Abstract
Individuals’ dependence on nicotine, primarily through cigarette smoking, is a major source of morbidity and mortality worldwide. Many smokers attempt but fail to quit smoking, motivating researchers to identify the origins of this dependence. Because of the known heritability of nicotine-dependence phenotypes, considerable interest has been focused on discovering the genetic factors underpinning the trait. This goal, however, is not easily attained: no single factor is likely to explain any great proportion of dependence because nicotine dependence is thought to be a complex trait (i.e., the result of many interacting factors). Genomewide association studies are powerful tools in the search for the genomic bases of complex traits, and in this context, novel candidate genes have been identified through single nucleotide polymorphism (SNP) association analyses. Beyond association, however, genetic data can be used to generate predictive models of nicotine dependence. As expected in the context of a complex trait, individual SNPs fail to accurately predict nicotine dependence, demanding the use of multivariate models. Standard approaches, such as logistic regression, are unable to consider large numbers of SNPs given existing sample sizes. However, using Bayesian networks, one can overcome these limitations to generate a multivariate predictive model, which has markedly enhanced predictive accuracy on fitted values relative to that of individual SNPs. This approach, combined with the data being generated by genomewide association studies, promises to shed new light on the common, complex trait nicotine dependence.
Keywords: addiction, nicotine, genetics, prediction
Cigarette smoking is the most prevalent form of tobacco use and is a major contributor to worldwide morbidity and mortality (DHHS, 2004), including being the single largest preventable cause of lung cancer (Murray, 2006). Nevertheless, 21.6% of adult Americans smoke (Centers for Disease Control and Prevention, 2005). Many want to stop smoking, as evidenced by the finding that 42.5% of American smokers reported having quit for at least 1 day in the previous year (Centers for Disease Control and Prevention, 2006). Most of these attempts fail: A meta-analysis of clinical trials demonstrated that an average of only 10% of individuals achieve abstinence for 6 months or more (Hughes et al., 2007). Nicotine dependence contributes to the failure rate, with more highly dependent smokers having greater difficulty in attaining abstinence (John et al., 2004; Xian et al., 2007). It has been established that nicotine dependence and associated characteristics are highly heritable phenotypes (Agrawal et al., 2006; Broms et al., 2007; Broms et al., 2006; Haberstick et al., 2007; Lessov et al., 2004; Maes et al., 2004; Pergadia et al., 2006; True et al., 1999; Vink et al., 2005; Xian et al., 2003, 2005). Despite its clear heritability, the search for the genetic variations underlying this trait is challenging. There have been issues with consistent replication of findings of association (Lerman & Swan, 2002; Vandenbergh et al., 2002), and no individual single-nucleotide polymorphism (SNP) is a sound predictor of nicotine dependence. This paper will address the challenges to identifying accurate genomic prognostic models of nicotine dependence and will review Bayesian networks, a novel proposed solution.
NICOTINE DEPENDENCE AS A COMPLEX PHENOTYPE
Despite the established heritability of nicotine-dependence phenotypes (Lessov-Schlaggar et al., 2008; M. R. Munafo & Johnstone, 2008), identifying replicable underlying genetic variations has proven difficult. Genomewide association studies (GWASs) hold promise and, indeed, novel associations have already been identified from using this approach. However, even the most strongly associated SNPs identified in these studies fail to predict nicotine dependence. This is likely due to the fact that nicotine dependence is a complex and potentially multidimensional trait, that is, a result of complicated interactions of multiple genetic and environmental factors (Cardon & Bell, 2001; M. R. Munafo & Johnstone, 2008; Rice et al., 2001; Risch, 2000).
Defining Nicotine Dependence
Careful phenotype definition is essential to achieving the phenotypic clarity required to identify valid, meaningful genetic predictors of complex human disorders (Rice et al., 2001). Several measures exist for defining nicotine dependence, the predominant measures being (1) diagnostic nicotine dependence as defined by the Diagnostic and Statistical Manual for Mental Disorders, 3rd edition Revised and 4th edition (DSM-III-R and DSM-IV) (APA, 1987, 1994) and (2) the Fagerström Tolerance Questionnaire (FTQ) and the Fagerström Test for Nicotine Dependence (FTND). Because of its widespread use (Hatsukami et al., 2008) and pertinence to the analyses presented, this report will focus on the FTND; in-depth descriptions of the other instrument can be found in the review articles (Piper et al., 2006; Lessov-Schlaggar et al., 2008).
