (DIVA) [32] and weighted ancestral area analysis (WAAA) [33].
Methods
Taxon sampling and sequence data
We used two separate datasets: matrix A (for higher-level analyses) and matrix B (for species-level analyses within Pseuduvaria). Both sets of analyses were based on chloroplast DNA sequences from five regions (psbA-trnH spacer, trnL-F, matK, rbcL and atpB-rbcL spacer). The materials and methods for DNA extraction, PCR amplification and sequencing of Pseuduvaria are detailed in earlier publications [25,26]. The sequences for the outgroups were downloaded from GenBank. The accession numbers for all sequences are presented in Additional File 1.
Matrix A comprised a total of 83 species. The ingroup included 22 of the total 56 Pseuduvaria species, representing all five main clades identified in our previous phylogenetic study [25], and 56 species from 46 different Annonaceae genera (inclusive of the basal grade, LBC and SBC). The outgroups included were Coelocaryon preussii (Myristicaceae), Eupomatia bennettii (Eupomatiaceae), Liriodendron chinense (Magnoliaceae), Magnolia kobus (Magnoliaceae), and Persea americana (Lauraceae). Matrix B consisted of 69 taxa, with 54 Pseuduvaria species. The selection of outgroups was based on the phylogeny generated by BEAST using matrix A (Figure 2A). The SBC genera selected as outgroups were Alphonsea, Haplostichanthus, Miliusa, Monocarpia, Neo-uvaria, Orophea, Polyalthia, and Sapranthus, representing 15 species.
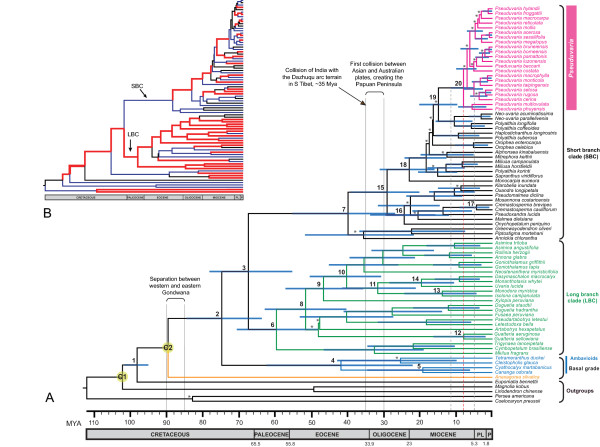
Figure 2.
Chronograms of Annonaceae: maximum clade credibility trees from the BEAST analysis of matrix A. (A). Posterior estimates of divergence times were inferred using partitioned analyses based on five chloroplast regions, a UCLD model, and two fossils as minimum age constraints: an Archaeanthus fossil date of 98 Mya (calibration C1), and a Futabanthus fossil date of 89 Mya (calibration C2). Nodes are posterior mean ages (Mya), with blue node bars representing the 95% HPD intervals (see Table 2 for details). Bayesian PP < 0.95 are indicated by asterisks above branches. Geological time-scale abbreviations: PL, Pliocene; P, Pleistocene. The red dashed line represents the estimated tMRCA of Pseuduvaria crown, with a 95% HPD indicated by the grey vertical lines. (B). Rate of evolution across all branches. Broad red lines indicate lineages where the posterior rate is greater than the mean rate; narrow blue lines indicate lineages where the posterior rate is lower than the mean rate.
For matrices A and B, sequences were edited and assembled in SeqManPro using DNAStar Lasergene 8 (DNAStar), and aligned manually using BioEdit ver. 7.09 [34] and Se-Al ver. 2.0a11 [35]. Ambiguously aligned positions were excluded from the analyses. The combined five-region data sets in matrices A and B are composed of 4860 and 4361 characters, respectively (see Table 1 for data on individual gene regions).
Table 1.
Length and best-fitting model for data partition analyses based on two different datasets (matrices A and B).
| Aligned length (bp) | Sequence evolutionary model | |||
| Matrix A | Matrix B | Matrix A | Matrix B | |
| psbA-trnH spacer | 572 | 441 | GTR+G | GTR+G |
| trnL-F | 1064 | 937 | GTR+G | GTR+I |
| matK | 823 | 811 | GTR+G | GTR+G |
| rbcL | 1344 | 1344 | GTR+I+G | HKY+I+G |
| atpB-rbcL spacer | 1057 | 828 | GTR+G | GTR+I |
| Combined data | 4860 | 4361 | GTR+I+G | GTR+I+G |
GTR, general time-reversible model; HKY, the Hasegawa, Kishino, and Yano model; I, proportion of invariant sites; G, gamma distribution.
Divergence time estimation
MrModelTest ver. 2.3 [36] was used to determine the appropriate DNA substitution model and gamma rate heterogeneity using the Akaike Information Criterion (AIC). For both matrices, MrModelTest was performed for each gene region and combined regions (Table 1). For the combined five chloroplast DNA regions in matrices A and B, the GTR+I+G was determined as the best-fitting statistical model.
The tree topology, node ages and substitution rates were simultaneously estimated using a Bayesian MCMC (Markov chain Monte Carlo) approach as implemented in BEAST ver. 1.4.8 [24,37]. For matrix A analyses, exponential priors were selected for the two calibration points (see discussion below): the Archaeanthus calibration used 98 Mya (labelled C1 in Figure 2A) as zero offset, with an exponential mean of 1; and the Futabanthus calibration used 89 Mya (labelled C2 in Figure 2A) as zero offset, with an exponential mean of 1. A Yule speciation tree prior was furthermore specified; this prior has been recommended for species-level phylogenies, and assumes a constant rate of speciation per lineage [24]. An uncorrelated lognormal distributed relaxed clock (UCLD) model was employed, which allows evolutionary rates to vary along branches within lognormal distributions [38]. The five chloroplast regions were partitioned (see Table 1 for the different models of substitution), allowing the incorporation of different models of substitution for each gene independently; the xml input file based on matrix A was edited manually for gene region partition following the tutorial by T.L.P. Couvreur [11,39].
Two independent MCMC runs were performed, each run of 50 million generations, with sampling every 5000 generations. The two separate runs were then combined (following the removal of 10% burn-in) using LogCombiner ver. 1.4.8 [24,37]. Adequate sampling and convergence of the chain to stationary distribution were confirmed by inspection of MCMC samples using Tracer ver. 1.4 [40]. The effective sample size (ESS) values of all parameters were greater than 200, which was considered a sufficient level of sampling. The sampled posterior trees were summarized using TreeAnnotator ver. 1.4.8 [24,37] to generate a maximum clade credibility tree (maximum posterior probabilities) and calculate the mean ages, 95% highest posterior density (HPD) intervals, posterior probabilities (PP) and substitution rates for each node. The BEAST topology (Figure 2A) was visualized with FigTree ver. 1.2 [41]. The MCMC analysis based on matrix A was also performed without sequence data; the prior distribution can therefore be compared with the posterior distribution in order to examine the influence of the data and prior, showing that the results are not influenced by the chosen priors alone [38].
The time to most recent common ancestor (tMRCA) of Monocarpia and Pseuduvaria inferred from the MCMC runs of matrix A were used as prior information in the subsequent BEAST analyses using matrix B in order to estimate the divergence times within Pseuduvaria lineages. A normal probability distribution was employed as priors for matrix B analyses, since the 95% HPD intervals from previous estimation (based on matrix A) can also be accommodated as uncertainty for this analysis. The normal probability distribution is thought to reflect uncertainty in secondary calibration points [11,42,43]. Normal distribution priors were therefore applied to calibration points at two nodes: the MRCA of Monocarpia, with a mean age of 23 Mya and a standard deviation of 3.78 Mya (labelled C3 in Figure 3); and the MRCA of Pseuduvaria, with a mean age of 8 Mya and a standard deviation of 1.64 Mya (labelled C4 in Figure 3). The xml input file based on matrix B was edited manually for partitioned chloroplast sequence data. A UCLD model with Yule speciation tree prior was employed. Two independent MCMC runs of 30 million generations were performed, with sampling every 3000 generations. The two separate runs were then combined (following removal of 10% burn-in) using LogCombiner ver. 1.4.8 [24,37]. The runs were checked for convergence using Tracer ver. 1.4 [40], and the ESS values of all parameters shown to be greater than 500, which was considered sufficient. The BEAST topology (Figure 3) was summarized and visualized as described above.
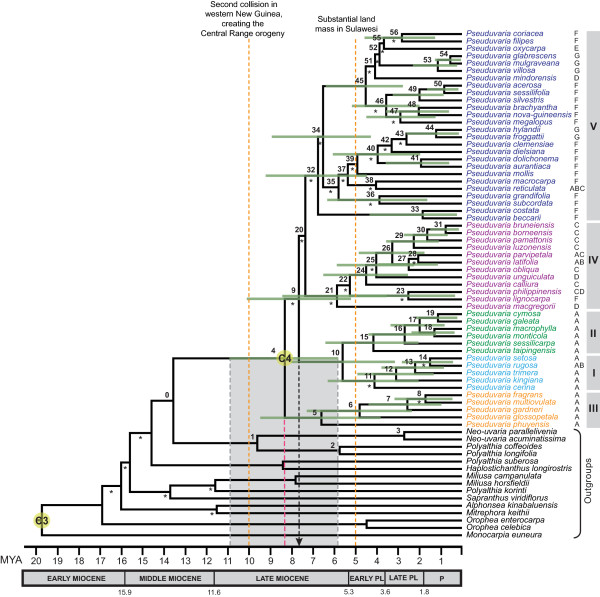
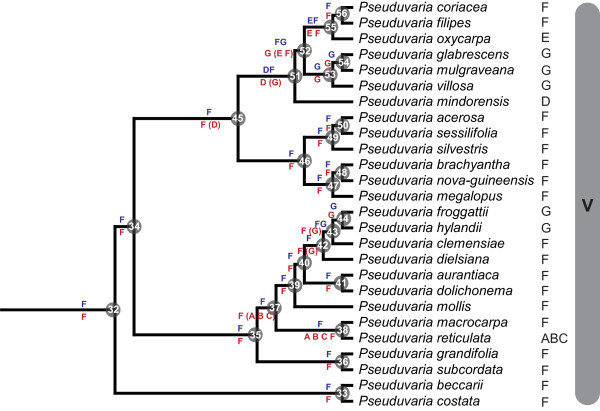
Figure 3.
