Summary
The dynamic ability of genomes to interact with discrete nuclear compartments appears to be essential for chromatin function. However, the extent to which structural nuclear proteins contribute to this level of organization is largely unresolved. To test the links between structure and function, we evaluated how nuclear lamins contribute to the organization of a major functional compartment, the nucleolus. HeLa cells with compromised expression of the genes encoding lamins were analyzed using high-resolution imaging and pull-down assays. When lamin B1 expression was depleted, inhibition of RNA synthesis correlated with complex structural changes within the nucleolar active centers until, eventually, the nucleoli were dispersed completely. With normal lamin expression, the nucleoli were highly plastic, with dramatic and freely reversible structural changes correlating with the demand for ribosome biogenesis. Preservation of the nucleolar compartment throughout these structural transitions is shown to be linked to lamin B1 expression, with the lamin B1 protein interacting with the major nucleolar protein nucleophosmin/B23.
Keywords: Nuclear organization, Nuclear compartments, Nucleoskeleton, Nuclear lamins, Nucleoli, Fibrillarin, Nucleophosmin/B23
Introduction
Chromatin function is regulated by the way in which genomes are folded in the nucleus. In part, this reflects the ability of chromatin to interact dynamically with nuclear compartments where specific nuclear functions are performed (reviewed by Spector, 1993; Lamond and Earnshaw, 1998; Misteli, 2001; Mateos-Langerak et al., 2007). The dynamic properties of the relevant components are particularly important during transcription, where both the regulatory and synthetic factors (Phair and Misteli, 2000) and the chromatin template (reviewed by Belmont, 2003; Rafalska-Metcalf and Janincki, 2007) must interact within the synthetic nuclear compartment. Although molecular mechanisms that drive nuclear dynamics remain largely unexplored, recent studies have shown that changes in gene structure and position that are linked to gene expression are dependent on nuclear actin-myosin motor function (Chuang et al., 2006; Dundr et al., 2007). It is unknown, however, how nuclear actin and myosin are organized and it is still unclear whether a structural framework exists that might facilitate chromatin repositioning.
If actin-dependent repositioning of chromatin is required during gene expression, the nuclear lamin-based nucleoskeleton would be a good candidate to support this role (Worman and Courvalin, 2005; Broers et al., 2006; Mounkes et al., 2003; Gruenbaum et al., 2003). Nuclear lamin proteins are type V intermediate filaments. In mammals, lamins A and C (derived from LMNA by alternative splicing) associate with lamins B1 and B2 (encoded by LMNB1 and LMNB2) to form a structural nuclear framework. The lamin-based filaments form both the nuclear lamina on the nucleoplasmic face of the nuclear membrane and a linked nucleoskeleton (Jackson and Cook, 1988; Hozak et al., 1995; Barboro et al., 2002) that spreads throughout the nucleus (Gruenbaum et al., 2003; Shumaker et al., 2003). In extracted nuclei, the peripheral nuclear lamina and diffuse internal network (Goldman et al., 2002; Gruenbaum et al., 2003; Broers et al., 2006) form a nuclear matrix that also contains actin and myosin (Goldman et al., 2002; de Lanerolle et al., 2005). The nuclear lamin plays an essential role in maintaining the structure of the nucleus, whereas the nucleoskeleton supports major nuclear functions (Cook, 1999) (reviewed by Dechat et al., 2008) such as DNA replication (Moir et al., 2000; Shumaker et al., 2008) and transcription (Spann et al., 2002; Kumaran et al., 2002; Tang et al., 2008). However, despite the potential importance of the structure-function links, only three studies have implicated lamin proteins in the regulation of the long-range dynamics of chromatin (Shumaker et al., 2006; Malhas et al., 2007; Tang et al., 2008).
The regulatory properties of nuclear lamins are demonstrated by the complex phenotypes associated with mutation in LMNA (Worman and Courvalin, 2005; Broers et al., 2006) and the related defects in knockout mice that are null for LMNA expression (Mounkes et al., 2003; Gruenbaum et al., 2003). Defects in the expression of B-type lamins appear to have more severe effects, and very few defects have been described in the expression of B-type lamins with established pathologies. Interestingly, transgenic mice that express mutated lamin B1 (Vergnes et al., 2004) have more severe pathology than LMNA-null mice and die at birth from respiratory failure. Although the isolation of mouse embryonic fibroblasts (MEFs) from such animals implies that lamin B1 is not essential for cell proliferation, the depletion of lamin B1, using RNA interference (RNAi), has shown that cells with reduced expression of LMNB1 have very limited proliferative potential (Harborth et al., 2001).
We have surprisingly little knowledge of the molecular mechanisms that link nuclear function to structural nuclear elements such as the lamin-containing nucleoskeleton. To explore this, we evaluated how the nuclear lamin proteins contribute to the functional plasticity of a well-characterized nuclear compartment – the nucleolus. The nucleolus is committed principally to polymerase I-dependent transcription of ribosomal genes and the assembly of pre-ribosomal particles (Raska et al., 2006). Nucleoli are built around nucleolar-organizing regions (NORs) that form on the ribosomal DNA (rDNA) gene loci (Scheer and Hock, 1999; Carmo-Fonseca et al., 2000). During interphase, nucleoli display clearly defined subcompartments (reviewed by Sirri et al., 2008). NORs, the associated transcription factors, synthetic machinery and nascent ribonucleoprotein (RNP) are located within the fibrillar centers/dense fibrillar component complexes where rRNA synthesis takes place. These active centers are embedded within a granular component, which is dedicated to biogenesis of ribosome particles. Despite their highly structured appearance, nucleoli are extremely dynamic. Indeed, most nucleolar proteins diffuse freely throughout nucleoli, typically displaying residence times of a minute or less (Misteli, 2001). At least in part, this explains the remarkable plasticity of the nucleolar structure that is seen when synthesis is inhibited (Haaf and Ward, 1996; Louvet et al., 2005) and during mitosis, when ribosomal RNA (rRNA) synthesis is switched off and the nucleoli disassemble before cell division (Savino et al., 2001).
Although the architecture of nucleoli is defined by the steps of ribosome biogenesis, the molecular mechanisms that are responsible for their formation and maintenance remain a matter of debate (Raska et al., 2006). A key organizational feature undoubtedly reflects the self-assembly properties of the major nucleolar proteins (Misteli, 2007); Cajal bodies provide another excellent example of this organizational principle (Kaiser et al., 2008). Nevertheless, it is unclear whether this property alone can account for the dynamic properties of nucleoli. In this regard, it is interesting to note that the nucleoli of somatic cells incubated in Xenopus egg extract can be almost completely disrupted without loss of pre-rRNA synthesis (Gonda et al., 2003). The nucleolar disassembly that occurs under these conditions is regulated by the germ cell-specific proteins FRGY2a and FRGY2b, and is dependent on the interaction of these proteins with the major nucleolar protein nucleophosmin/B23 (Gonda et al., 2006).
Nucleolar organization provides an excellent opportunity to explore links between a fundamental nuclear activity – transcription of rRNA – and the lamin-dependent nucleoskeleton. Using high-resolution imaging and biochemical assays on HeLa cells with compromised expression of the genes encoding lamins, we demonstrate that the normal expression of B-type lamins is required to maintain the architecture and functional plasticity of nucleoli. We show that an interaction between the lamin proteins and the major nucleolar protein nucleophosmin/B23 provides a molecular connectivity that links the lamin-dependent nuclear networks and nucleoli.
Results
Nucleoli undergo dramatic rearrangements during lamin B1 depletion
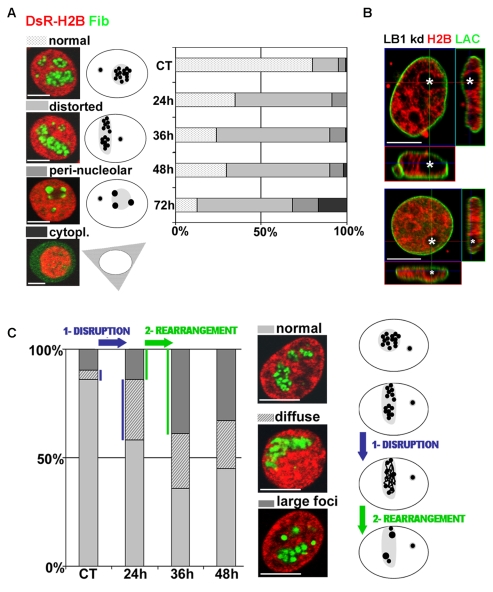
In mammalian cells, nuclear structure and function are linked inextricably. However, it is unknown whether structure is a fundamental determinant of function or a passive by-product. To address this, we evaluated how the major structural nuclear proteins, the nuclear lamins, contribute to the structure and dynamic behavior of the most obvious nuclear compartment, the nucleoli. Using a vector-based RNA interference protocol (Tang et al., 2008), LMNB1 gene expression was depleted in HeLa cells and the organization of the nucleoli was analyzed by immunostaining for fibrillarin, the main component of the active transcription centers. With this system, transfection efficiency remains at around 30%; however, in each cell expressing the control vector, the level of depletion is close to 90%, allowing efficient single cell analysis (Tang et al., 2008). Confocal analysis revealed four distinct phenotypes (Fig. 1A) that correlate with the severity of lamin B1 depletion. Initially, after 24 hours or less of depletion, nucleoli adopted a `distorted' phenotype with the majority of cells containing a single elongated nucleolus. The structure of the distorted nucleoli is particularly evident after 48 hours, when elongated nucleoli clearly fuse with the nuclear periphery (Fig. 1B, see the z-orthogonal sections of representative nucleoli in two cells with typically distorted nucleoli). Throughout the period of analysis, this dominant phenotype was four- to fivefold more prevalent in lamin B1-depleted cells than in controls. Two further abnormal patterns were seen with increased lamin B1 depletion. First, large round nucleoli were seen with large `peri-nucleolar' aggregates of fibrillarin that formed along their periphery (Fig. 1A). Second, about 20% of the treated cells showed a pronounced deconstruction of the nucleolar compartment, with fibrillarin dispersed throughout the cell (Fig. 1A).