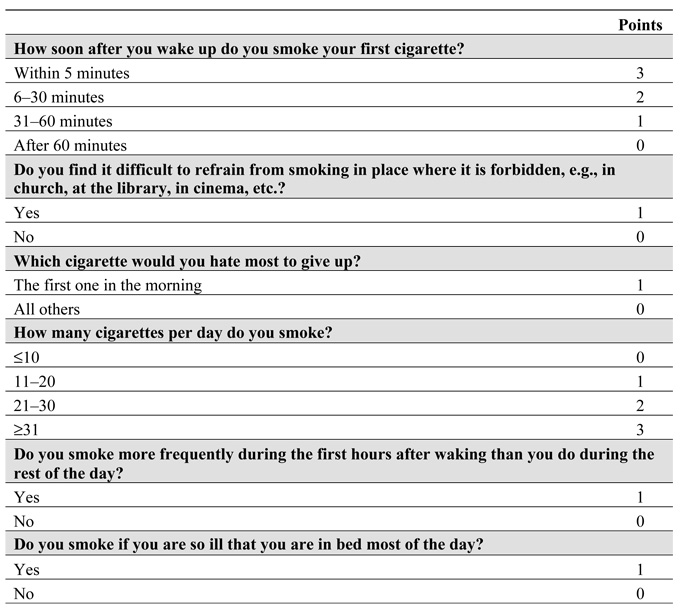
The FTND is a revision of the FTQ, designed to improve the psychometric properties of the scale (Heatherton et al., 1991; Payne et al., 1994). The FTND (Figure 1) is a six-question instrument, with a minimum score of 0 and a maximum score of 10. This scale focuses primarily on physiological dependence (Bierut et al., 2007), as opposed to other behavioral and psychological dimensions of dependence, for example, the subjective view of nicotine use being a “problem” in some way (Lessov-Schlaggar et al., 2008).
Figure 1.
The Fagerström Test for Nicotine Dependence.
Heritability of Nicotine-Dependence Phenotypes
Studies of twins have shown that a substantial proportion of the phenotypic variance in nicotine dependence is heritable. Indeed, studies of the FTND score, in particular, have shown it to be markedly heritable, with estimates ranging from 40 to 75% (Broms et al., 2006; Maes et al., 2004; Vink et al., 2005). Further, component measures of the FTND are substantially heritable. The daily cigarette quantity and time to first cigarette have been estimated to be 45.0–70.0% heritable (Broms et al., 2006; Carmelli et al., 1990; Haberstick et al., 2007; Hettema et al., 1999; Lessov et al., 2004; Pergadia et al., 2006; Prescott & Kendler, 1995; Swan et al., 1996, 1997; Swan et al., 1990) and 55.0–68.0% heritable (Haberstick et al., 2007; Lessov et al., 2004), respectively. Although the nicotine-dependence phenotype has a clear genetic component, it has been difficult to identify the causative variations, or even consistently associated variations, because of the complex nature of the phenotype (Lerman & Swan, 2002; M. R. Munafo et al., 2004; M. R. Munafo & Johnstone, 2008; Vandenbergh et al., 2002).
Genomewide Association Studies of Nicotine Dependence
It is thought that complex traits are influenced by multiple loci across the genome (Cardon & Bell, 2001; Risch, 2000). Thus, to identify the set of variations that contribute to vulnerability to nicotine dependence, GWASs have been conducted.