Chronogram of Pseuduvaria: maximum clade credibility tree from the BEAST analysis of matrix B. Posterior estimates of divergence times were inferred using partitioned analyses based on five combined chloroplast regions, a UCLD model, and two secondary calibration points (labelled C3 and C4). Nodes are posterior mean ages (Mya), with green node bars representing the 95% HPD intervals (see Table 3 for details). Bayesian PP < 0.95 are indicated by asterisks below branches. Letters to the right of taxon names represent current geographical distributions (defined in Figure 1). Geological time scale abbreviations: PL, Pliocene; P, Pleistocene. The red dashed line represents the estimated tMRCA of Pseuduvaria crown, with a 95% HPD indicated by the grey bar; black dashed line represents the earliest possible migration event for Pseuduvaria out of Sundaland.
Fossil calibrations
The most commonly used calibration point in previous studies of the Annonaceae has been the fossil Archaeanthus [7,8] which provides an age of 98 Mya for the stem of the Magnoliaceae. Other calibration points previously employed for the Annonaceae are 120 Mya for the node of the core Magnoliales using the pollen fossil Lethomasites [9], 112 Mya for the split between the Eupomatiaceae and Annonaceae using the Endressinia fossil [15], and 68 Mya for the common ancestor of the ambavioids and combined LBC-SBC clade using the Maastrichtian seed genus Anonaspermum from Nigeria and Sierra Leone [14].
Lethomasites was not used in the present study since its placement within the Magnoliales is doubtful [5]. The inclusion of Lethomasites would also require broader taxonomic sampling of other families within the Magnoliales (including Degeneriaceae and Himantandraceae), and the large resultant dataset would require considerably longer computational time. The Endressinia fossil was not reliable as its relationship with the Eupomatiaceae is doubtful [11]. The Anonaspermum seeds were similarly avoided as calibration points in the present study as the position of this fossil within the Annonaceae is uncertain [44].
The inclusion of a greater number of fossil calibration points can reduce bias and result in more accurate age estimates, assuming that the chosen points are mutually consistent [45]. The fossil record of the Annonaceae is very limited, however, and only two fossils, Archaeanthus and Futabanthus, were regarded as sufficiently reliable. The phylogenetic position of Archaeanthus [12] appears to be reliable as it shares several characters with the Magnoliaceae, including distinctive stipules, elongated receptacle and fruits with numerous follicles [5,11,12]. The phylogenetic position of the recently discovered Late Cretaceous fossil flower, Futabanthus asamigawaensis [31], is similarly reliable as it shares many morphological similarities with the Annonaceae, including hermaphroditic, hypogynous flowers with six tepals in two whorls of three, an androecium consisting of numerous stamens with flattened connectives that extend above the thecae, and a gynoecium of numerous free carpels. Takahashi et al. [[31]: 914] postulated that Futabanthus has "a position within crown-group Annonaceae, perhaps as sister or near the base of all extant taxa except Anaxagorea." Inner staminodes are absent in Futabanthus but are present in the basal genus Anaxgaorea; the lack of inner staminodes is therefore possibly synapomorphic for the rest of the Annonaceae (plesiomorphic in Anaxagorea). We set Futabanthus as a minimum age constraint of 89 Mya for the split between Anaxagorea and the combined ambavioid-LBC-SBC clade (i.e., the crown age of the Annonaceae, labelled as C2 in Figure 2A).
Previous dating studies furthermore utilized secondary calibration points that were extracted from earlier estimates, viz.: the age of 82 Mya for the stem of the Annonaceae based on Wikström et al. [17] using the NPRS method [7,10]; and the age of 90.93 Mya for the stem of the Annonaceae based on unpublished estimations by M.D. Pirie. Secondary calibration points generated by these previous Annonaceae studies were avoided as a different molecular dating method and calibration points were employed in this study.
Historical biogeography
Ancestral biogeographical areas occupied by Pseuduvaria were inferred using two methods: dispersal-vicariance analysis (DIVA) [32] and weighted ancestral area analysis (WAAA) [33]. Eight main biogeographical areas represented in the current distributions of Pseuduvaria species were used for the ancestral area reconstructions (Figure 1): (A) southern China, Indochina, Thailand, Burma, Peninsular Malaysia, and Sumatra; (B) Java and Bali; (C) Borneo and Palawan (identified as a single biogeographical unit following [46]); (D) Philippines (excluding Palawan); (E) Sulawesi; (F) New Guinea and the Moluccas; (G) Australia; and also the outgroup distribution (H) India and Sri Lanka (not shown in Figure 1). Species distribution data were derived from our monograph of Pseuduvaria [27].
DIVA was implemented using ver. 1.2 of the program [32], in which ancestral distributions are inferred by minimizing the number of dispersal and extinction events. DIVA assumes vicariance is the default mode of speciation, thus it does not assign a cost for vicariance but counts steps for dispersal and extinction events. The tree topology resulting from the BEAST analyses based on matrix B was used as DIVA requires a fully resolved tree. The sister clade with four species (Neo-uvaria acuminatissima, Neo-uvaria parallelivenia, Polyalthia coffeoides and Polyalthia longifolia) was also included in the biogeographical analysis. The geographical distributions estimated for internal nodes were optimized by constraining the maximum number of unit areas to two, hence restricting the number of unit areas in ancestral distributions.
WAAA estimates ancestral areas using reversible parsimony, and weights areas in plesiomorphic branches more than apomorphic branches [33]. Probability indices (PI) were calculated for each area at each node by counting the number of weighted gain steps (GSW) and weighted loss steps (LSW) on the cladogram resulting from the BEAST analyses (also based on matrix B). The PI is the ratio of LGW and LSW; biogeographical areas with a value of less than 0.2 are not considered as part of the ancestral area.
Results
Phylogeny and age estimations
The BEAST analyses of matrix A resulted in a robust phylogeny of the Annonaceae (Figure 2A), which is largely consistent with the topologies of previous phylogenetic analyses [7] and unpublished phylogenies using MrBayes (based on two and seven genes: L.W. Chatrou, pers. comm.). The mean ages of major nodes, with 95% HPD intervals and Bayesian PP values, are indicated in the chronogram (Figure 2A) and Table 2. The mean rate of evolution is 0.0006 substitutions per site per million years (95% HPD: 5.23E-4-7.10E-4). The 'birth rate' (i.e., speciation rate) indicated by the Yule prior is 0.041 (95% HPD: 0.031–0.052). The coefficient of variation is 0.69 (95% HPD: 0.56–0.84), indicating that substitution rate heterogeneity across the tree and that a relaxed clock model is most appropriate [47].
Table 2.
Prior and posterior age distributions of major nodes of Annonaceae based on matrix A using BEAST analyses.
| Node | Posterior | Prior | Previous estimates | Bayesian PP | ||
| Mean (Mya) | 95% HPD (Mya) | Mean (Mya) | 95% HPD (Mya) | |||
| C1: Magnoliaceae stem age | 102.10 | 106.20-98.0 | 98.89 | 100.09-98.00 | - | 1.00 |
| 1: Annonaceae stem age | 98.01 | 101.46-94.91 | - | - | 90.6 ± 1.3 by NPRS [7], 82 by PL [9] | 1.00 |
| C2: Annonaceae crown age | 89.46 | 90.41-89.0 | 90.05 | 92.20-89.00 | - | 1.00 |
| 2: split between Cananga- Tetrameranthus and both LBC-SBC clade | 74.66 | 84.4-63.62 | 82.58 | 91.88-69.53 | - | 1.00 |
| 3: split between LBC and SBC | 67.33 | 78.08-55.22 | - | - | 66.7-56.6 ± 2.3 by NPRS [7] | 1.00 |
| 4: Cananga-Tetrameranthus | 41.80 | 62.35-22.25 | 39.09 | 71.91-5.75 | 51.3-43.8 ± 5.1 by NPRS[7] | 1.00 |
| 5: Cananga-Cyathocalyx | 19.45 | 34.52-6.42 | - | - | - | 1.00 |
| 6: LBC crown node | 59.59 | 70.5-48.07 | 61.95 | 85.26-38.61 | 60.2-51.1 ± 2.3 by NPRS [7] | 1.00 |
| 7: SBC crown node | 39.84 | 55.17-26.86 | 70.82 | 88.09-52.69 | 62.5-53.1 ± 3.6 by NPRS[7], 58.76 by PL [8] | 1.00 |
| 8: Guatteria-Asimina | 51.54 | 62.81-40.32 | - | - | - | 1.00 |
| 9: Xylopia-Asimina | 46.62 | 57.1-35.68 | - | - | - | 0.97 |
| 10: Isolona-Asimina | 40.23 | 50.41-30.71 | - | - | - | 1.00 |
| 11: Isolona-Dasymaschalon | 31.65 | 41.45-21.18 | - | - | - | 1.00 |
| 12: Guatteria | 7.94 | 16.3-1.91 | - | - | 11.4 ± 1.4 by PL[10] | 1.00 |
| 13: Isolona-Monodora | 13.85 | 24.32-4.43 | - | - | Mean = ca. 14 by BEAST [11] | 1.00 |
| 14: Uvaria-Dasymaschalon | 19.61 | 28.42-10.59 | - | - | 14.8-12.4 ± 2.4 by NPRS[7] | 1.00 |
| 15: Onychopetalum-Pseuduvaria | 29.08 | 39.19-20.18 | - | - | - | 1.00 |
| 16: Onychopetalum-Karobelia | 24.36 | 37.75-15.77 | - | - | 37.65 by NPRS[8], 24.76 by PL[8], 31.81 by Multidivtime [8] | 1.00 |
| 17: Crematosperma | 4.44 | 9.09-0.90 | - | - | 22.33 by NPRS, 7.17 by PL[8], 16.49 by Multidivtime [8] | 1.00 |
| 18: Monocarpia-Pseuduvaria | 22.97 | 31.14-16.01 | - | - | - | 1.00 |
| 19: Neo-uvaria-Pseuduvaria | 14.9 | 20.2-9.7 | - | - | - | < 0.95 |
| 20: Pseuduvaria | 8.01 | 11.5-5.0 | 52.11 | 74.09-29.04 | ca. 16 by NPRS[7] | 1.00 |
Node numbers refer to Figure 2A. Mean ages and 95% HPD intervals of divergence times are in millions of years before the present. Age estimates were compared with earlier dating studies for the Annonaceae. Bayesian PP are also indicated. C1 and C2 represent the calibration nodes (see texts for details).