Fig. 1.
Nucleoli are reorganized in cells with reduced lamin B1 expression. HeLa cells were co-transfected with the LB1-RNAi vector and vector expressing DsRed-H2B (red), fixed at selected times during lamin B1 depletion and immunoprocessed for fibrillarin (A,C) or lamin A/C (B) using Alexa Fluor 488-conjugated secondary antibody (green). (A) Confocal analysis shows four distinct phenotypes (normal, distorted, peri-nucleolar and cytoplasmic) at 24-72 hours post-transfection (left). Alterations in nucleolar structure are depicted schematically (center) and the distribution of the major forms is shown graphically (right). (B) After staining for lamin A/C, optical sections of two representative nuclei at 48 hours after transfection show abnormal nucleolar stretching and pronounced association of the nucleoli (white asterisks) with the nuclear periphery (see also the z-orthogonal sections). (C) Changes in the texture of fibrillarin were analyzed using confocal microscopy. Compared with controls (normal), two distinct abnormal patterns (diffuse and large foci) appeared progressively between 24-48 hours post-transfection. The relative distribution of these forms is shown graphically (left). The cartoon (right) depicts the sequential disorganization: elongated nucleoli that retain visibly normal active centers become disrupted (blue arrow) and then rearranged (green arrow) to form large peri-nucleolar aggregates. In all experiments, controls (CT) were transfected with the DsRed-H2B vector only. For each time point, ∼100 nuclei were scored (see supplementary material Table S1). Scale bars: 5 μm.
These data show that depletion of lamin B1 expression correlates with a two-phase deconstruction of nucleoli, with distinct morphological alterations (elongation) followed by more profound structural remodeling. To evaluate if these patterns represent distinct structural transitions, we evaluated whether more subtle changes could be defined as lamin B1 expression was reduced. For this analysis we used high-resolution 3D image reconstruction techniques that focused on the texture of fibrillarin as a marker for nucleolar activity (Fig. 1C). In contrast to the classical `grape-like' organization of the fibrillarin-rich centers that are seen in ∼90% of untreated cells (Fig. 1C, top), lamin B1 depletion correlated with the appearance of elongated nucleoli with diffuse fibrillarin that dispersed throughout the nucleolar volume (Fig. 1C, middle). As lamin B1 expression fell, fibrillarin was seen to coalesce into larger spherical foci (Fig. 1C, bottom). Hence, fibrillarin rearranges according to a two-step process (Fig. 1C, cartoon) in which: (1) disruption of the active centers first leads to disorganization of fibrillarin throughout the distorted nucleolar compartment, and (2) aggregates of fibrillarin form large foci that disperse throughout the distorted nucleoli.
To ensure that the observed rearrangements were not a product of unpredictable off-target effects, cells were also co-transfected with both the lamin B1-RNAi vector and a rescue vector that complements the depletion by expressing an RNAi-immune RNA that preserves the natural protein sequence (Tang et al., 2008). A vector-expressing yellow fluorescent protein (YFP)-tubulin was used as a reporter and a control experiment was performed by transfecting only the reporter and the RNAi vector. Nucleoli were assessed as before (Fig. 1), and the proportion of cells displaying an aberrant nucleolar phenotype (peri-nucleolar, distorted or diffuse) (supplementary material Fig. S1A) were counted. The appearance of all abnormal patterns was fully reversed by expression of the rescue vector (supplementary material Fig. S1), confirming that the structural rearrangements within nucleoli, and the associated disruption of their active centers, correlate with the depletion of lamin B1 expression.
Nucleolar disruption mediated by the loss of lamin B1 is mimicked by specific RNA polymerase inhibitors
To better characterize the nucleolar rearrangements linked to lamin B1 depletion, we also monitored ultrastructural changes that are described classically using electron microscopy. However, although the vector-based system provides efficient depletion within single cells, the analysis of cell populations is compromised by the levels of transfection, which are typically ∼30% (Tang et al., 2008). Hence, for experimental approaches that cannot define transfected cells, or that require large cell populations, alternative strategies are required. Since our previous studies showed that lamin B1 depletion correlated directly with reduced transcription by both polymerases I and II (Tang et al., 2008), we hypothesized that the changes in structure and function might be linked directly. Moreover, the peri-nucleolar structures that are seen at late stages of lamin B1 depletion are reminiscent of the `nucleolar caps' that have been described during transcriptional arrest (Shav-Tal et al., 2005; Junéra et al., 1995).
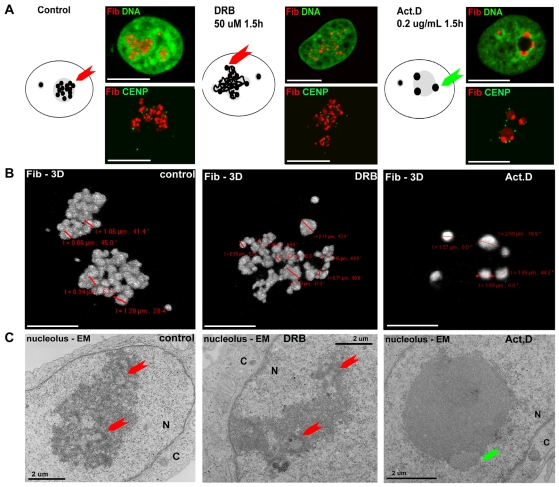
To confirm any correlation between the structural changes during LMNB1 depletion and the inhibition of transcription, we next analyzed the organization of fibrillarin in cells with reduced transcription, using actinomycin D and DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole) to inhibit synthesis by RNA polymerase I and II, respectively (Fig. 2). Following inhibition, characteristic changes mimicked those observed following depletion of lamin B1 (Fig. 1). During RNA polymerase II inhibition, the nucleoli appeared elongated, usually in contact with the nuclear periphery, and contained active centers that were noticeably disorganized, open and diffuse relative to controls (Fig. 2A-C, center). By contrast, when RNA polymerase I activity was inhibited, nucleoli adopted a spherical structure that located to the center of the nucleus and associated with a small number of dense fibrillarin aggregates at their periphery (Fig. 2A-C, right). Each of these phenotypes is reminiscent of defects seen at different times during lamin B1 depletion (compare the cartoons and pictures in Fig. 2A with those in Fig. 1), with the same sequence of changes occurring in all cells. Hence, the efficient manipulation of nucleolar structure using transcriptional inhibition provides an analytical strategy that is amenable to techniques such as proteomics and electron microscopy that require large populations of cells with a similar structure. Using this approach, 3D reconstruction of fibrillarin organization in individual nucleoli emphasizes the extent of the structural changes (Fig. 2B). In untreated cells, active centers were homogenous in size, ranging from 0.9-1.2 μm (Fig. 2B, left). By contrast, when RNA polymerase II activity was inhibited, fibrillarin localized into foci that were structurally heterogeneous, ranging from 0.4-2.0 μm (Fig. 2B, center), whereas loss of RNA polymerase I activity correlated with more pronounced aggregation of the fibrillarin-rich compartment to give sites of 1.6-2.6 μm (Fig. 2B, right).
Fig. 2.