Bierut and colleagues first performed a pooled analysis of smokers who were nicotine dependent and smokers who were not nicotine dependent at over 2.4 million SNPs and, on this basis, selected 31,960 SNPs for genotyping in individuals (Bierut et al., 2007). Both cases and controls reported having smoked at least 100 cigarettes in their lifetime. Cases were individuals who had a FTND score of 4 or more when smoking the most, which is a commonly employed definition of nicotine dependence. Controls’ status was defined as having a lifetime FTND of 0. Although none of the individual findings was statistically significant after correcting for multiple tests, they identified 35 SNPs with P-values less than 10−4. Some of these SNPs were in intergenic regions and the β3 nicotinic receptor (CHRNB3), a known candidate gene, but many SNPs were in genes not previously associated with nicotine dependence. The novel genes have been associated with the following biological processes: protein transport (CLCA1 and VPS13A), protein catabolism (RNF5, FBXL17), regulation of transcription (PBX2), signal transduction (GPSM3), ion transport (TRPC7), lipid metabolism (FTO, AGPAT1), cell adhesion (NRXN1, AGER, CTNNA3), and developmental processes (NOTCH4) (Lessov-Schlaggar et al., 2008). In the same case-control sample, a targeted analysis of 348 known candidate genes was conducted (S. F. Saccone et al., 2007). The authors identified 39 SNPs with the greatest evidence for association with nicotine dependence, based upon the false discovery rate. One of these SNPs (rs6474413, CHRNB3) was shared with the 35 reported by the Bierut study. The majority of these SNPs were in cholinergic nicotinic receptor genes, including CHRNB3 and CHRNA5, the α5 nicotinic receptor subunit gene.
Uhl and colleagues (Uhl et al., 2007) conducted a case-control study at 520,000 SNPs in pools of DNA, with a special interest in SNPs in genes that have overlap with dependence on other substances. To identify variants associated with nicotine dependence, cases were European-American active smokers (mean FTND = 6.4, average carbon monoxide = 34.7) recruited into a smoking cessation study, and controls were European-American people with no substantial lifetime histories of use of any addictive substance. The authors identified SNPs in 32 genes that were both: (1) significant at P<0.0005 between cases and controls in their study and (2) had been significantly associated with polysubstance abuse, alcohol dependence, and methamphetamine dependence in previous studies. Like the study by Bierut and colleagues (Bierut et al., 2007), this investigation identified genes involved in cell adhesion, signal transduction, and transport functions, revealing convergence in the findings of these studies, despite the difference in their study populations and methods (Lessov-Schlaggar et al., 2008).
More recently, the associations reported by (Saccone et al., 2007) between nicotine dependence and variants in the CHRNA5-CHRNA3-CHRNB4 gene cluster have been replicated in several additional samples studying smoking quantity and nicotine dependence (Berrettini et al., 2008; Bierut et al., 2008; Thorgeirsson et al., 2008). Further, the nonsynonymous CHRNA5 SNP, rs16969968, or its r2 proxies (SNPs highly correlated with it) have now been associated with lung cancer (Amos et al., 2008; Hung et al., 2008; Thorgeirsson et al., 2008). These findings provide strong confirmation of the association between these SNPs and nicotine dependence. However, the challenge of moving beyond individual SNP associations to identify predictive models of phenotype remains.
Association with versus Prediction of Nicotine Dependence
The genomewide data that have been and will be collected hold the promise for uncovering the genetic origins of nicotine dependence, and the association analyses of individual SNPs have revealed new possible suspects. In addition, these analyses help to define overall risk classes, for example, a man with the G allele at SNP rs2791480 in CLCA1 may be at higher risk for being nicotine dependent (Bierut et al., 2007). Beyond association, the genomewide data can be used to identify predictors of complex traits such as nicotine dependence. Predictive modeling is a logical complement to the association-based approach, because predictive measures are both amenable to translation into clinical practice and are able to address some of the challenges of the analysis and interpretation of genomewide data. In particular, measures of predictive accuracy can address the issues of replication and generalizability of genetic association studies (Heidema et al., 2006; M. R. Munafo et al., 2004; M. R. Munafo & Johnstone, 2008). Even when the basis for the predictive measure is a multivariate model, the predictive accuracy can be used as a single, comprehensive measure of the extent of the generalizability of the model among different studies. Further, measuring the predictive accuracy is directly responsive to concerns that genetic tests will have a poor disease-predictive ability (Holtzman, 1992; Khoury et al., 1985). Indeed, couching results in the language of prediction poises them for translation into clinical practice, where they can be used in risk communication and counseling.