The BEAST chronogram (Figure 2A) is composed of four major clades: Anaxagorea (sister to all other members of the Annonaceae); a clade consisting of Cananga, Cleistopholis, Cyathocalyx and Tetrameranthus; the long branch clade (LBC); and the short branch clade (SBC). Each of these clades received strong statistical support (Table 2: PP = 1.00). BEAST analyses based on matrix A gave an estimated tMRCA for Pseuduvaria of 8.0 Mya (95% HPD: 11.5-5.0 Mya, PP = 1.00; Figure 2A: node 20). The MCMC result of the posterior distribution for the tMRCA of Pseuduvaria (Table 2: node 20) is shifted ca. 44 Mya later, indicating that the sequence data strongly influences date estimation.
The BEAST analyses of matrix B generated a well-resolved phylogeny of Pseuduvaria. The mean ages, 95% HPD intervals and Bayesian PP values of all nodes within Pseuduvaria, are indicated in the chronogram (Figure 3) and Table 3. The mean rate of evolution is 0.0005 substitutions per site per million years (95% HPD: 3.57E-4-7.024E-4). The birth rate indicated by the Yule prior is 0.188 (95% HPD: 0.122–0.270). The coefficient of variation is 0.543 (95% HPD: 0.365–0.723), indicating that substitution rate heterogeneity across the tree and that a relaxed clock model is appropriate [47]. The phylogeny of Pseuduvaria is segregated into five clades (Figure 3: clades I-V, numbering follows [25]). The BEAST topology of Pseuduvaria is largely consistent with those resulting from earlier ML and Bayesian analyses [[25]: Figure 3].
Table 3.
Posterior estimates of divergence time estimates (Mya) in Pseuduvaria based on matrix B using BEAST analyses, with results of ancestral area reconstructions using dispersal-vicariance analysis (DIVA) and weighted ancestral area analysis (WAAA).
| Node | Age estimates | DIVA | WAAA | Bayesian | ||||||||
| Mean (Mya) | 95% HPD (Mya) | A | B | C | D | E | F | G | H | PP | ||
| 0 | 13.6 | 18.49-8.85 | AH | 0.40 | 0.08 | 0.40 | 0.17 | 0.03 | 0.43 | 0.06 | 0.33 | 1.00 |
| 1 | 9.64 | 14.10-5.29 | CH | - | - | 1 | - | - | - | - | 1 | 1.00 |
| 2 | 5.76 | 9.37-2.41 | H | - | - | - | - | - | - | - | ∞ | 1.00 |
| 3 | 2.72 | 5.34-0.59 | C | - | - | ∞ | - | - | - | - | - | 1.00 |
| 4 | 8.34 | 10.95-5.82 | A | 0.93 | 0.09 | 0.25 | 0.29 | 0.04 | 0.52 | 0.07 | - | 1.00 |
| 5 | 6.62 | 9.50-3.83 | A | ∞ | - | - | - | - | - | - | - | 0.99 |
| 6 | 4.82 | 7.31-2.36 | A | ∞ | - | - | - | - | - | - | - | 1.00 |
| 7 | 2.57 | 4.40-0.97 | A | ∞ | - | - | - | - | - | - | - | 1.00 |
| 8 | 1.72 | 3.18-0.44 | A | ∞ | - | - | - | - | - | - | - | < 0.95 |
| 9 | 7.68 | 10.13-5.23 | AC AD AF | 0.55 | 0.11 | 0.30 | 0.38 | 0.05 | 0.68 | 0.08 | - | < 0.95 |
| 10 | 5.62 | 8.06-3.15 | A | ∞ | 0.09 | - | - | - | - | - | - | 0.99 |
| 11 | 4.12 | 6.32-2.04 | A | ∞ | 0.12 | - | - | - | - | - | - | < 0.95 |
| 12 | 3.10 | 4.93-1.33 | A | ∞ | 0.18 | - | - | - | - | - | - | 0.99 |
| 13 | 2.20 | 3.79-0.85 | A | ∞ | 0.33 | - | - | - | - | - | - | 0.96 |
| 14 | 1.50 | 2.83-0.42 | A | ∞ | 1 | - | - | - | - | - | - | < 0.95 |
| 15 | 4.17 | 6.43-2.05 | A | ∞ | - | - | - | - | - | - | - | 0.99 |
| 16 | 2.70 | 4.40-1.22 | A | ∞ | - | - | - | - | - | - | - | 1.00 |
| 17 | 1.99 | 3.36-0.82 | A | ∞ | - | - | - | - | - | - | - | 0.97 |
| 18 | 1.30 | 2.37-0.34 | A | ∞ | - | - | - | - | - | - | - | 0.99 |
| 19 | 1.13 | 2.17-0.22 | A | ∞ | - | - | - | - | - | - | - | 0.98 |
| 20 | 7.38 | - | F CF DF | 0.2 | 0.09 | 0.41 | 0.62 | 0.05 | 1.13 | 0.09 | - | < 0.95 |
| 21 | 5.88 | 8.46-3.54 | D CD DF | 0.07 | 0.05 | 0.6 | 1.7 | - | 0.18 | - | - | < 0.95 |
| 22 | 5.28 | - | C D CD CF | 0.09 | 0.07 | 1.31 | 0.71 | - | 0.33 | - | - | < 0.95 |
| 23 | 2.52 | 5.34-0.32 | CF DF | - | - | 1 | 1 | - | 1 | - | - | < 0.95 |
| 24 | 4.51 | 6.53-2.54 | C CD | 0.12 | 0.09 | 1.79 | 0.48 | - | - | - | - | 1.00 |
| 25 | 4.04 | 5.89-2.25 | D CD | 0.18 | 0.12 | 0.58 | 1.6 | - | - | - | - | < 0.95 |
| 26 | 3.28 | 4.86-1.76 | C AD CD | 0.33 | 0.18 | 1.6 | 0.33 | - | - | - | - | 1.00 |
| 27 | 2.50 | 3.94-1.13 | C AC | 1 | 0.33 | 3 | - | - | - | - | - | 0.99 |
| 28 | 2.06 | - | A AC BC | ∞ | 1 | 1 | - | - | - | - | - | < 0.95 |
| 29 | 2.27 | 3.57-1.09 | CD | - | - | 1 | 1 | - | - | - | - | 1.00 |
| 30 | 1.63 | 2.73-0.67 | C | - | - | ∞ | - | - | - | - | - | 0.99 |
| 31 | 0.77 | 1.49-0.15 | C | - | - | ∞ | - | - | - | - | - | 1.00 |
| 32 | 6.78 | 9.24-4.47 | F | 0.09 | 0.09 | 0.09 | 0.12 | 0.07 | 2.84 | 0.11 | - | < 0.95 |
| 33 | 1.85 | 4.35-0.23 | F | - | - | - | - | - | ∞ | - | - | 0.96 |
| 34 | 6.54 | 8.96-4.29 | F | 0.12 | 0.12 | 0.12 | 0.18 | 0.09 | 1.88 | 0.15 | - | < 0.95 |
| 35 | 5.81 | - | F | 0.18 | 0.18 | 0.18 | - | - | 4.56 | 0.07 | - | < 0.95 |
| 36 | 3.88 | 6.34-1.63 | F | - | - | - | - | - | ∞ | - | - | < 0.95 |
| 37 | 5.37 | - | F | 0.33 | 0.33 | 0.33 | - | - | 2.54 | 0.09 | - | < 0.95 |
| 38 | 4.05 | - | F | 1 | 1 | 1 | - | - | 1 | - | - | < 0.95 |
| 39 | 4.93 | - | F | - | - | - | - | - | 8.32 | 0.12 | - | < 0.95 |
| 40 | 3.97 | 6.08-1.97 | F | - | - | - | - | - | 5.55 | 0.18 | - | < 0.95 |
| 41 | 1.92 | 3.59-0.60 | F | - | - | - | - | - | ∞ | - | - | 1.00 |
| 42 | 3.31 | - | F | - | - | - | - | - | 3 | 0.33 | - | < 0.95 |
| 43 | 2.61 | - | FG | - | - | - | - | - | 3 | 0.33 | - | < 0.95 |
| 44 | 1.22 | 2.61-0.15 | G | - | - | - | - | - | - | ∞ | - | 1.00 |
| 45 | 4.53 | 6.42-2.78 | F | - | - | - | 0.33 | 0.12 | 1.16 | 0.18 | - | 1.00 |
| 46 | 3.57 | 5.18-2.00 | F | - | - | - | - | - | ∞ | - | - | < 0.95 |
| 47 | 2.90 | 4.50-1.24 | F | - | - | - | - | - | ∞ | - | - | < 0.95 |
| 48 | 2.03 | 3.57-0.61 | F | - | - | - | - | - | ∞ | - | - | 0.98 |
| 49 | 2.00 | 3.26-0.86 | F | - | - | - | - | - | ∞ | - | - | 1.00 |
| 50 | 0.83 | 1.64-0.16 | F | - | - | - | - | - | ∞ | - | - | 1.00 |
| 51 | 4.09 | - | DF | - | - | - | 1.00 | 0.18 | 0.18 | 0.33 | - | < 0.95 |
| 52 | 3.88 | - | FG | - | - | - | - | 0.33 | 0.33 | 1 | - | < 0.95 |
| 53 | 1.13 | 2.31-0.22 | G | - | - | - | - | - | - | ∞ | - | 1.00 |
| 54 | 0.55 | 1.27-0.04 | G | - | - | - | - | - | - | ∞ | - | 0.99 |
| 55 | 3.67 | - | EF | - | - | - | - | 1 | 1 | - | - | < 0.95 |
| 56 | 2.82 | 4.59-1.26 | F | - | - | - | - | - | ∞ | - | - | 0.99 |
Node numbers refer to Figure 3. Posterior mean ages and 95% HPD intervals of divergence times are in millions of years before the present. DIVA reconstructions are shown for maxareas = 2. WAAA reconstructions are shown with probability indices (PI). Biogeographical areas are indicated by capital letters (as defined in Figure 1). Bayesian PP are also indicated.