Three-dimensional ultrastructure of nucleoli in cells with reduced RNA polymerase activity. HeLa cells were treated to inhibit specifically RNA polymerase II (middle column, DRB for 1.5 hours) or RNA polymerase I (right column, actinomycin D for 1.5 hours; Act. D), and the architecture and composition of the nucleoli was assessed by comparison with untreated controls (left column). (A) Optical sections of the predominant structures seen in each population when nuclei were immunostained to visualize fibrillarin (red) and either DNA-counterstained with Sytox green (DNA, green) or co-labeled for centromeres (CENP, green). Cartoons depicting the predominant structures in each population are shown. (B) High-resolution 3D views of fibrillarin (Fib-3D) were constructed from optical scans of whole nuclei. These images were used to measure the separation of individual FCs from the centers of the masses of fibrillarin foci; coordinates generated by imaging software are shown in red on the individual panels. (C) The ultrastructure of the nucleoli revealed by electron microscopy (nucleolus-EM). Active centers (red arrows) displayed a classical structure in controls but were much more heterogeneous in size (generally bigger) and found in lower numbers following DRB treatment. The structural changes seen in cells treated with actinomycin D were more dramatic, with fibrillarin accumulated in dense lentil-shaped aggregates (green arrow) at the periphery of the now spherical and amorphous residual nucleoli. The images shown focus on a central, densely staining nucleolus within a nucleus (N) that is separated from the cytoplasm (C) by a clearly defined nuclear membrane. Scale bars: 5 μm (A,B); 2 μm (C).
Using the inhibitor strategy, we performed an ultrastructural analysis of nucleoli using electron microscopy of ultrathin sections to characterize the three major nucleolar compartments (Hozak et al., 1994; Ploton et al., 1987) (for a review, see Sirri et al., 2008). Typically, nucleoli were seen to contain several fibrillar centers (FC) that were surrounded by a dense fibrillar component (DFC), which together form sites of polymerase I activity and RNA synthesis (the active centers are marked by red arrows in Fig. 2C). These active centers were surrounded by a more dispersed granular component (GC), where processing and maturation of the ribosomal particles occurs.
All three nucleolar compartments were maintained following DRB treatment (Fig. 2C, center), although their relative arrangement was clearly distorted in line with observations made by light microscopy. Importantly, FC-DFC complexes were still present and maintained a characteristic architecture, although generally the density and intensity of the DFC was reduced. Again in line with the light microscopy, the active centers marked by the FC-DFC complexes were clearly less abundant and more heterogeneous in size. By clear contrast, when RNA polymerase I activity was inhibited, the typical organization of the nucleolar compartment was lost completely (Fig. 2C, right). After this treatment, classical FC-DFC centers were absent and, instead, fibrillarin aggregated into large lentil-like structures that formed along the periphery of an amorphous GC.
Nucleoli display highly plastic properties that are maintained after the depletion of lamin A/C
Nucleoli clearly undergo characteristic morphological changes when the status of RNA synthesis is altered (Fig. 2); these changes mimic those seen during lamin B1 depletion (Fig. 1) and, importantly, provide large populations of cells with similar structural characteristics. Using this system, we asked whether the changes in the nucleolar subcompartments that occur in response to either depletion of lamin B1 expression or transcriptional inhibition might generate global changes in genome organization that would reflect the general plasticity of the nucleus. A key feature of this analysis is to define the efficiency with which the specific changes that are seen under different conditions revert to the organization seen in untreated cells. For these recovery experiments, we focused on inhibition of RNA polymerase II using DRB, as the inhibition of transcription with this drug is both efficient and freely reversible.
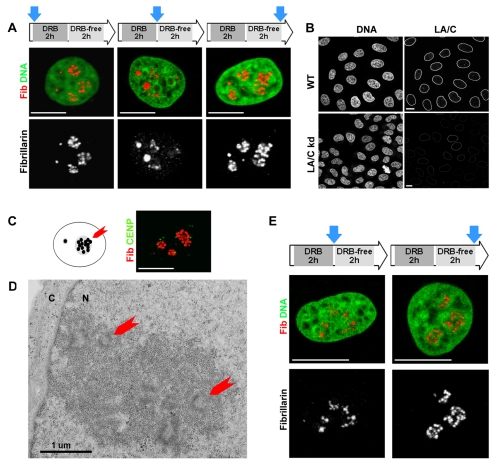
HeLa cells were treated with DRB and allowed to recover by incubation in medium without inhibitor; the organization of the active centers in nucleoli was then monitored by immunostaining (Fig. 3). As before, the DRB treatment resulted in a clear alteration in the organization of the fibrillarin-rich nucleolar sites, with a dispersed reticulate network of staining and a small number of larger condensed foci (Fig. 3A, center) when compared with untreated controls (Fig. 3A, left). Remarkably, despite the dramatic nature of these structural changes, removal of the inhibitor correlated with efficient and complete recovery of the normal nucleolar organization in all cells (Fig. 3A, right).
Fig. 3.
Changes in nucleolar structure are freely reversible in wild-type and lamin A/C-depleted cells. The plasticity of nucleoli was assessed by evaluating the reversibility of nucleolar organization in HeLa cells treated with DRB. (A) HeLa cells grown on coverslips were treated with DRB for 2 hours to inhibit RNA polymerase II and then grown for 2 hours in medium without DRB. Samples were immunoprocessed for fibrillarin (red) and DNA-counterstained with Sytox green (green) at the times indicated (blue arrows) in the incubation scheme (top). Confocal analysis shows that fibrillarin, which is massively disorganized when RNA polymerase II is inhibited, quickly recovered the normal structure when transcription resumed. The same structural transitions were seen in HeLa cells with reduced expression of A-type lamins (B-E). In contrast to the dramatic loss in nucleolar structure seen when lamin B1 was depleted, a HeLa cell line that constitutively expresses only 5% of the natural level of lamin A/C (LA/C kd; B) maintained normal nucleolar architecture, as shown by immunostaining for fibrillarin (Fib, red; C), and a normal distribution of centromeric repeats (CENP, green; C). Nucleoli with classical morphology (D; red arrows indicate active centers that are embedded with the granular component) were clearly seen in the nuclei (N) of lamin A/C-deficient cells and, when transcription was inhibited using DRB and later allowed to recover, these nucleoli showed the same structural transitions (E) as control cells (A). Scale bars: 5 μm.
These experiments emphasize the plasticity of the nucleolar compartment and also demonstrate a remarkable correlation between the changes seen during transcriptional inhibition and the structural changes that arise during lamin B1 depletion (Fig. 1). This implies that lamin-based structures might provide a framework that contributes to the regulation of nucleolar organization and plasticity in normal proliferating cells. To confirm the extent of lamin involvement in nucleolar plasticity, we evaluated whether lamin A and C proteins contribute to the structure of nucleoli.
As A-type lamins are not essential for proliferation, we were able to generate a short hairpin RNA (shRNA)-expressing cell line with LMNA expression reduced to only ∼5% of the natural levels (Fig. 3B). In this cell line, nucleolar structure and ultrastructure were assessed by confocal microscopy after fibrillarin immunostaining (Fig. 3C) and by electron microscopy (Fig. 3D). In marked contrast to cells with depleted lamin B1 expression, cells with very little expression of LMNA displayed normal nucleolar organization with classical FC, DFC and GC compartments. Structurally, fibrillarin showed a normal distribution in active centers that were homogeneous in size and texture (Fig. 3C,D), as seen in untreated controls (Fig. 2). The nucleoli in lamin A/C-depleted cells showed the same ability as normal HeLa cells to remodel nucleoli during recovery from RNA polymerase II inhibition (Fig. 3E), showing that expression of LMNA makes little or no direct contribution to the structural plasticity of nucleoli.
Nucleoli contain both A- and B-type lamins but are enriched in lamin B1
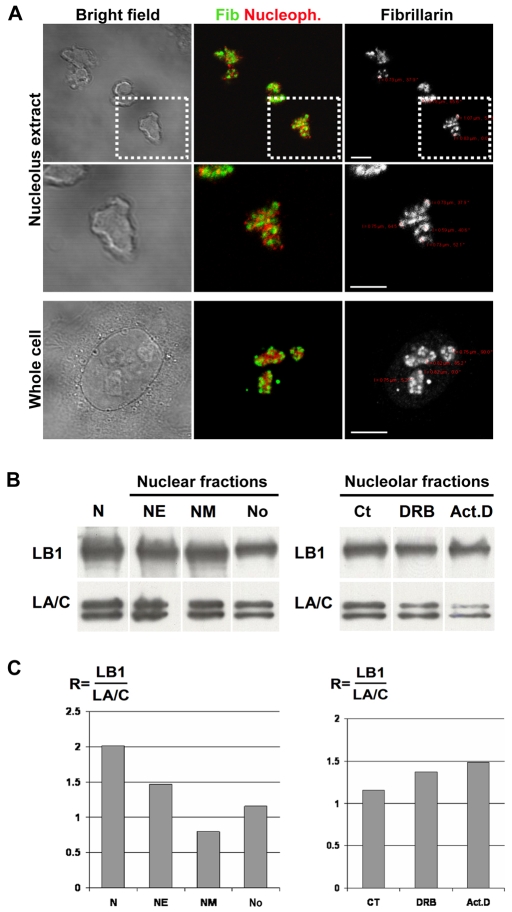
To reinforce the possibility that lamin B1 contributes to the maintenance of nucleolar structure, we next evaluated the composition of lamin proteins in isolated nucleoli (Fig. 4A). In fact, despite their evident plasticity, nucleoli that retain a normal architecture can be purified using well-established protocols, which allow analysis of the nucleolar proteome under different experimental conditions (Andersen et al., 2005; Trinkle-Mulcahy and Lamond, 2007). Isolated nucleoli were clearly discrete, phase dark entities that retained the same structural features (Fig. 4A, bright field, nucleolus extract) as nucleoli in situ (Fig. 4A, bright field, whole cell). Using recognized markers – fibrillarin to stain the FC/DFC and nucleophosmin to stain the surrounding GC – purified nucleoli maintained the natural distribution of size, shape and structure of the active centers, even though the isolated structures were freely dispersed within the sample (Fig. 4A). Moreover, by both phase and fluorescent microscopy, dispersed nucleoli were free of other particulate debris and appeared as discrete ovoid structures of 2-5 μm diameter with sharply delineated boundaries (Fig. 4A). In the isolated nucleoli, fibrillarin was maintained as discrete foci, with an average diameter of 0.7 μm, embedded in a structural compartment that was rich in nucleophosmin (Fig. 4A; see high-power views). Crucially, this organization of fibrillarin and nucleophosmin in isolated nucleoli mimics the organization of these components in intact cells (Fig. 4A; compare high-power views of nucleolus extract with those of whole cells), demonstrating that the structure of the nucleolar subcompartment is preserved during isolation.