Predictive Accuracy of Individual SNPs and Demographic Factors
Because of the utility of predictive measures, the predictive accuracies of the 73 SNPs identified by Bierut (Bierut et al., 2007) and Saccone (S. F. Saccone et al., 2007), as well as the demographic factors, age and gender, were calculated. The predictive accuracy of the single-SNP model was computed as the area under the receiver-operator characteristic curve (AUROC) (DeLong et al., 1988) of the prediction of the fitted values. The probability of nicotine dependence, given the genotype of an individual subject, was calculated by using the clique algorithm (S. L. Lauritzen, & Spiegelhalter, 1988).
As shown in AUROC: area under the receiver operator characteristic curve, the highest predictive accuracy demonstrated by an individual SNP was 54.4% (P = 0.002), while gender and quartiles of age achieved predictive accuracies of 54.5 (P = 0.001) and 54.7% (P = 0.001), respectively. In these and the following cases, the P-value tests the hypothesis that the AUROC is significantly better than random (i.e., AUROC = 50%). The highest AUROC attained by an SNP in CHRNB3, a known candidate gene that was identified as highly associated in both the Bierut (Bierut et al., 2007) and Saccone (S. F. Saccone et al., 2007) papers, was 51.9% (P = 0.093), a performance not significantly better than random assignment. Because of the complex nature of nicotine dependence, the low predictive accuracies of individuals SNPs are perhaps not surprising, given that the effect of any single SNP on the trait is likely to be very small (Lerman & Swan, 2002). This issue can be addressed only by considering the polygenic nature of nicotine dependence (Lerman & Swan, 2002). Additional benefits accrue via the multivariate approach, including responsiveness to the issues of confounding (Cardon & Bell, 2001), linkage disequilibrium (Cardon & Bell, 2001), and genetic heterogeneity (Heidema et al., 2006). It has been noted, however, that a significant obstacle to accomplishing this goal is the large sample sizes required by standard multivariate methods such as logistic regression (Heidema et al., 2006; Lerman & Swan, 2002; M. R. Munafo & Johnstone, 2008). There has, therefore, been an interest in exploring alternative analytical methods that are more efficient. One such approach with a demonstrated track record in the analysis of complex phenotypes is the Bayesian network.
USING BAYESIAN NETWORKS TO BUILD PREDICTIVE MODELS OF COMPLEX PHENOTYPES
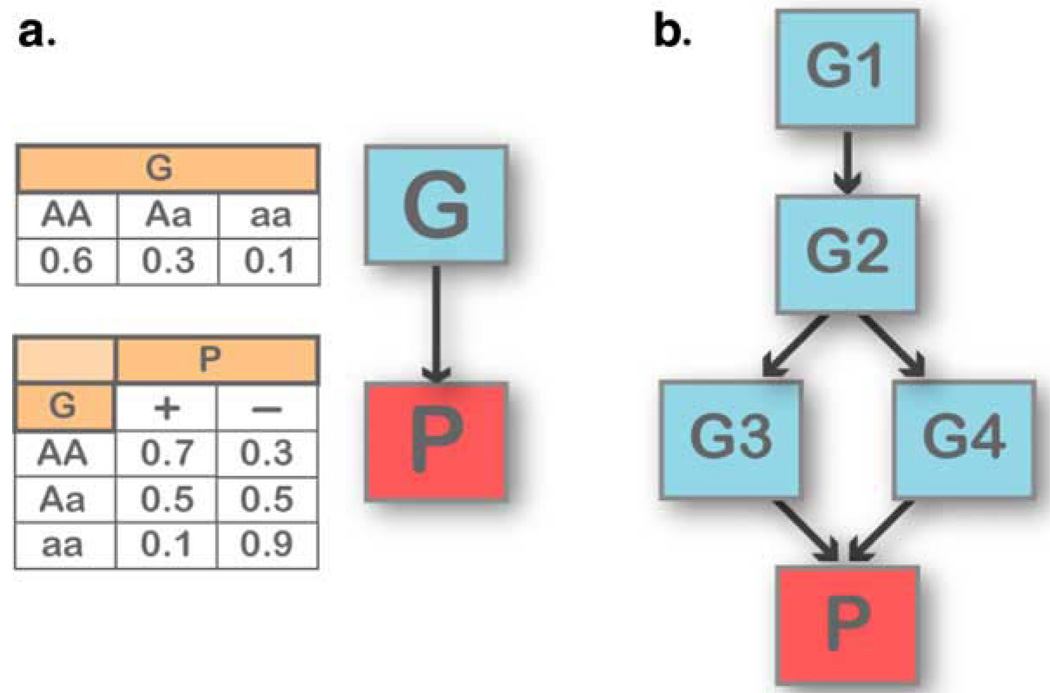
Bayesian networks are multivariate dependency models that account for simultaneous associations and interactions among multiple SNPs. A Bayesian network is a directed acyclic graph in which nodes represent random variables and arcs (arrows) define directed stochastic dependencies quantified by probability distributions. When an arc connects two nodes, the node at the point of the arrow is called a “child” of the “parent” node at the end of the arrow. Figure 2 depicts two Bayesian networks, starting with a simple network describing the dependency of a phenotype P on a single SNP G (Figure 2A). The graph decomposes the joint probability distribution of the two variables, P and G, into the product of