Historical biogeography of Pseuduvaria
The BEAST tree generated using matrix B was used as input for DIVA and WAAA (results summarized in Table 3). The DIVA and WAAA results were largely congruent: the inferred ancestral distributions are shown on the tree (Figs 4, 5). The DIVA reconstructions required at least 20 dispersal events to explain the present-day distribution when the maximum number of areas was restricted to two at each node.
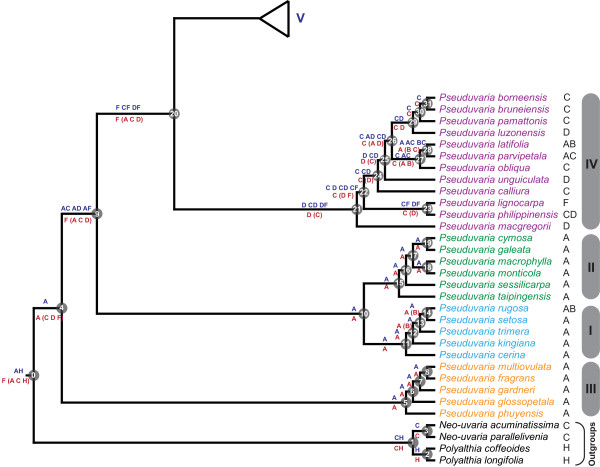
Figure 4.
Mapping of ancestral distributions on Pseuduvaria. Blue letters (above lines) represent DIVA results, and red letters (below lines) are WAAA results. For the latter results, red letters in brackets represent areas with lower probability indices. Letters to the right of the species names represent current species distributions (as defined in Figure 1).
Figure 5.
Mapping of ancestral distributions on Pseuduvaria (continued). Blue letters (above lines) represent DIVA results, and red letters (below lines) are WAAA results. For the latter results, red letters in brackets represent areas with lower probability indices. Letters to the right of the species names represent current species distributions (as defined in Figure 1).
Both DIVA and WAAA indicate that the most likely ancestral area for Pseuduvaria was Sundaland (Figure 4: node 4). Diversification of Pseuduvaria within Sundaland gave rise to clades I, II and III. The inferred ancestral area of the combined clade IV-V was ambiguous, however (Figure 4: node 20): DIVA indicated that the ancestral area was either New Guinea, Borneo-New Guinea, or Philippines-New Guinea; whereas WAAA suggested that it was either New Guinea (with the highest PI of 1.13; Table 3) or the Philippines (PI = 0.62). It is noteworthy, however, that node 20 lacks statistical support.
The inferred ancestral area for the common ancestor of clade IV (Figure 4: node 21) is also ambiguous. DIVA indicated the inferred distribution was either the Philippines, Borneo-Philippines, or Philippines-New Guinea; whereas WAAA indicated that the Philippines was the most probable ancestral area, with the highest PI of 1.70 (Table 3). Within clade IV, a series of dispersal events could have occurred between Borneo and the Philippines, and also possibly between the Philippines and New Guinea. Furthermore, reverse westwards movements of Pseuduvaria from Borneo and/or the Philippines can also be inferred (Figure 4: node 28), as shown by the distribution of two species, P. latifolia and P. parvipetala, in Java and Sumatra.
The DIVA and WAAA results were congruent regarding the inferred ancestral distribution of clade V, both indicating that New Guinea was the most likely region (Figure 5: node 32). Subsequent speciation events occurred within New Guinea, which became the secondary centre of diversity in Pseuduvaria. Within clade V, dispersal events presumably occurred from New Guinea to Australia, although a westward migration also seems to have occurred from New Guinea to western Malesia, as evident in one species (P. reticulata).
Discussion
Historical biogeography of major Annonaceae clades
The divergence times within the Annonaceae differ slightly from previous age estimates based on NPRS and PL methods as implemented in r8s (see Table 2). Our results are comparable, however, with previous BEAST analyses: the Isolona-Monodora clade is dated at 13.9 Mya (95% HPD: 24.3-4.4 Mya; Figure 2A: node 13) in this study, consistent with estimates of ca. 14 Mya in a previous study using BEAST [11].
Previous studies have suggested that the Annonaceae are likely to have originated in western Gondwana [7,9]. The break-up of the Gondwanan supercontinent into western Gondwana (Africa and South America) and eastern Gondwana (Australia, Antarctica, Madagascar and India) began 180-150 Mya [48,49], but it was not until 90-85 Mya that the two became fully separated [50]. Biotic interchange between South America and Africa had essentially ceased by the latter half of the Late Cretaceous (80-65 Mya) [48,49], although it has been suggested that limited connection between the two continents was possible for sometime afterwards via the island chains of the Rio Grande Rise-Walvis Ridge and Sierra Leone Rises [51]. The earliest divergence in the Annonaceae appears to have occurred between 98.0 Mya (95% HPD: 101.5-94.9 Mya; Figure 2A: node 1) and 89.5 Mya (Figure 2A: calibrated node C2 at a minimum age of 89 Mya for the oldest known crown group fossil, Futabanthus), probably after the separation of the two landmasses.
Anaxagorea is shown to be sister to all other members of the Annonaceae. The age of the split between the Cananga-Cyathocalyx-Cleistopholis-Tetrameranthus clade and the combined LBC-SBC clade is estimated at 74.7 Mya (95% HPD: 84.4-63.6 Mya, PP = 1.00; Figure 2A: node 2). In previous studies this node has been used as a minimum age constraint of 68 Mya. The split age estimated here may correspond to the origin of seeds of the Anonaspermum type, and our results furthermore indicate that the age of this node could be older than 68 Mya. The tMRCA of the Cananga-Cyathocalyx-Cleistopholis-Tetrameranthus clade is estimated at 41.8 Mya (95% HPD: 62.4-22.3 Mya, PP = 1.00; Figure 2A: node 4). The genera in this clade are currently distributed in geographically disparate regions: Cananga occurs in Southeast Asia and Australia [52]; Cyathocalyx in Southeast Asia, extending east to Fiji [53,54]; Cleistopholis in Africa [52]; and Tetrameranthus in South America [55]. The ancestors of this clade may have dispersed between Africa, Asia and South America from 74.7 Mya (95% HPD: 84.4-63.6 Mya, PP = 1.00; Figure 2A: node 2) to 41.8 Mya (95% HPD: 62.4-22.3 Mya, PP = 1.00; Figure 2A: node 4). This disjunct distribution may have been the result of Eocene and early Oligocene cooling [7,56].
The LBC and SBC constitute over 90% of all Annonaceae genera. The age of the split between the LBC and SBC lineages is estimated at 67.3 Mya (95% HPD: 78.1-55.2 Mya, PP = 1.00; Figure 2A: node 3), and the mean tMRCA of the LBC at 59.6 Mya (95% HPD: 70.5-48.1 Mya, PP = 1.00; Figure 2A: node 6). The ancestors of the majority of LBC genera (Guatteria-Asimina) appear to have evolved ca. 51.5 Mya (95% HPD: 62.8-40.3 Mya, PP = 1.00; Figure 2A: node 8). These genera are mainly distributed in South America and Africa, although several occur in Asia, including Artabotrys, Goniothalamus, Uvaria and Xylopia; this suggests that the LBC originated in South America and/or Africa-Madagascar [[5]: Figure 5]. The wide geographical distribution of boreotropical taxa could be associated with the combined effects of plate tectonics and global climatic changes. The warming period during the late Paleocene-early Eocene thermal maximum, which peaked around the early Eocene Climatic Optimum (52-50 Mya) [57], may have promoted the northward dispersal of tropical plants [51]. This coincides with the suggested date for the clade consisting of all members (node 8) of the LBC except the basal Mkilua-Trigynaea lineage in the early Eocene. In a recent paleontological review, most modern subgroups of mammalian orders were shown to have appeared during the Paleocene-Eocene thermal maximum, suggesting that rapid environmental changes may have evolutionary significance [58]. During this period (ca. 56 Mya: [59]), a Greenland land bridge may have connected North America and Eurasia, enabling plant dispersals between the two continents [51]. Another possible dispersal route was across the Tethys seaway between Africa-Arabia and Asia, allowing tropical plant dispersal until the late Eocene-early Oligocene [56].
The mean tMRCA of the SBC is estimated at 39.8 Mya (95% HPD: 55.2-26.9 Mya, PP = 1.00; Figure 2A: node 7). The ancestral biogeographical origin of the SBC cannot be postulated here without a more extensive sampling of genera, although the inferred areas could be Africa-Madagascar as shown in the Annickia-Piptostigma-Greenwayodendron clade [[5]: Figure 5]. Most evolutionary diversifications within the SBC seemed to have initiated ca. 29.1 Mya (95% HPD: 39.2-20.2 Mya, PP = 1.00; Figure 2A: node 15). The rate of molecular evolution during the early divergence of the SBC (arrowed in Figure 2B, indicated by blue line) is about four times slower than that during the early divergence of the LBC (arrowed in Figure 2B, shown as thick red line).
The SBC, as shown in Figure 2A, consists of small clades from Africa (a clade consisting Annickia, Piptostigma, and Greenwayodendron) and South to Central America (Onychopetalum-Cremastosperma), with all other members from Asia. Diversification within the SBC ca. 29.1 Mya (Figure 2A: node 15) resulted in a South American clade consisting of Cremastosperma, Klarobelia, Malmea, Mosannona, Onychopetalum, Oxandra, Pseudomalmea and Pseudoxandra. The descendents of SBC genera could have dispersed from South America into Asia (since Monocarpia euneura occurs in Borneo [60]) via Africa and the Indian Plate; this presumably occurred between 29.1 (95% HPD: 39.2-20.2 Mya, PP = 1.00; Figure 2A: node 15) and 23.0 Mya (95% HPD: 31.1-16.0 Mya, PP = 1.00; Figure 2A: node 18). The alternative dispersal scenario (from Africa into Asia via Europe) is less likely because most Asian Annonaceae occur in warm and moist tropical rainforests. It is therefore more probable that the SBC genera dispersed from Africa to Asia via India, and that the dispersal between Africa and India occurred before the collision between Asia and India. Many past biogeographical studies have postulated that the collision between Asia and India occurred 50–55 Mya [61-63]. More recent work [64], however, suggests that this Early Eocene event marks the collision of India's northern margin with a sub-equatorially located island arc (remnants of which are today preserved as the Dazhuqu arc terrain in southern Tibet), with the main continent-continent collision taking place ca. 34 Mya. However, the passage of the subcontinent may have taken the north-eastern corner of the block [65] close to western South-east Asia, and thus faunal and floral exchanges between the two geographical regions may have been possible from the late Paleocene (ca. 57 Mya) onwards [50].