Fig. 4.
Lamin proteins are retained in purified nucleoli. (A-C) Immunoblotting was used to monitor the relative concentration of the A- and B-type lamin proteins in nuclear fractions isolated from HeLa cells. Different nuclear fractions (N, nucleus; NE, nuclear envelope; NM, nuclear matrix; No, nucleolus) were isolated using established protocols. (A) As a quality control, isolated nucleoli (nucleolus extract) were shown to retain the same size, structure and organization as the nucleoli of untreated cells (whole cell) when analyzed by indirect immunostaining. Bright-field images (left) and optical sections (right) are shown together with a 2× magnification of a typical nucleolus (middle row, from boxed areas in the top row) to show that the fibrillarin-stained active centers (Fib, green) and nucleophosmin-stained granular component (Nucleoph, red) are maintained throughout isolation; the separation of active sites was determined as described (see Fig. 2 legend). (B,C) Following purification, nuclear fractions (7 μg/lane) were separated by gel electrophoresis and the presence of A- and B-type lamins was determined by immunoblotting (left). The same approach was applied to isolated nucleoli after treating cells for 2 hours with DRB or actinomyicn D (Act. D), using untreated controls (Ct) for comparison (right). The changes in the lamin protein concentration (C) were assessed by calculating the intensity ratio of lamin B1 (LB1) relative to lamin A and C (LA/C). Note that although this analysis shows the relative content of A- and B-type lamins in different fractions, it is not designed to provide a quantitative distribution of the proteins during fractionation (see text). Based on the cell numbers used and the product yields, nucleoli contain <2% of the lamin polypeptides in HeLa cells. Scale bars: 5 μm.
We compared the composition of the lamin proteins in the nucleoli (No) with different nuclear compartments by using established protocols to prepare purified nuclei (N), nuclear envelope (NE) and nuclear matrix (NM) fractions. Both lamin B1 and lamin A/C were detected readily in all fractions using routine immunoblotting (Fig. 4B, left blot). During this analysis, we found that it was impossible to avoid the slightly variable yields that result from recovery efficiencies of the different purification protocols. To accommodate this experimental variability, intensity measurements were used to calculate a lamin content ratio, which defines the relative intensity of lamin B1 immunostaining with respect to lamin A/C [R=(LB1)/(LA/C)] in the different fractions. This approach rules out the uncertainty of absolute measurements, which require that identical cell equivalents be compared for the different fractions. Crucially, this analysis confirms the retention of lamin proteins within purified nucleoli, even though different fractions are seen to retain slightly different lamin protein compositions (Fig. 4C). Although these slight differences in the lamin content ratio might be of functional importance, it is premature to speculate on these differences here, other than to note that the relative enrichment of lamin B1 in intact nuclei is consistent with the differential loss of a small fraction of lamin B1 during nuclear extraction.
To confirm the structure-function links, we also assessed the lamin composition of purified nucleoli during inhibition of RNA synthesis (Fig. 4B, right blot). Nucleoli were isolated from untreated cells as well as cells treated with the transcriptional inhibitors DRB and actinomycin D, and the composition of their lamin proteins was determined (Fig. 4). During transcriptional inhibition, the relative content of lamin B1 in the nucleoli was seen to increase as the normal, function-dependent architecture of nucleoli became disrupted. Notably, this relative enrichment of lamin B1 correlated with a notable loss of lamin A/C (especially lamin A) (Fig. 4B, right blot, Act.D lane). Taken together, these experiments highlight the presence of lamin B1 in nucleoli and show that the content of lamin B1, relative to lamin A/C, is enriched when nucleolar components become rearranged or disorganized as a consequence of transcriptional inhibition. This reinforces the idea that lamin B1 plays a role in nucleolar plasticity, which might be of particular relevance under conditions of morphological stress.
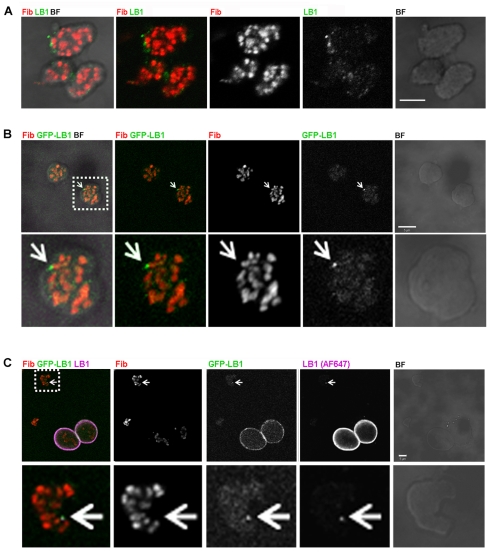
Microscopy was also used to confirm the presence of lamin B1 in nucleoli. Extracted nucleoli were used for this analysis to allow the best possible sensitivity and antibody accessibility. Following indirect immunodetection, confocal microscopy revealed distinct lamin B1-containing foci that were heterogeneous in size and dispersed throughout the GC but in close proximity to the active centers (Fig. 5A). Analysis of samples from cells expressing green fluorescent protein (GFP)-LB1 confirmed the localization of lamin B1, within the nucleolar GC, in close proximity to the active centers (Fig. 5B, enlargement). As a control, purified nucleoli and intact nuclei were mixed and lamin B1 detected using both the GFP-LB1 signal and lamin B1 immunostaining. Nuclei showed the expected strong staining of the nuclear periphery (Fig. 5C) and clear co-localization of GFP-LB1 with lamin B1 was determined by immunodetection (Fig. 5C). This confirms the association of lamin B1 with nucleoli following biochemical fractionation (Fig. 4) and shows that lamin B1 in nucleoli is distributed throughout the GC in close proximity to the FC-DFC complexes.
Fig. 5.
Distribution of lamin B1 in extracted nucleoli. Nucleoli were extracted, deposited on glass slides and immunoprocessed. (A-C) Immunolocalization shows lamin B1 (LB1, green in A and cyan in C) to be dispersed throughout the GC as small discrete foci that localize to the periphery of the FC-DFC complexes that have been stained with fibrillarin (Fib, red). (B) The localization seen by immunostaining was confirmed in nucleoli isolated from cells expressing GFP-conjugated lamin B1. In both cases, the majority of foci have a similar structure, although a small number of bright foci (indicated by arrows in B and C) also localize to the FC-DFC borders. (C) As a staining control, isolated nucleoli and nuclei from GFP–LB1-expressing cells were mixed before immunostaining. In both nucleoli in situ and after isolation, the sites with GFP-LB1 co-localized with lamin B1-containing sites, as defined by immunostaining (LB1, cyan); note the very bright staining of the nuclear lamina in the nuclei, which was never seen in preparations of purified nucleoli. Magnified images (lower panels in B and C show a 2.5× magnification of box regions in the upper panels) emphasize the distribution of labeled sites. Scale bars: 5 μm.