the marginal distribution of G (the parent node) and the conditional distribution of P (the child node) given G. The marginal and conditional probability distributions are sufficient to define the association between P and G because their product determines the joint probability distribution. The property persists when we invert the direction of the arc in the graph, and when we expand the graphical structure to include several variables (Figure 2B): the overall association is measured by the joint probability distribution that is still defined by the product of each child-parent conditional distribution. This modular nature of a Bayesian network is due to the conditional independences among the variables encoded by the directed acyclic graph (Cowell et al., 1999): each node is independent of its predecessors given its parent nodes. This modular representation captures complex dependency models that are able to integrate associations between SNPs and phenotype; associations between SNPs due to linkage disequilibrium or evolutionary patterns (Chakravarti, 1999); and interaction processes linking SNPs, phenotypes, and modulating factors (Hoh & Ott, 2003) with a small number of parameters. Reducing the number of parameters allows us to “learn” large dependency networks from comparatively small datasets, and well-established techniques exist to develop Bayesian networks from data in an almost automated manner (Cowell et al., 1999).
Figure 2.
Examples of Bayesian network structures. (A) A simple Bayesian network with two nodes, representing a SNP (G) and a phenotype (P). The probability distribution of G represents the genotype distribution in the population, and the conditional probability distribution of P describes the distribution of each phenotype given each genotype. The direction of the association between G and P can be reversed by using Bayes theorem. (B) A Bayesian network linking four SNPs (G1–G4) to a phenotype P. The phenotype is independent of G1 and G2 given G3 and G4. The joint probability distribution of the network is fully specified by the five distributions representing the distribution of G1 (two parameters), of G2 given G1 (six parameters), of G3 given G2 (six parameters), of G4 given G2 (six parameters), and of P given G3 and G4 (nine parameters). The full probability distribution requires 81 × 2 − 1 = 161 parameters, while this network requires only 29.
The Bayesian network approach has been used to analyze several types of genomic data, including gene expression (Friedman, 2004; Friedman et al., 2000; Sebastiani et al., 2004), protein-protein interactions (Jansen et al., 2003), and pedigree analysis (S. L. Lauritzen & Sheehan, 2004). In the context of SNP data, Bayesian networks have been used to model the multigenic risk of stroke in sickle cell anemia (Sebastiani et al., 2005). While sickle cell anemia is a paradigmatic monogenic disease, the occurrence of stroke in individuals with the disorder is, like nicotine dependence, complex. This study demonstrated that Bayesian networks can generate a model of a complex trait that has a high predictive accuracy. Indeed, the stroke in the sickle cell anemia model demonstrated a predictive accuracy of 98.2% in an independent population.