Phylogeny and historical biogeography of Pseuduvaria
The BEAST topology of Pseuduvaria (Figure 3) is similar to those resulting from previous ML and Bayesian analyses (see Figure 3 in [25]). Although the phylogenetic positions of Pseuduvaria species in clades I, II and III remain identical, there are nevertheless some minor topological discrepancies between the trees. In the present study, clade III is shown to be sister to all other representatives of Pseuduvaria, whereas in previous analyses [25] this position was occupied by clade I. This is unlikely to have a significant impact on biogeographical interpretations, however, because the species in clades I, II and III are all distributed in geographical regions that were formerly part of the Sunda landmass. Another difference between the present and previous analyses relates to the phylogenetic position of two New Guinea species, P. becarrii and P. costata. These species are shown to be sister to all other members of clade V in the present study, but were basal within clade IV in the previous analyses [25]; the position of these two species is not statistically supported, although we suggest that they are more likely to belong to clade V as this clade is predominantly composed of New Guinea species. Several species in clade V (P. oxycarpa, P. filipes, P. coriacea, P. mindorensis, P. reticulata, and P. mollis) are furthermore located in different positions in different analyses, but their positions lack statistical support. Again, this may have little effect on biogeographical interpretations as most species in the clade are from New Guinea (except P. mindorensis from the Philippines, P. oxycarpa from Sulawesi, and P. reticulata, which is widespread in Malesia).
Late Miocene origin in Sundaland
Biogeographical reconstructions using DIVA and WAAA both suggest that Pseuduvaria originated in Sundaland (Figure 4: node 4). Initial BEAST analyses based on matrix A (with a broad taxonomic sampling) gave an estimated tMRCA for the Pseuduvaria lineage of 8.0 Mya (95% HPD: 11.5-5.0 Mya, PP = 1.00; Figure 2A: node 20); this node was subsequently selected as one of the two secondary molecular calibration points for Pseuduvaria in analyses based on matrix B (Figure 3: C4). These later analyses suggest a late Miocene origin, with a mean tMRCA of 8.3 Mya (95% HPD: 11.0-5.8 Mya, PP = 1.00; Figure 3: node 4). The former estimation from matrix A analyses, with a mean age of 8.0 Mya, will be used in all subsequent discussions presented here since this age was assigned as a calibration prior to the tMRCA of Pseuduvaria for matrix B analyses.
Early divergence of Pseuduvaria could have occurred between 14.9 Mya (95% HPD: 20.2-9.7 Mya, PP < 0.95; Figure 2A: node 19) and 8.0 Mya (95% HPD: 11.5-5.0 Mya; Figure 2A: node 20) during the Miocene, possibly associated with the 'mid-Miocene climatic optimum'. This warming phase occurred ca. 17-15 Mya [[57]: Figure 2], and led to the extensive growth of megathermal vegetation throughout most regions in Sundaland [66]. Subsequent global cooling during the middle Miocene climate transition (14.2-13.8 Mya) gradually resulted in the reduction of sea surface temperatures and the expansion of the Antarctic ice-sheets [67], which continued until ca. 6 Mya, in the early Pliocene [57]. These cold global temperatures were associated with a decline in CO2 levels, affecting the productivity of terrestrial vegetation [68], and resulted in the contraction of moist megathermal vegetation to the tropics and the concomitant expansion of grasslands [66]. This was followed by a warming phase that continued until ca. 3.2 Mya [57]. This climatic change was reflected by a steady increase in rainforest diversity in Southeast Asia until the mid Pliocene [66,69]. Subsequent diversifications of Pseuduvaria gave rise to clade III at ca. 6.6 Mya (95% HPD: 9.5-3.8 Mya; Figure 3: node 5) and clades I-II collectively at ca. 5.6 Mya (95% HPD: 8.1-3.2 Mya; Figure 3: node 10); this might be linked to these global climatic oscillations, with evolutionary divergence promoted by the significantly warmer and wetter climate.
Diversification in Sundaland
Biogeographical reconstructions of the ancestral distributions of clades I-III using DIVA and WAAA suggest that all three clades originated in Sundaland (Figure 4: nodes 5, 11, 15). Clade III consists of five species, P. phuyensis, P. glossopetala, P. gardneri, P. multiovulata, and P. fragrans; the tMCRA is estimated at ca. 6.6 Mya (node 5 in Table 3 and Figure 3). Clades I and II originated at a similar time, with tMRCA estimates of 4.1 and 4.2 Mya, respectively (nodes 11 and 15 in Table 3 and Figure 3).
Species in clades I and II are mostly endemic to Peninsular Malaysia or Thailand, with the exception of four comparatively widespread species, viz.: P trimera (southern China, Vietnam, Myanmar and Thailand), P. setosa (Thailand and Peninsular Malaysia), P. rugosa (Myanmar, Thailand, Peninsular Malaysia, Sumatra and Java), and P. macrophylla (Peninsular Malaysia and Sumatra) [25]. Pseuduvaria rugosa is estimated to have diverged around 1.5 Mya (95% HPD: 2.8-0.4 Mya, PP < 0.95; Table 3: node 14), at a similar time to P. macrophylla at ca. 1.3 Mya (95% HPD: 2.4-0.3 Mya, PP = 0.99; Table 3: node 18). In the middle and late Miocene, land connected Sumatra-Java to mainland Southeast Asia [70]. Further reduction of sea levels during the Pleistocene (to a maximum depth of 120 m below the present level) exposed additional land area that connected Peninsular Malaysia, Sumatra, Java, and Borneo [71]. Tropical forests also gradually expanded, as shown by the arrival of orangutans in Java [72]. At that time, P. macrophylla and P. rugosa were likely to have been able to disperse unimpeded between Sumatra, Java and Peninsular Malaysia; P. trimera would similarly have been able to advance northwards into southern China. Seed dispersal of P. rugosa is known to be facilitated by orangutans [27].
Dispersal out of Sundaland
DIVA and WAAA indicated several possible ancestral distributions for clades IV and V collectively (Figure 4: node 20). According to DIVA, New Guinea, Borneo-New Guinea, or Philippines-New Guinea are potential ancestral areas, whereas WAAA highlighted New Guinea (PI = 1.13) and the Philippines (PI = 0.62). The age of node 9 is estimated at ca. 7.7 Mya, although this node is not statistically supported (PP < 0.95; Table 3). This age estimate (indicated by a dotted black line in Figure 3) could represent the earliest migration event for Pseuduvaria out of Sundaland to the Philippines and/or New Guinea (Figure 6).
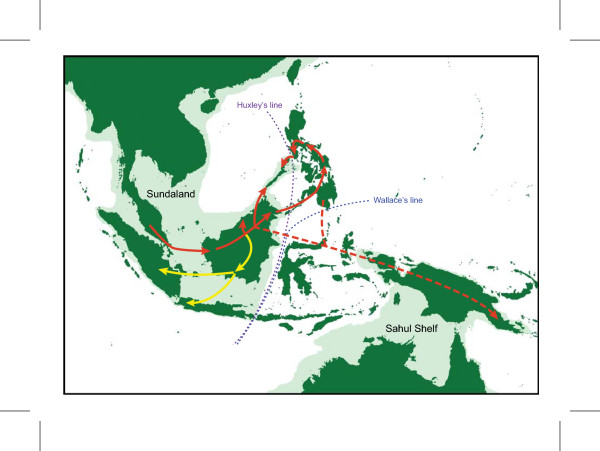
Figure 6.
Proposed biogeographical scenarios for Pseuduvaria clade IV. Arrows represent possible migration routes (see text for details).
The tMRCA of clade IV is estimated at ca. 5.9 Mya (95% HPD: 8.5-3.5 Mya, PP < 0.95; Figure 3: node 21). WAAA indicated that the Philippines is the most likely ancestral area, with the highest PI value of 1.7 (Table 3 and Figure 4: node 21), although Borneo also returned a relatively high PI of 0.6. Similar results were obtained using DIVA, with the Philippines or Borneo-Philippines as the most likely ancestral areas (the third suggested scenario, with New Guinea-Philippines as the ancestral area, is improbable since the two areas are not contiguous). The timing of dispersal events from Sundaland to the Philippines may be associated with the docking of the Philippine Sea Plate with the Sunda Block, which was initiated in the late Miocene-Pliocene [73].
The WAAA and DIVA results are complex within clade IV, but appear to show probable dispersals from Borneo and/or the Philippines eastwards into New Guinea (dotted red arrows in Figure 6) and westwards into continental Southeast Asia, Sumatra and/or Java (yellow arrows in Figure 6). The eastward migration into New Guinea may have occurred between node 22 (mean tMRCA: 5.3 Mya; without HPD, PP < 0.95; Table 3) and node 23 (mean tMRCA: 2.5 Mya; 95% HPD: 5.3-0.3 Mya, PP < 0.95; Table 3). These dispersals from Sundaland to New Guinea may have been achieved by three routes: island hopping after the collision of the Australian Plate with the Philippine Plate; stochastic dispersals across the Makassar Straits; or dispersal of montane taxa via island-hopping following the uplift of New Guinea and other islands [51]. The dispersal of Dacrycarpus (Podocarpaceae) is a suggested example of the third route: Dacrycarpus dispersed to New Guinea from Australia in the mid-Miocene and then dispersed to Borneo ca. 3.5 Mya in the mid-Pliocene [51]. Pseuduvaria species generally occur at low altitudes, suggesting that this third route is less likely to be important, although P. lignocarpa can occur at altitudes over 1000 m [27].