Lamin B1 is associated with nucleoli through an interaction with the major GC protein nucleophosmin
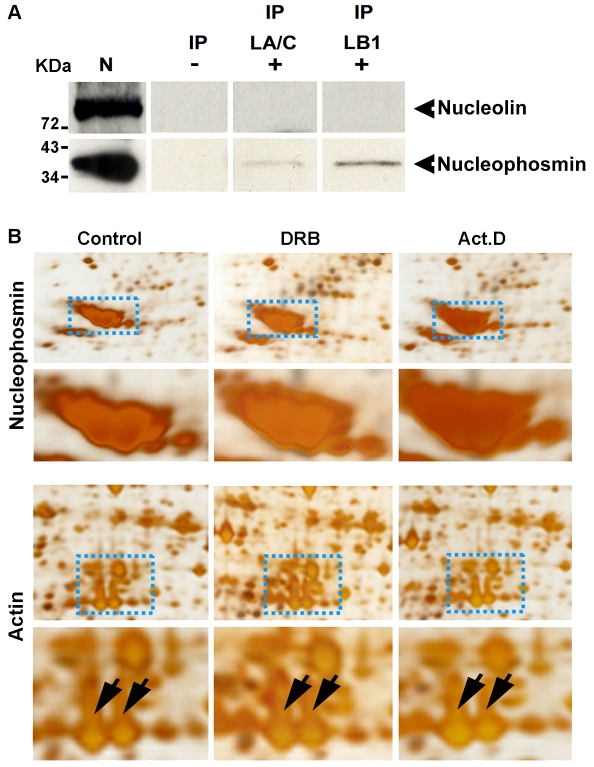
Finally, we wanted to evaluate possible interactions between nucleoli and the lamin protein-based nuclear networks. With this in mind, we note preliminary evidence that the major GC protein nucleophosmin/B23 might contribute to the organization of nucleoli in somatic cells (Gonda et al., 2003; Gonda et al., 2006). To explore this possibility, we used immunoprecipitation (IP) to probe the interaction of nucleophosmin and another major GC protein, nucleolin, with the nuclear lamin proteins (Fig. 6). Nuclear extracts were prepared by sonication and control experiments were performed to ensure that nucleolar components, as well as nuclear lamin proteins, were retained in the soluble fraction before IP. Using these fractions and standard IP protocols, proteins interacting with lamin B1 or lamin A/C were isolated and the protein complexes separated by gel electrophoresis. Interactions between nucleophosmin or nucleolin and the lamin proteins were then assessed by immunoblotting using specific antibodies (Fig. 6). Using this approach, nucleophosmin displayed a clear and reproducible interaction with lamin B1 and immunoprecipitated to a lesser extent using antibodies to lamin A/C, whereas nucleolin showed no evidence of interacting with either lamin protein (Fig. 6A). This provides clear evidence for an interaction between the nuclear lamin proteins and nucleophosmin/B23 without ruling out the possibility that other nucleolar proteins might also interact with the lamin-based network.
Fig. 6.
Lamin B1 preferentially associates with nucleophosmin within the nucleolar compartment. HeLa cells were processed for IP and protein complexes linked to lamin B1 or lamin A/C were separated by electrophoresis. Two major GC proteins (nucleolin and nucleophosmin) were then detected by immunoblotting (A). In three separate experiments, nucleophosmin showed a clear association with the nuclear lamin IP fractions, particularly lamin B1 (exposure 40 minutes). No detectable signal was seen when immunoblotting was performed using antibodies to nucleolin (A). IP, IP fraction with either lamin A/C (LA/C) or lamin B1 (LB1); N, nuclear extract (positive control for blotting); (–), IgG (negative control for IP). (B) Proteins from nucleoli isolated after drug treatment to inhibit RNA polymerase I (Act. D) or II (DRB) were inspected after separation by 2D gel electrophoresis. The intensity of the major nucleolar protein nucleophosmin (upper panel, enlargement below) increased by ∼1.5-fold after DRB and by ∼2.5-fold after Act. D treatment. As a loading control, actin spots from the same gels do not show any intensity variations (lower panel, actin indicated by the arrows on the corresponding enlargement). Images shown are representative of typical gels from experiments performed on at least three occasions.
The demonstration of a link between lamin B1 and nucleophosmin (Fig. 6), and the eventual disassembly of the nucleolar compartment when lamin B1 expression is reduced (Fig. 1), suggests that interactions between lamin proteins and nucleophosmin in the nucleolar GC contribute to the global organization of the nucleolar compartment in mammalian cells. Interestingly, under conditions of transcriptional inhibition, when the relative concentration of lamin B1 protein in nucleoli was increased (Fig. 4), a concomitant 1.5-fold to twofold increase in nucleolar nucleophosmin was seen (Fig. 6). Taken together, these data support the interaction of lamin B1 and the major nucleolar protein nucleophosmin/B23, and propose a model in which the dynamic behavior of these components is a fundamental determinant of nucleolar function and plasticity in mammalian cells (Fig. 7).
Fig. 7.
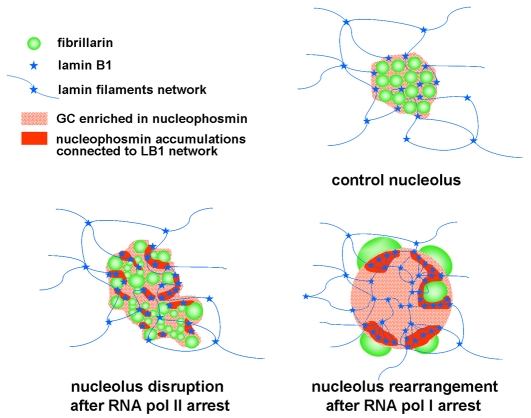
Model for a lamin B1-nucleophosmin network maintaining nucleolar architecture. The model depicts a schematic view of nucleolar architecture and changes during transcriptional stress. Under physiological conditions (control), the active sites of rDNA synthesis are contained within fibrillarin-rich FC-DFC complexes (green beads) and surrounded by a granular component (GC) that is rich in nucleophosmin (dotted red). This component interacts with a lamin-containing filament network (blue) through an interaction between nucleophosmin and lamin B1 (blue stars). When RNA polymerase II activity is inhibited and the demand for rRNA falls, the normally highly structured active centers unfold and global nucleolar structure is disrupted. Clusters of rDNA genes that normally occupy a single FC-DFC complex are now distributed throughout the GC. Quite different structural changes are seen when RNA polymerase I activity is inhibited. When rRNA synthesis is completely switched off, the FC-DFC complexes are no longer present and fibrillarin accumulates in large peri-nucleolar aggregates. This structural change correlates with increased retention of nucleophosmin and lamin B1 in residual nucleoli, suggesting that the concentration of these components is normally regulated by the dynamic properties of nucleolar proteins. The model explains how reduced lamin B1 expression leads to loss of nucleolar structure so that fibrillarin can diffuse throughout the cell. Under these conditions, the spherical residual nucleoli seen during RNA polymerase I arrest are not seen, suggesting that this level of organization is dependent on lamin B1-nucleophosmin interactions in the GC. In the presence of NORs that position the rRNA genes, the structural framework formed by the interaction of lamin B1 with nucleophosmin would form an underlying architecture on which the localization and concentration of nucleophosmin is sufficient to drive the assembly of nucleoli as a consequence of the self-organization properties of the major nucleolar proteins. The changes seen when transcription is compromised imply that the underlying architecture is also locally dynamic. Hence, our model supports the dynamic properties of nucleolar components based on their biophysical characteristics, and explains how the nuclear localization and organization of nucleoli respond to any changing demand for RNA synthesis.
Discussion
This study highlights, for the first time, how the structural nuclear protein lamin B1 contributes to the functional plasticity of a major nuclear compartment – the nucleolus. In a previous study, we showed that reduced expression of lamin B1 correlated with a gradual decline in transcription by RNA polymerase II. At the later stages of lamin B1 depletion, a clear loss of ribosomal gene transcription within nucleoli was also seen, although it is unknown whether this reflects a corresponding fall in the demand for ribosomal biogenesis or an essential requirement for lamin B1-containing structures within the nucleus (Tang et al., 2008). We now show that reduced synthetic activity within nucleoli coincides with a series of organizational changes that reflect declining transcription (Fig. 1). Interestingly, morphological changes seen in cells with compromised lamin B1 expression can be mimicked using inhibitors of RNA synthesis. Moreover, despite the seemingly dramatic structural changes that are seen, the major changes are freely reversible if inhibition of transcription is relieved, reinforcing the remarkable structural plasticity of nucleoli in accommodating different levels of rRNA synthesis (Figs 2 and 3).
In a previous study (Haaf and Ward, 1996), a general dispersion of nucleolar FCs in cells treated with DRB supported clear links between global nuclear organization and function. However, despite possible roles for global nuclear structure in regulating nucleolar plasticity, little is known about the structural context through which the nucleolus and nuclear structure might interact. Our analysis extends the present view of organizational plasticity within nucleoli and, in addition, provides a molecular framework in which interactions between lamin B1 in the nucleoskeleton and the structural nucleolar protein nucleophosmin/B23 form a basis for maintaining the nucleolar compartment (Figs 4, 5, 6). The role of nuclear lamins in nucleolar plasticity is also supported by changes in the relative amounts of B- and A-type lamins in nucleoli under conditions of transcriptional stress. Together, these observations emphasize how different structural steady states can arise in nucleoli with different functional properties, and support a model (Fig. 7) in which lamin B1-nucleophosmin interactions could contribute to the maintenance of the global organization of nucleoli and regulate the dynamics within the compartment under conditions of transcriptional stress.