APPLICATION OF BAYESIAN NETWORKS TO NICOTINE-DEPENDENCE PREDICTION
Bayesian networks are able to generate SNP-based models of complex traits by using relatively small sample sizes. Combining this method with the existing large-scale genotyping data affords a powerful, prognostic vantage on nicotine dependence.
Study Population
This study was based upon the genetic data and FTND phenotype definitions of the Collaborative Genetic Study of Nicotine Dependence (COGEND) (Bierut et al., 2007; S. F. Saccone et al., 2007). The 73 SNPs reported as associated to nicotine dependence in this population were subjected to Bayesian network analysis.
Statistical Analysis
All of the SNPs considered in the Bayesian network analysis were tested for deviation from the Hardy-Weinberg equilibrium (as described in Bierut et al., 2007; S. F. Saccone et al., 2007). The analysis was conducted by using Bayesware Discoverer (Bayesware, Boston, Massachusets, USA). The dependencies in the model of CASE (nicotine dependent) status were identified by calculating the Bayes Factors corresponding to the ratio of the marginal log likelihood of the statusdetermining genotype to the marginal log likelihood of status being independent of genotype. In the absence of an independent validation set, we assessed the goodness-of-fit of the model by calculating the AUROC of the prediction of the fitted values. The probability of nicotine dependence given the genotype of an individual subject was calculated by using the clique algorithm implemented in Bayesware Discoverer; the predictive accuracy of the model was estimated as the AUROC curve convex hull, using the trapezoidal rule (DeLong et al., 1988).