WAAA determined that the most probable ancestral areas at node 24 (Figure 4) are either Borneo (with the highest PI of 1.79; Table 3) or the Philippines (PI = 0.48; Table 3); similar results were obtained using DIVA. The ancestral distributions for node 25 (Figure 4) were the same as those for node 24, although node 25 is not statistically supported. A westward migration from Borneo-Philippines into mainland Southeast Asia, Sumatra and/or Java may have occurred by nodes 26, 27 and/or 28 (yellow arrows in Figure 6).
Colonization of New Guinea via long-distance dispersal
Clade V contains almost half of the genus, with most species currently distributed in New Guinea. The earliest suggested date for the colonization of New Guinea by Pseuduvaria species is 7.7 Mya (95% HPD: 10.1-5.2 Mya, PP < 0.95; Figure 4: node 9), although this clade lacks statistical support (Table 3). If colonization of New Guinea by node 9 is accepted, it is necessary to invoke a subsequent reverse westward dispersal for clades I and II (Figure 4: node 10). A more conservative scenario involves a later colonization of New Guinea after 7.4 Mya (Figure 4: node 20), although this node is also unsupported.
A recent plate tectonic modeling of New Guinea during the Cenozoic [[74]: Figure 8] has shown major changes in the motion of the Pacific Plate beginning around 43 Mya (Eocene), with the initiation of two subduction systems, generating the Inner and Outer Melanesian arcs. During the Oligocene (ca. 35-30 Mya), the first significant collision event began and the northern part of the Australia plate was subducted underneath the Inner Melanesian arc, creating the 'Peninsular orogeny' and generating the Papuan Peninsula [74]. As a result of a drop in sea level (ca. 90 m) during the Oligocene, central and western New Guinea became largely sub-aerial.
In the late Miocene (ca. 10 Mya), a second collision event initiated in western New Guinea, creating the 'Central Range orogeny' [74]. At that time, the Sunda and the Sahul Shelves may have reached closest proximity, although floristic migration may have been unlikely since New Guinea was still largely submerged, and deep ocean barriers existed with no land bridges [75,76]. The best opportunity for biota to 'island hop' across Wallacea appears to have been during the last 5 million years [75], due to: (1) the formation of a substantial landmass in Sulawesi from ca. 5 Mya [75]; (2) the connection between New Guinea and Sundaland via the Banda and Sunda arcs [[70], p. 122]; (3) the increasing number of volcanic islands in East Indonesia [77]; (4) the exposure of the Sunda and Sahul Shelves caused by falling sea level (up to 120 m lower than present levels) during the Pleistocene glaciations [71]; and (5) the accretion of microcontinental island arc fragments at the northern edge of the Australian craton portion of New Guinea [75,77]. Migration of Pseuduvaria from Sundaland to New Guinea may therefore have been facilitated by a series of stepping stones, possibly via Borneo (dotted red arrow in Figure 7) or the Lesser Sunda Islands (dotted orange arrow in Figure 7). Pseuduvaria also occurs in the Aru islands (P. aurantiaca) and New Britain (P. macrocarpa). Similar biogeographical patterns are found in other plant groups such as the tribe Aglaieae (Meliaceae) which originated in western Malesia and subsequently dispersed eastwards to the Aru and Kai islands, New Guinea, New Britain, Australia, Fiji and other Pacific islands [78]; migration between the Sunda and Sahul shelves was likely to have been achieved via long-distance dispersal by birds [78].
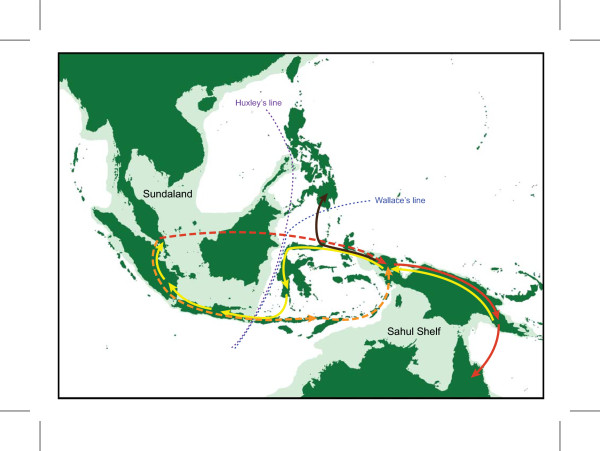
Figure 7.
Proposed biogeographical scenarios for Pseuduvaria clade V. Arrows represent possible migration routes (see text for details).
Diversification patterns in New Guinea and Australia
Pseuduvaria may have begun to diversify in New Guinea after node 34 from ca. 6.5 Mya (95% HPD: 9.0-4.3 Mya, PP < 0.95; Figure 3), resulting in two main lineages with tMRCAs of ca. 5.8 Mya (Table 3: node 35) and ca. 4.5 Mya (Table 3: node 45). Coincident with this timeframe, there was a gradual transition between global cooling and warming phases: the cooling phase lasted until ca. 6 Mya, followed by a warming phase with moist climate until ca. 3.2 Mya [57]. A cooling period subsequently started again with a drier climate in the late Pliocene [79], followed by several cycles of glacial-interglacial episodes from ca. 1.8 Mya during the Pleistocene [80,81]. These climatic oscillations, combined with the changes in sea level, may have promoted Pseuduvaria speciation in New Guinea.
Pseuduvaria exhibits a high level of endemism (95%) in New Guinea [27]. This could be associated with the Central Range orogeny in New Guinea, which developed from ca. 8 Mya; this collisional orogenesis led to the formation of a ca. 1300 km-long mountainous backbone from the Bird's Neck to the Papuan Peninsula [74] with some peaks over 5000 m [82]. Speciation within New Guinea may therefore have been promoted by mountain ranges over 4000 m acting as physical barriers and influencing temperatures, and a continuously hot and wet climate with rainfalls of over 2500 mm per annum [83]. Pseuduvaria species in New Guinea diversified morphologically in response to these physical and climatic barriers, with evolutionary shifts from unisexual into bisexual flowers, elongation of flowering and fruiting peduncles (favouring fruit-bat frugivory), and the evolution of distinct pollination systems [25,27]. The New Guinea orogeny is analogous to the Miocene uplift of the Andean range, where different elevations (lowland or highland habitats) have impacted species diversification in the Neotropics [84].
There is also evidence of dispersal of Pseuduvaria species from New Guinea to northern Australia (Figs 5, 7), as shown in node 44 (P. froggattii and P. hylandii) and node 53 (P. villosa, P. mulgraveana and P. glabrescens). These clades are estimated to have evolved after 2.6 Mya (Table 3: node 43) and 3.9 Mya (Table 3: node 52), respectively. Pseuduvaria would have been able to disperse between New Guinea and Australia during the Pleistocene (red arrows in Figure 7): glaciations caused a lowering of oceans to at least 120 m below the present level [85-87], and as a result New Guinea and northern Australia formed a continuous land mass [76,86] with open forest vegetation growing across the Torres Strait land bridge (between eastern New Guinea and northern Australia) between 17,000 and 8,000 years ago [88].
Although clade V is largely composed of New Guinea and Australian species, there are three significant exceptions. Pseuduvaria reticulata is widespread in Peninsular Malaysia, Sumatra, Java, Lesser Sunda islands and Borneo, indicating an apparent westwards recolonization of Sundaland between 5.4 Mya (Table 3: node 37) and 4.1 Mya (Table 3: node 38). The most likely migration route from the Sahul Shelf to Sundaland (Java) may have been be achieved via Sulawesi [79] (yellow arrows in Figure 7); dispersal across the wide Makassar Straits seems unlikely since these straits have been shown to be an effective barrier between eastern and western Malesia [89].
A northward migration from New Guinea to the Philippines, possibly via Sulawesi (brown arrow in Figure 7), is shown by P. mindorensis after ca. 4.1 Mya (Table 3: node 51), although this node lacks statistical support. Another westwards reversal event is evident in P. oxycarpa, which may have migrated from New Guinea to Sulawesi between 3.9 (Table 3: node 52) and 3.7 Mya (Table 3: node 55). A substantial landmass was present in the Sulawesi region between ca. 10-5 Mya [75], but it was not until the last 5 Ma that the three distinct fragments (peninsular NE, mainland NE, and W Sulawesi) became united [90], facilitating the migration of plants and animals across Wallacea [75].
Conclusion
The divergence times of the main clades within the Annonaceae (derived here using BEAST) deviate slightly from those previously determined using different calibration points and dating methods. In particular, our estimate for the SBC crown is 39.8 Mya (95% HPD: 55.2-26.9 Mya), which is much younger than previous estimates of 62.5-53.1 ± 3.6 Mya [7] and 58.76 Mya [8]. The present study differs from previous research as it utilizes an uncorrelated lognormal relaxed clock model and is calibrated using the recently described fossil, Futabanthus; these factors are likely to enable more precise divergence time estimates within the Annonaceae.
Pseuduvaria is shown to have originated in Sundaland in the late Miocene, ca. 8 Mya (95% HPD: 11.5-5.0 Mya). Subsequent migration events were predominantly eastwards towards New Guinea and Australia, although several migratory reversals are also postulated. The suggested dispersal episodes are broadly consistent with the available geological data.
The migration of Pseuduvaria species into New Guinea from Sundaland and/or the Philippines is of particular biogeographical interest. DIVA and WAAA reconstructions of the most probable ancestral areas at each node suggest that the earliest migration of Pseuduvaria species into New Guinea may have occurred as early as 7.7 Mya (95% HPD: 10.1-5.2 Mya), although this node is not statistically supported. If such a scenario is correct, it is necessary to invoke long-distance dispersal events since the geological data suggest that biotic migration between the two landmasses would have been unlikely: New Guinea was largely submerged during this period and the Sunda and Sahul landmasses were separated by deep ocean barriers without land bridges [75,76]. It was only within the past 5 Ma that significant opportunities may have existed for biotic migration into New Guinea from Sundaland [75], and significantly all the other postulated migrations between New Guinea and Sundaland occurred within this period: from Sundaland to New Guinea between nodes 22 and 23 in clade IV (i.e., after 5.3 Mya); from New Guinea to Sundaland between nodes 37 and 38 in clade V (i.e., after 5.4 Mya); and from New Guinea to Sulawesi between nodes 52 and 55 in clade V (i.e., after 3.9 Mya).