In recent studies, new insights into nucleolar function have been developed by comparing nucleolar morphology with the corresponding proteome (Andersen et al., 2005; Trinkle-Mulcahy and Lamond, 2007). Based on liquid chromatography-tandem mass spectrometry after stable isotope labeling, these authors identified 489 nucleolar proteins that were differentially expressed during transcription inhibition, only 11 of which were seen to migrate into the nucleolus (Andersen et al., 2002). Interestingly, this analysis shows that changes in nucleolar morphology that result from altered patterns of transcription are predominantly a consequence of the redistribution of pre-existing components. Our high-resolution 3D and ultrastructural study (Fig. 2), together with a proteomic analysis based on 2D gel electrophoresis (Fig. 6), also show that the nucleolar compartment remains largely intact even during profound morphological rearrangements that are induced during transcriptional inhibition.
In proliferating cells, the most dramatic demonstration of nucleolar plasticity is seen during mitosis, when nucleoli disassemble so that most of their components disperse throughout the cytoplasm. Notably, during this process, RNA synthesis is switched off and the nuclear lamin filaments are disassembled. As daughter nuclei reform in the following interphase, nucleoli reassemble around NORs, which are retained on rDNA gene clusters throughout mitosis, to generate prenucleolar bodies (PNBs) that become functional nucleoli in early G1 (Dundr et al., 2000; Jimenez-Garcia et al., 1994; Savino et al., 2001; Leung et al., 2004) (for a review, see Hernandez-Verdun and Louvet, 2004). In functional nucleoli, structural plasticity also correlates with the high mobility of nucleolar proteins that are required for rRNA transcription and processing, such as fibrillarin, upstream binding factor (UBF), nucleophosmin, nucleolin and Nop52 (Phair and Misteli, 2000; Dundr et al., 2004; Chen and Huang, 2001; Louvet et al., 2005). In addition, nucleoli have been shown to move inside living cells in a way that allows their fusion and dissociation; this explains the observed heterogeneity in the numbers of nucleoli in cell culture (Leung et al., 2004; Dundr et al., 2000).
In regulating nucleolar plasticity, a lamin B1-nucleophosmin framework could reflect a fundamental mechanism by which the B-type lamins contribute to mammalian cell viability. Supporting this role, B-type lamins are present throughout development, whereas A-type lamins are expressed only following the onset of cell differentiation (Stewart and Burke, 1987; Rober et al., 1989; Lehner et al., 1987). Since the zygotic genome establishes specific epigenetic marks that correlate with embryo-specific transcriptional activities before A-type lamins are expressed (reviewed by Santos and Dean, 2004), it is reasonable to propose that B-type lamins contribute to nuclear remodeling at this time. In particular, the assembly of functional nucleoli in mice is a sequential process in which PNBs that form soon after fertilization are maintained through early cell cycles before becoming active nucleoli (Zatsepina et al., 2003). The configuration of these PNBs is reminiscent of the peri-nucleolar patterns that are seen at late stages of lamin B1 depletion in cultured cells and following inhibition of RNA polymerase I activity (Fig. 1A; Fig. 2). Since PNBs turn into functional nucleoli only after polymerase I-dependent transcription has begun (Zatsepina et al., 2000; Zatsepina et al., 2003), it is clear that distinct morphologies correlate with nucleoli that are either inert or engaged in RNA synthesis (Shav-Tal et al., 2005; Junéra et al., 1995).
Although our data suggest a role of lamin B1 in contributing to the structure-function links in mammalian cells, some experiments appear to contradict this possibility. Most notably, mice with a disruption of Lmnb1 are able to develop to term and embryonic fibroblasts isolated from the developing embryos are able to proliferate in culture (Vergnes et al., 2004). However, this mutant is not a genetic null mutant and although insertion of a βgeo gene into intron 5 of Lmnb1 prevents the normal assembly of lamin B1 filaments, it does not alter the promoter function and yields a fusion protein that still locates to the nuclear periphery. Hence, although cells lacking the lamin B1 carboxyl-terminal extremity display altered structural properties (Vergnes et al., 2004), altered patterns of gene expression and abnormal chromosome territory distribution (Malhas et al., 2007), it is not entirely clear how these phenotypes relate to changes in natural lamin B1-containing nuclear structures.
Finally, our data support the existence of a nucleoskeleton that contributes to spatial architecture in mammalian nuclei. Nuclear lamin proteins are known to provide an intermediate filament network that maintains nuclear structure (Worman and Courvalin, 2005; Broers et al., 2006; Mounkes et al., 2003; Gruenbaum et al., 2003) and our present study suggests that natural levels of lamin B1 expression contribute to nucleolar structure and also function. This is consistent with the idea that transcription occurs at a lamin B1-containing nucleoskeleton in human cells (Jackson and Cook, 1985) and that this network contributes to the organization of nucleoli (Hozak et al., 1994). If this is true, it is interesting to speculate how the nucleoskeleton and biophysical features such as protein self-assembly (Misteli, 2001) contribute to the overall structure of eukaryotic nuclei. Our experiments support the idea that the nucleoskeleton-dependent interactions dictate the long-range organization of active centers, whereas the properties of self-assembly maintain the structure of the active compartments locally (Fig. 7). In the nucleolus, our model proposes that the interaction between nucleoskeleton-associated lamin B1 and the major nucleolar protein nucleophosmin defines the spatial architecture of the compartment to allow remodeling, which occurs during mitosis or under conditions of transcription stress, while maintaining the dynamic properties that are essential for functional efficacy.
Materials and Methods
Cell culture
HeLa cells were grown in DMEM supplemented by 10% fetal bovine serum and antibiotics (37°C, 5% CO2). To inhibit transcription, cells plated on glass coverslips were incubated with 50 μM DRB (Sigma) or 0.2 μg/ml actinomycin D (Sigma) and added to the culture media for 1.5 or 2 hours. For DRB recovery experiments, after 2 hours, DRB treatment cells were washed and grown in drug-free medium for 2 hours.
RNAi protocols
LB1-RNAi and RNAi-rescue vectors were described by Tang et al. (Tang et al., 2008). DsRed-H2B and YFP-tubulin expression were used as reporters to identify RNAi-treated cells. Vectors (1 μg in OptiMEM) were complexed with Polyfect (12 μl, Qiagen) for 15 minutes at 20°C and then added to fresh DMEM; this media was added to HeLa cells that had already been plated in 3 cm diameter Petri dishes (75% confluence). After 20 hours of incubation (37°C, 5% CO2), cells were seeded on coverslips and grown in DMEM for various times, as indicated in individual experiments.
Nuclear fractionation
Nucleoli were extracted as described by Lam and Lamond (Lam and Lamond, 2006). Briefly, nuclei were isolated after hypotonic stress and sonication, and pelleted by centrifugation (4°C). Nucleoli were isolated by centrifugation through a sucrose gradient (0.25 M, 0.35 M and 0.88 M) containing 0.5 mM MgCl2. For immunofluorescence, isolated nucleoli were resuspended in PBS, fixed in PBS-PFA 2% [10 minutes, room temperature (RT)] and deposited (30 × g, 15 minutes) on to glass slides (Superfrost) before processing, as described below.
Nuclear matrices were isolated as described by Comerford et al. (Comerford et al., 1986). Briefly, nuclei were pelleted by centrifugation, and washed and resuspended in 20 μg/ml DNAase I for 20 minutes (37°C). After spinning through a sucrose gradient in 2 M NaCl, nuclear matrices were recovered at the interface between the 40% and 50% sucrose steps.
Nuclear envelopes were extracted following the procedure described by Kaufmann et al. (Kaufmann et al., 1983). After treatment with 250 μg/ml DNAase I and 250 μg/ml boiled RNAase for 1 hour (4°C), nuclei in phenylmethylsulfonyl fluoride (PMSF) were lyzed and resuspended in four volumes of 2 M NaCl for 15 minutes to remove chromatin. Nuclear envelopes were then pelleted by centrifugation at 1600 × g for 30 minutes (4°C).
Immunoprecipitation
Cells were washed in PBS, resuspended in RIPA buffer containing protease inhibitors (Roche) and lyzed by sonication [3×10-second bursts, 30% power output (amp)]. After spinning (10,000 × g, 10 minutes, 4°C), supernatant was collected and incubated overnight with LA/C or LB1 antibodies (1/50, 4°C). Beads of protein G sepharose were then added and incubated for 1.5 hours. After three washes in RIPA (without proteases inhibitors), the bead pellet was resuspended in SDS sample buffer and boiled at 90-100°C for 5 minutes. Proteins were finally resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and nucleolin and nucleophosmin were then detected by western blotting.