Predictive Accuracy of the Multivariate Model
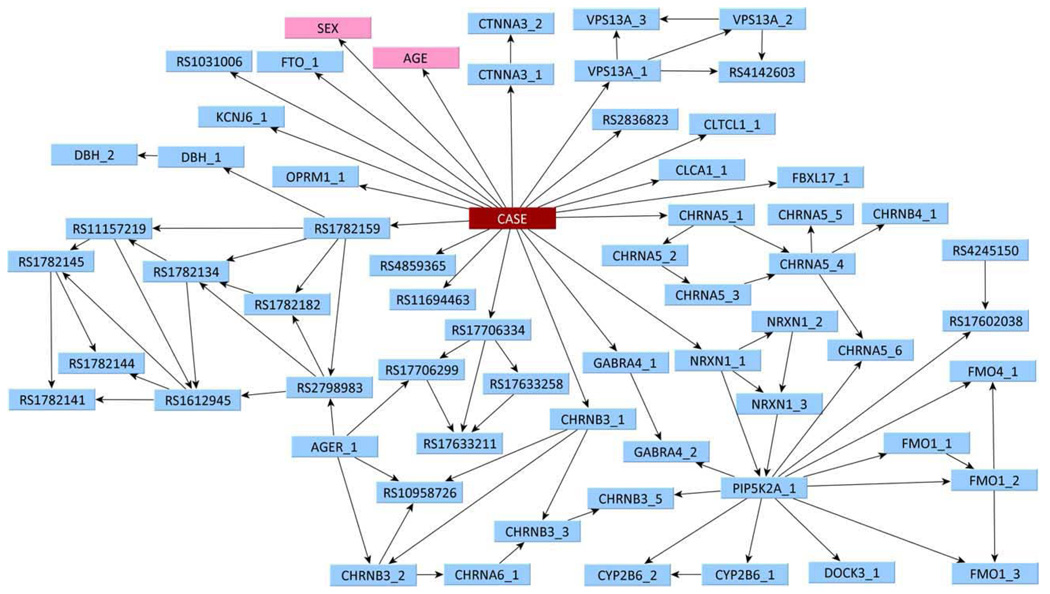
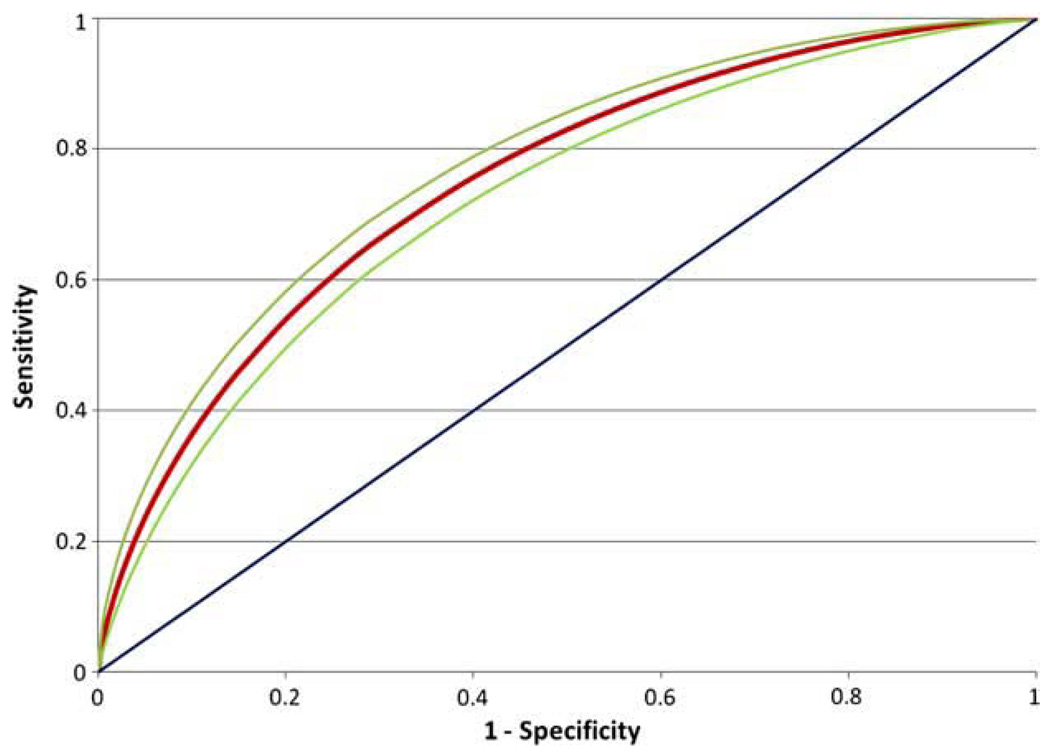
The multivariate model generated by the Bayesian network analysis is shown in Figure 3. Out of the original 73 SNPs, 60 were incorporated into the model, along with the two demographic factors (SEX and AGE). However, to predict nicotine-dependence status, it is not always necessary to have information about all 60 SNPs. If one has information on all of the nodes that are children of CASE, as well as any other parents of these nodes, one does not need any information about any other nodes. This subset of nodes is called the Markov blanket of CASE. In this network shown in Figure 3, there are 20 nodes in the Markov blanket: 18 SNPs and both demographic factors. The SNPs directly connected to CASE are in genes CHRNA3, CHRNB3, CLCA1, CLTCL1, CTNNA3, FBXL17, FTO, GABRA4, KCNJ6, NRXN1, OPRM1, and VPS13A, with the remaining SNPs being in intergenic regions. Individually, the best-per-forming of these SNPs, rs2836823, had a predictive accuracy on fitted values of 54.4% (P = 0.002; Table 1). While this is statistically significantly different from randomness, such a low AUROC indicates a lack of predictive ability. By contrast, the network as a whole (Figure 4) was able to achieve a predictive accuracy on fitted values of 75.0% (P<0.0001), as shown in Figure 4.
Figure 3.
Bayesian network of nicotine dependence. The Bayesian network describing the joint association of 60 SNPs (blue) and two demographic factors (pink) with nicotine dependence. CASE represents nicotine-dependence status. The RS numbers corresponding to the codes in the nodes are shown in Table 1.
Table 1.