Our previous study of morphological character evolution within Pseuduvaria [25] indicated that the ancestors of clade V were likely to have had fruits with large monocarps (individual fruit segments derived from the maturation of individual carpels within the flower); this inference holds true irrespective of whether the New Guinea species P. beccarii and P. costata are included in clade IV (as indicated in our previous study [25]) or included in clade V (present analysis). There is a dearth of observational reports on frugivory and seed dispersal of Pseuduvaria species [27], although it appears that seeds of smaller-fruited species are bird-dispersed whereas those of larger-fruited species are dispersed by primates or fruit bats. It is probable that the long-distance dispersal events hypothesised to explain the early colonization of New Guinea by Pseuduvaria species may have been facilitated by fruit bats. Significantly, the majority of New Guinea species of Pseuduvaria possess elongate flowering peduncles and/or pedicels [25]: separation of fruits from the foliage (thereby ensuring that the fruits are conspicuous) is a common feature of plants adapted for fruit-bat frugivory [90].
Authors' contributions
YCFS collected and analysed the molecular data. RMKS conceived the study and obtained funding. Both authors wrote the paper and have read and approved the final manuscript.
Supplementary Material
Specimens and GenBank information. Information regarding species sampling, localities, and GenBank accession numbers for sequences used in BEAST analyses.
Acknowledgments
Acknowledgements
This research was supported by a grant from the Hong Kong Research Grants Council (7358/03M). We thank Lars Chatrou for sending unpublished DNA sequences of two outgroups, Justin Bahl, Dhanesekaran Vijaykrishna, Alexei Drummond, Thomas Couvreur, and Gavin Smith for helpful advice and discussions on using BEAST, Jason Ali for discussions on the geological history of Southeast Asia, and Jim Doyle and two anonymous reviewers for their valuable comments on the manuscript.
Contributor Information
Yvonne CF Su, Email: ycfsu@hkucc.hku.hk.
Richard MK Saunders, Email: saunders@hkucc.hku.hk.
References
- Angiosperm Phylogeny Group (APG) An update of the Angiosperm phylogeny group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 2003;141:399–436. [Google Scholar]
- Doyle JA, Le Thomas A. Cladistic analysis and pollen evolution in Annonaceae. Acta Bot Gall. 1994;141:149–170. [Google Scholar]
- Doyle JA, Le Thomas A. Phylogenetic analysis and character evolution in Annonaceae. Bull Mus Hist Nat, Paris, B, Adansonia. 1996;18:279–334. [Google Scholar]
- Doyle JA, Le Thomas A. Significance of palynology for phylogeny of Annonaceae: experiments with removal of pollen characters. Pl Syst Evol. 1997;206:133–159. [Google Scholar]
- Doyle JA, Sauquet H, Scharaschkin T, Le Thomas A. Phylogeny, molecular and fossil dating, and biogeographic history of Annonaceae and Myristicaceae (Magnoliales) Int J Pl Sci. 2004;165:S55–S67. [Google Scholar]
- Mols JB, Gravendeel B, Chatrou LW, Pirie MD, Bygrave PC, Chase MW, Keßler PJA. Identifying clades in Asian Annonaceae: monophyletic genera in the polyphyletic Miliuseae. Amer J Bot. 2004;91:590–600. doi: 10.3732/ajb.91.4.590. [DOI] [PubMed] [Google Scholar]
- Richardson JE, Chatrou LW, Mols JB, Erkens RHJ, Pirie MD. Historical biogeography of two cosmopolitan families of flowering plants: Annonaceae and Rhamnaceae. Phil Trans Roy Soc Lond, B. 2004;359:1495–1508. doi: 10.1098/rstb.2004.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirie MD, Chatrou LW, Mols JB, Erkens RHJ, Oosterhof J. 'Andean-centred' genera in the short-branch clade of Annonaceae: testing biogeographical hypotheses using phylogeny reconstruction and molecular dating. J Biogeogr. 2006;33:31–46. [Google Scholar]
- Scharaschkin T, Doyle JA. Phylogeny and historical biogeography of Anaxagorea (Annonaceae) using morphology and non-coding chloroplast sequence data. Syst Bot. 2005;30:712–735. [Google Scholar]
- Erkens RHJ, Chatrou LW, Maas JW, Niet T van der, Savolainen V. A rapid diversification of rainforest trees (Guatteria; Annonaceae) following dispersal from Central to South America. Mol Phylogenet Evol. 2007;44:399–411. doi: 10.1016/j.ympev.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Couvreur TLP, Chatrou LW, Sosef MSM, Richardson JE. Molecular phylogenetics reveal multiple tertiary vicariance origins of the African rain forest trees. BMC Biology. 2008;6:54. doi: 10.1186/1741-7007-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilcher DL, Crane PR. Archaeanthus: an early angiosperm from the Cenomanian of the Western Interior of North America. Ann Missouri Bot Gard. 1984;71:351–361. [Google Scholar]
- Gröcke DR, Ludvigson GA, Witzke BL, Robinson SA, Joeckel RM, Ufnar DF, Ravn RL. Recognizing the Albian-Cenomanian (OAE1d) sequence boundary using plant carbon isotopes: Dakota Formation, Western Interior Basin, USA. Geology. 2006;34:193–196. [Google Scholar]
- Chesters KIM. Some plant remains from the Upper Cretaceous and Tertiary of West Africa. Ann Mag Nat Hist. 1955;12:489–504. [Google Scholar]
- Mohr BAR, Bernardes-de-Oliveira MEC. Endressinia brasiliana, a magnolialean angiosperm from the Lower Cretaceous Crato Formation (Brazil) Int J Plant Sci. 2004;165:1121–1133. [Google Scholar]
- Ward JV, Doyle JA, Hotton CL. Probable granular magnoliid angiosperm pollen from the Early Cretaceous. Pollen Spores. 1989;31:101–120. [Google Scholar]
- Wikström N, Savolainen V, Chase MW. Evolution of the angiosperms: calibrating the family tree. Proc Roy Soc Lond, B. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. r8s version 171. 2004. http://loco.biosci.arizona.edu/r8s/
- Sanderson MJ. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol Biol Evol. 1997;14:1218–1231. [Google Scholar]
- Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Pirie MD, Chatrou LW, Erkens RHJ, Maas JW, Niet T van der, Mols JB, Richardson JE. Phylogeny reconstruction and molecular dating in four Neotropical genera of Annonaceae: the effect of taxon sampling in age estimations. In: Bakker FT, Chatrou LW, Gravendeel B, Pelser PB, editor. Plant species-level systematics: new perspectives on pattern and process Regnum Vegetabile. Vol. 143. Germany, Koenigstein: Koeltz; 2005. pp. 150–174. [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comp Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST ver 148. 2008. http://beast.bio.ed.ac.uk/
- Su YCF, Smith GJD, Saunders RMK. Phylogeny of the basal angiosperm genus Pseuduvaria (Annonaceae) inferred from five chloroplast DNA regions, with interpretation of morphological character evolution. Mol Phylogenet Evol. 2008;48:188–206. doi: 10.1016/j.ympev.2008.03.028. [DOI] [PubMed] [Google Scholar]
- Su YCF, Chaowasku T, Saunders RMK. An extended phylogeny of Pseuduvaria (Annonaceae) with descriptions of three new species and a reassessment of the generic status of Oreomitra. Syst Bot.
- Su YCF, Saunders RMK. Monograph of Pseuduvaria (Annonaceae) Syst Bot Monogr. 2006;79:1–204. [Google Scholar]
- Hall R. Extension during late Neogene collision in east Indonesia and New Guinea. J Virtual Explorer. 2001;4:1–13. [Google Scholar]
- Wiffin T. Plant Biogeography of the SE Asian-AustralianRegion. In: Kershaw P, David B, Tapper N, Penny D, Brown J, editor. Bridging Wallace's Line: the environmental and cultural history and dynamics of the SE-Asian-Australian Region Advances in Geoecology. Vol. 34. Germany, Reiskirchen: Catena Verlag; 2002. pp. 61–82. [Google Scholar]
- Hall R, Morley CK. Sundaland Basins. In: Clift P, Wang P, Kuhnt W, Hayes DE, editor. Continent-Ocean Interactions within the East Asian Marginal Seas Geophysical Monograph,) Vol. 149. USA, Washington DC: American Geophysical Union; 2004. pp. 55–85. [Google Scholar]
- Takahashi M, Friis EM, Uesugi K, Suzuki Y, Crane PR. Floral evidence of Annonaceae from the Late Cretaceous of Japan. Int J Plant Sci. 2008;169:908–917. [Google Scholar]
- Ronquist F. DIVA (DIspersal Vicariance Analysis) HOMEPAGE. DIVA ver 12. 1996. http://www.ebc.uu.se/systzoo/research/diva/diva.html
- Hausdorf B. Weighted ancestral area analysis and a solution of the redundant distribution problem. Syst Biol. 1998;47:445–456. doi: 10.1080/106351598260815. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/95/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Rambaut A. Se-Al. Sequence Alignment Editor, ver 20a11. 1996. http://tree.bio.ed.ac.uk/software/seal/
- Nylander JAA. MrModeltest ver 23. 2008. http://www.abc.se/~nylander/
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:art. 214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:699–710. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreur TLP. BEAST partitioning: a 7 step tutorial. 2008. http://tlpcouvreur.googlepages.com/beastpartitioning
- Rambaut A, Drummond AJ. Tracer ver 14. 2007. http://tree.bio.ed.ac.uk/software/tracer/
- Rambaut A. FigTree ver 12. 2008. http://tree.bio.ed.ac.uk/software/figtree/
- Ho SYW. Calibrating molecular estimates of substitution rates and divergence times in birds. J Avian Biol. 2007;38:409–414. [Google Scholar]
- Bergh NG, Linder HP. Cape diversification and repeated out-of-southern Africa dispersal in paper daises (Asteraceae-Gnaphalieae) Mol Phylogenet Evol. 2009;51:5–18. doi: 10.1016/j.ympev.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Couvreur TLP, Richardson JE, Sosef MSM, Erkens RHJ, Chatrou LW. Evolution of syncarpy and other morphological characters in African Annonaceae: a posterior mapping approach. Mol Phylogenet Evol. 2008;47:302–318. doi: 10.1016/j.ympev.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Hug LA, Roger AJ. The impact of fossils and taxon sampling on ancient molecular dating analyses. Mol Biol Evol. 2007;24:1889–1897. doi: 10.1093/molbev/msm115. [DOI] [PubMed] [Google Scholar]
- Takhtajan A. Floristic regions of the world. California: University of California Press; 1986. [Google Scholar]
- Drummond AJ, Ho SYW, Rawlence N, Rambaut A. A rough guide to BEAST 1.4. Auckland: University of Auckland; 2007. [Google Scholar]
- Scotese CR, Gahagan LM, Larson RL. Plate tectonic reconstructions of the Cretaceous and Cenozoic ocean basins. Tectonophysics. 1988;155:27–48. [Google Scholar]
- Hallam A. An outline of Phanerozoic biogeography. Oxford: Oxford University Press; 1994. [Google Scholar]
- Ali JR, Aitchison JC. Gondwana to Asia: Plate tectonics, paleogeography and the biological connectivity of the Indian sub-continent from the Middle Jurassic through latest Eocene (166-35 Ma) Earth-Sci Rev. 2008;88:145–166. [Google Scholar]
- Morley RJ. Interplate dispersal paths for megathermal angiosperms. Persp Pl Ecol, Evol Syst. 2003;6:5–20. [Google Scholar]
- Heusden van ECH. Flowers of Annonaceae: morphology, classification, and evolution. Blumea. 1992. pp. 1–218.