Immunofluorescence and confocal microscopy
Cells were fixed at specific time points in PBS-PFA 2% (5 minutes, RT), permeabilized in PBS-Triton X-100 0.5% (15 minutes, RT) and blocked in PBS-BSA 2% (1 hour, 20°C). Primary antibodies against fibrillarin (1/200, Cytoskeleton; 1/400, Abcam), CENP-A (1/300, Abcam), lamin A/C (1/400, Santa Cruz) and nucleophosmin (1/500, Abcam) were diluted in PBS-BSA 2% and incubated overnight at 4°C. After successive washes in PBS-Tween 0.02%, secondary antibodies coupled to Cy3 (1/300, Jackson ImmunoResearch) or Alexa Fluor 488 (1/500, Molecular Probes) were added (1 hour, 20°C). Samples were washed in PBS-Tween and antibody complexes were briefly post-fixed in PBS-PFA 1%. DNA was counterstained with 0.25 μM Sytox green (Invitrogen) and finally mounted in Vectashield (Vector Laboratories).
Single optical sections were captured through a confocal microscope (Zeiss LSM 510) equipped with a plan apochromat 63× objective (DIC 1.4NA). For 3D views, coverslips were mounted on slides with tape spacer (to avoid cell squashing) and 300 nm z-stepped stacks were acquired. Images were processed using Zeiss and Imaris (Bitplane) software.
Electron microscopy
Cultured cells were fixed with 0.1% glutaraldehyde and 4% formaldehyde in 0.1 M cacodylate buffer (pH 7.4) containing 0.15 M sucrose and 2 mM CaCl2 for 2 hours at 20°C. Cells were then scraped and collected by centrifugation. Pellets were post-fixed for 1 hour in reduced osmium tetroxide solution [1% OsO4, 1.5% K4Fe(CN)6 in 0.1 M cacodylate buffer, pH 7.2] and for 15 hours in uranyl acetate (1% aqueous solution). Pellets were then dehydrated in ethanol, transferred to propylene oxide and infiltrated with low-viscosity resin (TAAB). Polymerization was performed for at least 24 hours and ∼70 nm sections were contrasted with aqueous uranyl acetate and lead citrate, and observed using an FEI Tecnai 12 electron microscope with an accelerating voltage of 100 kV.
Western blotting
Proteins from nuclear fractions (7 μg/well) or products from IP (20 μg/well) were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. Standard procedures were then applied for immunoblotting. Goat anti-mouse HRP (172-1011) and goat anti-rabbit HRP (172-1019) from Bio-Rad were used as secondary antibodies and detected with an ECL Western Blot Analysis System (RPN2109, GE Healthcare).
For detection of nucleolar partners in lamin A/C and lamin B1 IP products, rabbit anti-fibrillarin (1/250, Abcam), rabbit anti-nucleophosmin (1/1000, Abcam) and rabbit anti-nucleolin (1/100, Abcam) were used. For determination of lamin protein content in the different nuclear fractions, mouse anti-lamin A/C (1/750, Santa Cruz) and rabbit anti-lamin B1 (1/1000, Abcam) were used. To calculate the ratio R=(LB1)/(LA/C), band intensities were measured with the AlphaEaseFC software (ChemiImager 4400).
Two-dimensional electrophoresis and mass spectrometry
Protein extracts were dissolved in lysis buffer containing 9.5 M urea, 2.0 M thiourea, 4% (w/v) CHAPS, 1% (w/v) DTT, 2.5 mM EDTA and 2.5 mM EGTA. Proteins were separated by 2D gel electrophoresis and identified by liquid chromatography-tandem mass spectrometry, as described (Nirmalan et al., 2007). AlphaEaseFC software was used for analysis.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/10/1551/DC1
We thank Aleksandr Mironov for help with electron microscopy and the Wellcome Trust for providing equipment within the EM facility. We thank Chi Tang and Kang Zeng (of the author's group) for providing siRNA, DsRed-H2B and rescue vectors. This project was supported by the BBSRC (project grant D00327X). Deposited in PMC for release after 6 months.
References
- Andersen, J. S., Lyon, C. E., Fox, A. H., Leung K. L., Lam, Y. W., Steen, H., Mann, M. and Lamond, A. I. (2002). Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1-11. [DOI] [PubMed] [Google Scholar]
- Andersen, J. S., Lam, Y. W., Leung, A. K., Ong, S. E., Lyon, C. E., Lamond, A. I. and Mann, M. (2005). Nucleolar proteome dynamics. Nature 433, 77-83. [DOI] [PubMed] [Google Scholar]
- Barboro, P., D'Arrigo, C., Diaspro, A., Mormino, M., Alberti, I., Parodi, S., Patrone, E. and Balbi, C. (2002). Unraveling the organization of the internal nuclear matrix: RNA-dependent anchoring of NuMA to a lamin scaffold. Exp. Cell Res. 279, 202-218. [DOI] [PubMed] [Google Scholar]
- Belmont, A. (2003). Dynamics of chromatin, proteins, and bodies within the cell nucleus. Curr. Opin. Cell Biol. 15, 304-310. [DOI] [PubMed] [Google Scholar]
- Broers, J. L., Ramaekers, F. C., Bonne, G., Yaou, R. B. and Hutchison, C. J. (2006). Nuclear lamins: laminopathies and their role in premature ageing. Physiol. Rev. 86, 967-1008. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca, M., Mendes-Soares, L. and Campos, I. (2000). To be or not to be in the nucleolus. Nat. Cell Biol. 2, E107-E112. [DOI] [PubMed] [Google Scholar]
- Chen, D. and Huang, S. (2001). Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153, 169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, C. H., Carpenter, A. E., Fuchsova, B., Johnson, T., de Lanerolle, P. and Belmont, A.S. (2006). Long-range directional movement of an interphase chromosome site. Curr. Biol. 16, 825-831. [DOI] [PubMed] [Google Scholar]
- Comerford, S. A., Agutter, P. S. and McLennan, A. G. (1986). Nuclear matrices. In Nuclear Structures: Their Isolation and Characterization (ed. A. J. MacGillvray and G. D. Birnie), pp. 1-13. London: Butterworths.
- Cook, P. R. (1999). The organization of replication and transcription. Science 284, 1790-1795. [DOI] [PubMed] [Google Scholar]
- Dechat, T., Pfleghaar, K., Sengupta, K., Shimi, T., Shumaker, D. K., Solimando, L. and Goldman, R. D. (2008). Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 22, 832-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lanerolle, P., Johnson, T. and Hofmann, W. A. (2005). Actin and myosin I in the nucleus: what next? Nat. Struct. Mol. Biol. 12, 742-746. [DOI] [PubMed] [Google Scholar]
- Dundr, M., Misteli, T. and Olson, M. O. (2000). The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 150, 433-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr, M., Hebert, M. D., Karpova, T. S., Stanek, D., Xu, H., Shpargel, K. B., Meier, U. T., Neugebauer, K. M., Matera, A. G. and Misteli, T. (2004). In vivo kinetics of Cajal body components. J. Cell Biol. 164, 831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr, M., Ospina, J. K., Sung, M. H., John, S., Upender, M., Ried, T., Hager, G. L. and Matera, A. G. (2007). Actin-dependent intranuclear repositioning of an active gene locus in vivo. J. Cell Biol. 179, 1095-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, R. D., Gruenbaum, Y., Moir, R. D., Shumaker, D. K. and Spann, T. P. (2002). Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 16, 533-547. [DOI] [PubMed] [Google Scholar]
- Gonda, K., Fowler, J., Katoku-Kikyo, N., Haroldson, J., Wudel, J. and Kikyo, N. (2003). Reversible disassembly of somatic nucleoli by the germ cell proteins FRGY2a and FRGY2b. Nat. Cell Biol. 5, 205-210. [DOI] [PubMed] [Google Scholar]
- Gonda, K., Wudel, J., Nelson, D., Katoku-Kikyo, N., Reed, P., Tamada, H. and Kikyo, N. (2006). Requirement of the protein B23 for nucleolar disassembly induced by the FRGY2a family proteins. J. Biol. Chem. 281, 8153-8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum, Y., Goldman, R. D., Meyuhas, R., Mills, E., Margalit, A., Fridkin, A., Dayani, Y., Prokocimer, M. and Enosh, A. (2003). The nuclear lamina and its functions in the nucleus. Int. Rev. Cytol. 226, 1-62. [DOI] [PubMed] [Google Scholar]
- Haaf, T. and Ward, D. C. (1996). Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure, and dispersion of chromosomal domains. Exp. Cell Res. 224, 163-173. [DOI] [PubMed] [Google Scholar]
- Harborth, J., Elbashir, S. M., Bechert, K., Tuschl, T. and Weber, K. (2001). Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114, 4557-4565. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun, D. and Louvet, E. (2004). The nucleolus: structure, functions, and associated diseases. Med. Sci. (Paris) 20, 37-44. [DOI] [PubMed] [Google Scholar]
- Hozak, P., Cook, P. R., Schofer, C., Mosgoller, W. and Wachtler, F. (1994). Site of transcription of ribosomal RNA and intranucleolar structure in HeLa cells. J. Cell Sci. 107, 639-648. [DOI] [PubMed] [Google Scholar]
- Hozak, P., Sasseville, A. M., Raymond, Y. and Cook, P. R. (1995). Lamin proteins form an internal nucleoskeleton as well as a peripheral lamina in human cells. J. Cell Sci. 108, 635-644. [DOI] [PubMed] [Google Scholar]
- Jackson, D. A. and Cook, P. R. (1985). Transcription occurs at a nucleoskeleton. EMBO J. 4, 919-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D. A. and Cook, P. R. (1988). Visualization of a filamentous nucleoskeleton with a 23 nm axial repeat. EMBO J. 7, 3667-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Garcia, L. F., Segura-Valdez, M. L., Ochs, R. L., Rothblum, L. I., Hannan, R. and Spector, D. L. (1994). Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol. Biol. Cell 5, 955-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junera, H. R., Masson, C., Geraud, G. and Hernandez-Verdun, D. (1995). The three-dimensional organization of ribosomal genes and the architecture of the nucleoli vary with G1, S and G2 phases. J. Cell Sci. 108, 3427-3441. [DOI] [PubMed] [Google Scholar]
- Kaiser, T. E., Intine, R. V. and Dundr, M. (2008). De novo formation of a subnuclear body. Science 322, 1713-1717. [DOI] [PubMed] [Google Scholar]
- Kaufmann, S. H., Gibson, W. and Shaper, J. H. (1983). Characterization of the major polypeptides of the rat liver nuclear envelope. J. Biol. Chem. 258, 2710-2719. [PubMed] [Google Scholar]
- Kumaran, R. I., Muralikrishna, B. and Parnaik, V. K. (2002). Lamin A/C speckles mediate spatial organization of splicing factor compartments and RNA polymerase II transcription. J. Cell Biol. 159, 783-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, Y. W. and Lammond, A. I. (2006). Isolation of nucleoli. In Cell Biology, A Laboratory Handbook, 3rd edn (ed. J. Celis, N. Carter, K. Simons, J. Small, T. Hunter and D. Shotton), pp. 103-107. San Diego: Academic Press.
- Lamond, A. I. and Earnshaw, W. C. (1998). Structure and function in the nucleus. Science 280, 547-553. [DOI] [PubMed] [Google Scholar]
- Lehner, C. F., Stick, R., Eppenberger, H. M. and Nigg, E. A. (1987). Differential expression of nuclear lamin proteins during chicken development. J. Cell Biol. 105, 577-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, A. K., Gerlich, D., Miller, G., Lyon, C., Lam, Y. W., Lleres, D., Daigle, N., Zomerdijk, J., Ellenberg, J. and Lamond, A. I. (2004). Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 166, 787-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvet, E., Junera, H. R., Le Panse, S. and Hernandez-Verdun, D. (2005). Dynamics and compartmentation of the nucleolar processing machinery. Exp. Cell Res. 304, 457-470. [DOI] [PubMed] [Google Scholar]
- Malhas, A., Lee, C. F., Sanders, R., Saunders, N. J. and Vaux, D. J. (2007). Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J. Cell Biol. 176, 593-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Langerak, J., Goetze, S., Leonhardt, H., Cremer, T., van Driel, R. and Lanctot, C. (2007). Nuclear architecture: is it important for genome function and can we prove it? J. Cell Biochem. 102, 1067-1075. [DOI] [PubMed] [Google Scholar]
- Misteli, T. (2001). The concept of self-organization in cellular architecture. J. Cell Biol. 155, 181-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T. (2007). Beyond the sequence: Cellular organization of genome function. Cell 128, 787-800. [DOI] [PubMed] [Google Scholar]
- Moir, R. D., Spann, T. P., Herrmann, H. and Goldman, R. D. (2000). Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J. Cell Biol. 149, 79-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounkes, L., Kozlov, S., Burke, B. and Stewart, C. L. (2003). The laminopathies: nuclear structure meets disease. Curr. Opin. Genet. Dev. 13, 223-230. [DOI] [PubMed] [Google Scholar]
- Nirmalan, N., Flett, F., Skinner, T., Hyde, J. E. and Sims, P. F. G. (2007). Microscale solution isoelectric focusing as an effective strategy enabling containment of hemeoglobin-derived products for high-resolution gel-based analysis of the Plasmodium falciparum proteome. J. Proteome Res. 6, 3780-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair, R. D. and Misteli, T. (2000). High mobility of proteins in the mammalian cell nucleus. Nature 404, 604-609. [DOI] [PubMed] [Google Scholar]
- Ploton, D., Thiry, M., Menager, M., Lepoint, A., Adnet, J. J. and Goessens, G. (1987). Behaviour of nucleolus during mitosis: a comparative ultrastructural study of various cancerous cell lines using the Ag-NOR staining procedure. Chromosoma 95, 95-107. [DOI] [PubMed] [Google Scholar]
- Rafalska-Metcalf, I. U. and Janicki, S. M. (2007). Show and tell: visualizing gene expression in living cells. J. Cell Sci. 120, 2301-2307. [DOI] [PubMed] [Google Scholar]
- Raska, I., Shaw, P. J. and Cmarko, D. (2006). New insights into nucleolar architecture and activity. Int. Rev. Cytol. 255, 177-235. [DOI] [PubMed] [Google Scholar]
- Rober, R. A., Weber, K. and Osborn, M. (1989). Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development 105, 365-378. [DOI] [PubMed] [Google Scholar]
- Santos, F. and Dean, W. (2004). Epigenetic reprogramming during early development in mammals. Reproduction 127, 643-651. [DOI] [PubMed] [Google Scholar]
- Savino, T. M., Gebrane-Younes, J., De Mey, J., Sibarita, J. B. and Hernandez-Verdun, D. (2001). Nucleolar assembly of the rRNA processing machinery in living cells. J. Cell Biol. 153, 1097-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer, U. and Hock, R. (1999). Structure and function of the nucleolus. Curr. Opin. Cell Biol. 11, 385-390. [DOI] [PubMed] [Google Scholar]
- Shav-Tal, Y., Blechman, J., Darzacq, X., Montagna, C., Dye, B. T., Patton, J. G., Singer, R. H. and Zipori, D. (2005). Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell 16, 2395-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker, D. K., Kuczmarski, E. R. and Goldman, R. D. (2003). The nucleoskeleton: lamins and actin are major players in essential nuclear functions. Curr. Opin. Cell Biol. 15, 358-366. [DOI] [PubMed] [Google Scholar]
- Shumaker, D. K., Dechat, T., Kohlmaier, A., Adam, S. A, Bozovsky, M. R., Erdos, M. R., Eriksson, M., Goldman, A. E., Khuon, S., Collins, F. S. et al. (2006). Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA 103, 8703-8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker, D. K., Solimando, L., Sengupta, K., Shimi, T., Adam, S. A., Grunwald, A., Strelkov, S. V., Aebi, U., Cardoso, M. C. and Goldman, R. D. (2008). The highly conserved nuclear lamin Ig-fold binds to PCNA: its role in DNA replication. J. Cell Biol. 181, 269-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri, V., Urcuqui-Inchima, S., Roussel, P. and Hernandez-Verdun, D. (2008). Nucleolus: the fascinating nuclear body. Histochem. Cell Biol. 129, 13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann, T. P., Goldman, A. E., Wang, C., Huang, S. and Goldman, R. D. (2002). Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J. Cell Biol. 156, 603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector, D. L. (1993). Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9, 265-315. [DOI] [PubMed] [Google Scholar]
- Stewart, C. and Burke, B. (1987). Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell 51, 383-392. [DOI] [PubMed] [Google Scholar]
- Tang, C. W., Maya-Mendoza, A., Martin, C., Zeng, K., Chen, S., Feret, D., Wilson, S. A. and Jackson, D. A. (2008). The integrity of a lamin-B1-dependent nucleoskeleton is a fundamental determinant of RNA synthesis in human cells. J. Cell Sci. 121, 1014-1024. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy, L. and Lamond. A. I. (2007). Toward a high-resolution view of nuclear dynamics. Science 318, 1402-1407. [DOI] [PubMed] [Google Scholar]
- Vergnes, L., Peterfy, M., Bergo, M. O., Young, S. G. and Reue, K. (2004). Lamin B1 is required for mouse development and nuclear integrity. Proc. Natl. Acad. Sci. USA 101, 10428-10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman, H. J. and Courvalin, J. C. (2005). Nuclear envelope, nuclear lamina, and inherited disease. Int. Rev. Cytol. 246, 231-279. [DOI] [PubMed] [Google Scholar]
- Zatsepina, O. V., Bouniol-Baly, C., Amirand, C. and Debey, P. (2000). Functional and molecular reorganization of the nucleolar apparatus in maturing mouse oocytes. Dev. Biol. 223, 354-370. [DOI] [PubMed] [Google Scholar]
- Zatsepina, O., Baly, C., Chebrout, M. and Debey, P. (2003). The step-wise assembly of a functional nucleolus in preimplantation mouse embryos involves the cajal (coiled) body. Dev. Biol. 253, 66-83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.