- Wang RJ, Saunders RMK. The genus Cyathocalyx (Annonaceae) in the Philippines. Syst Bot. 2006;31:285–297. [Google Scholar]
- Wang RJ, Saunders RMK. A synopsis of Cyathocalyx species (Annonaceae) in Peninsular Malaysia, Sumatra, and Borneo, with descriptions of two new species. Bot J Linn Soc. 2006;152:513–532. [Google Scholar]
- Westra LYT. Studies in Annonaceae. IV. A taxonomic revision of Tetrameranthus R. E. Fries. Proc Kon Ned Akad Wetensch, C. 1985;88:449–482. [Google Scholar]
- Pennington RT, Dick CW. The role of immigrants in the assembly of the South American rainforest tree flora. Phil Trans Roy Soc Lond, B. 2004;359:1611–1622. doi: 10.1098/rstb.2004.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberration in global climate 65 Ma to present. Science. 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- Gingerich PD. Environment and evolution through the Paleocene-Eocene thermal maximum. Trends Ecol Evol. 2006;21:246–253. doi: 10.1016/j.tree.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Morley RJ, Dick CW. Missing fossils, molecular clocks, and the origin of the Melastomataceae. Amer J Bot. 2003;90:1638–1644. doi: 10.3732/ajb.90.11.1638. [DOI] [PubMed] [Google Scholar]
- Mols JB, Keßler PJA. The genus Monocarpia (Annonaceae) in Borneo including a new species Monocarpiaborneensis. Bot Jahrb Syst. 2000;122:233–240. [Google Scholar]
- Rowley DB. Age of initiation of collision between India and Asia: a review of stratigraphic data. Earth Planet Sci Lett. 1996;145:1–13. [Google Scholar]
- Zhu BD, Kidd WSF, Rowley DB, Currie BS, Shaffique N. Age of initiation of the India-Asia collision in the east-central Himalaya. J Geol. 2004;113:265–285. [Google Scholar]
- Leech ML, Singh S, Jain AK, Klemperer SL, Manickavasagam RM. The onset of India-Asia continental collision: early, steep subduction required by the timing of UHP metamorphism in the western Himalaya. Earth Planet Sci Letters. 2005;234:83–97. [Google Scholar]
- Aitchison JC, Ali JR, Davis AM. When and where did India and Asia collide? J Geophys Res. 2007;112B:Art. B05423. [Google Scholar]
- Ali JR, Aitchison JC. Greater India. Earth-Sci Rev. 2005;72:169–188. [Google Scholar]
- Morley RJ. Cretaceous and Tertiary climate change and the past distribution of megathermal rainforests. In: Flenley JR, Bush MB, editor. Tropical rainforest responses to climatic changes. Berlin: Springer; 2007. pp. 1–31. [Google Scholar]
- Shevenell AE, Kennett JP, Lea DW. Middle Miocene southern ocean cooling and Antarctic cryosphere expansion. Science. 2004;305:1766–1770. doi: 10.1126/science.1100061. [DOI] [PubMed] [Google Scholar]
- Kürschner WM, Kvaèek Z, Dilcher DL. The impact of Miocene atmospheric carbon dioxide fluctuations on climate and the evolution of terrestrial ecosystems. Proc Natl Acad Sci USA. 2008;105:449–453. doi: 10.1073/pnas.0708588105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney LR. A synopsis of climatic and vegetational change in Southeast Asia. Clim Change. 1991;19:53–61. [Google Scholar]
- Hall R. The plate tectonics of Cenozoic SE Asia and the distribution of land and sea. In: Hall R, Holloway JD, editor. Biogeography and geological evolution of SE Asia. Leiden: Backhuys Publishers; 1998. pp. 99–131. [Google Scholar]
- Inger RF, Voris HK. The biogeographical relations of the frogs and snakes of Sundaland. J Biogeogr. 2001;28:863–891. [Google Scholar]
- Bird MI, Taylor D, Hunt C. Paleoenvironments of insular Southeast Asia during the Last Glacial Period: a savanna corridor in Sundaland. Quaternary Sci Rev. 2005;24:2228–2242. [Google Scholar]
- Pubellier M, Ali J, Monnier C. Cenozoic Plate interaction of the Australia and Philippine Sea Plate: "hit-and-run" tectonics. Tectonophysics. 2003;363:181–199. [Google Scholar]
- Ufford AQ van, Cloos M. Cenozoic tectonics of New Guinea. Amer Assoc Petrol Geol Bull. 2005;89:119–140. [Google Scholar]
- Hall R. Cenozoic reconstructions of SE Asia and the SW Pacific: changing patterns of land and sea. In: Metcalfe I, Smith JMB, Morwood M, Davidson ID, editor. Faunal and floral migrations and evolution in SE Asia-Asutralasia. Lisse: Swets and Zeitlinger Publisher; 2001. pp. 35–56. [Google Scholar]
- Rowe KC, Reno ML, Richmond DM, Adkins RM, Steppan SJ. Pliocene colonization and adaptive radiations in Australia and New Guinea (Sahul): multilocus systematics of the old endemic rodents (Muroidea: Murinae) Mol Phylogenet Evol. 2008;47:84–101. doi: 10.1016/j.ympev.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Barby MF, Pierce NE. Systematics, biogeography anddiversification of the Indo-Australian genus Delias Hübner (Lepidoptera: Pieridae): phylogenetic evidence supports an 'out-of-Australia' origin. Syst Entomol. 2007;32:2–25. [Google Scholar]
- Muellner AN, Pannell CM, Coleman A, Chase MW. The origin and evolution of Indomalesian, Australasian and Pacific island biotas: insights from Aglaieae (Meliaceae, Sapindales) J Biogeogr. 2008;35:1769–1789. [Google Scholar]
- Morley RJ. Tertiary vegetational history of Southeast Asia, with emphasis on the biogeographical relationships with Australia. In: Kershaw P, David B, Tapper N, Penny D, Brown J, editor. Bridging Wallace's line: the environmental and cultural history and dynamics of the SE-Asian-Australian region. Reiskirchen: Catena Verlag; 2002. pp. 49–60. [Google Scholar]
- Johnson DJ. The geology of Australia. Cambridge: Cambridge University Press; 2004. [Google Scholar]
- Poulin E, Palma AT, Féral JP. Evolutionary versus ecological success in Antarctic benthic invertebrates. Trends Ecol Evol. 2002;17:218–222. [Google Scholar]
- Hill KC, Hall R. Mesozoic-Cenozoic evolution of Australia's New Guinea margin in a west Pacific context. In: Hillis RR, Müller RD, editor. Evolution and dynamics of the Australian Plate. Sydney: Geological Society of Australia Special Publication 22 and Geological Society of America Special Paper 372; 2003. pp. 259–283. [Google Scholar]
- Beebe NW, Cooper RD. Distribution and evolution of the Anopheles punctulatus group (Diptera: Culicidae) in Australia and Papua New Guinea. Int J Parasitol. 2002;32:563–574. doi: 10.1016/s0020-7519(01)00359-9. [DOI] [PubMed] [Google Scholar]
- Young KR, Ulloa CU, Luteyn JL, Knapp S. Plant evolution and endemism in Andean South America: an introduction. Bot Rev. 2002;68:4–21. [Google Scholar]
- Heaney LR. Biogeography of mammals in SE Asia: estimates of rates of colonization, extinction and speciation. Biol J Linn Soc. 1986;28:127–165. [Google Scholar]
- Voris HK. Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. J Biogeogr. 2000;27:1153–1167. [Google Scholar]
- Morley RJ, Flenley JR. Late Cainozoic vegetational and environmental changes in the Malay Archipelago. In: Whitmore TC, editor. Biogeographical Evolution of the Malay Archipelago. Vol. 4. Oxford Monographs on Biogeography, UK, Oxford: Oxford University Press; 1987. pp. 50–59. [Google Scholar]
- Rawlings LH, Barker D, Donnellan SC. Phylogenetic relationships of the Australo-Papuan Liasis pythons (Reptilia: Macrostomata), based on mitochondrial DNA. Austral J Zool. 2004;52:215–227. [Google Scholar]
- Van Tol J, Gassmann D. Zoogeography of freshwater invertebrates of Southeast Asia, with special reference to Odonata. In: Renema W, editor. Biogeography, Time and Place–Distributions, Barr iers and Islands. Dordrecht: Springer; 2007. pp. 45–91. [Google Scholar]
- Marshall AG. Bats, flowers and fruit: evolutionary relationships in the Old World. Biol J Linn Soc. 1983;20:115–135. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specimens and GenBank information. Information regarding species sampling, localities, and GenBank accession numbers for sequences used in BEAST analyses.