Predictive Accuracies (AUROC) of Individual SNPs and Demographic Factors on the Fitted Values.
| Gene | Label | SNP | AUROC | p-value |
|---|---|---|---|---|
| AVPR1A | AVPR1A_1 | 3021529 | 47.10% | 0.98 |
| CHRNA5/3 | CHRNA5_1* | 578776 | 54.00% | 0.01 |
| CHRNA5 | CHRNA5_2* | 16969968 | 50.30% | 0.40 |
| CHRNA5_3* | 1051730 | 49.60% | 0.62 | |
| CHRNA5_4* | 637137 | 48.90% | 0.79 | |
| CHRNA5_5* | 684513 | 49.20% | 0.72 | |
| CHRNA5_6* | 3743078 | 48.40% | 0.89 | |
| CHRNA6 | CHRNA6_1* | 2304297 | 47.10% | 0.98 |
| CHRNB3 | CHRNB3_1* | 13277254 | 51.90% | 0.09 |
| CHRNB3_2* | 6474413 | 48.10% | 0.91 | |
| CHRNB3_3* | 4953 | 48.80% | 0.82 | |
| CHRNB3_5* | 4952 | 47.90% | 0.93 | |
| CHRNB4 | CHRNB4_1* | 3813567 | 47.70% | 0.95 |
| CHRND | CHRNG_1 | 2767 | 47.00% | 0.98 |
| CHRNG_2 | 3791729 | 47.60% | 0.96 | |
| CLCA1 | CLCA1_1* | 2791480 | 54.20% | 0.00 |
| CLTCL1 | CLTCL1_1* | 1206549 | 52.20% | 0.06 |
| CTNNA3 | CTNNA3_1* | 4142041 | 53.10% | 0.02 |
| CTNNA3_2* | 9332406 | 49.00% | 0.75 | |
| CYP2B6 | CYP2B6_1* | 4802100 | 47.90% | 0.93 |
| CYP2B6_2* | 3760657 | 48.50% | 0.86 | |
| DAO | DAO_1 | 17041074 | 47.10% | 0.98 |
| DBH | DBH_1* | 4531 | 49.30% | 0.68 |
| DBH_2* | 3025382 | 47.10% | 0.98 | |
| DOCK3(GRM2) | DOCK3_1* | 6772197 | 44.80% | 1.00 |
| CHRNG | EIF4E2_1 | 6749955 | 48.80% | 0.82 |
| FBXL17 | FBXL17_1* | 10793832 | 52.50% | 0.04 |
| FMO1 | FMO1_1* | 7877 | 48.30% | 0.89 |
| FMO1_2* | 742350 | 47.10% | 0.98 | |
| FMO1_3* | 7517376 | 47.20% | 0.98 | |
| FMO4_1* | 16864387 | 48.10% | 0.92 | |
| FTO | FTO_1* | 2302673 | 52.60% | 0.03 |
| GABRA4 | GABRA4_1* | 3762611 | 50.70% | 0.30 |
| GABRA4_2 | 3762607 | 48.30% | 0.89 | |
| GPSM3, | AGER_1* | 999 | 47.50% | 0.97 |
| AGPAT1, | ||||
| NOTCH4, | ||||
| RNF5, | ||||
| AGER, | ||||
| PBX2, | ||||
| AGER | ||||
| HTR5A | HTR5A_1 | 6320 | 47.90% | 0.93 |
| HTR5A_2 | 1657273 | 48.60% | 0.85 | |
| KCNJ6 | KCNJ6_1* | 6517442 | 54.10% | 0.01 |
| NRXN1 | NRXN1_1* | 12623467 | 51.20% | 0.20 |
| NRXN1_2* | 10490162 | 47.60% | 0.95 | |
| NRXN1_3* | 12467557 | 47.10% | 0.98 | |
| OPRM1 | OPRM1_1* | 510769 | 53.60% | 0.01 |
| PDYN | PDYN_1 | 6045733 | 48.50% | 0.86 |
| PIP5K2A | PIP5K2A_1* | 10508649 | 46.00% | 1.00 |
| TRPC7 | TRPC7_1 | 2673931 | 47.70% | 0.95 |
| TRPC7_2 | 2546657 | 47.10% | 0.98 | |
| VPS13A | VPS13A_1* | 12380218 | 53.10% | 0.01 |
| VPS13A_2* | 2022443 | 47.00% | 0.98 | |
| VPS13A_3* | 11145381 | 48.30% | 0.88 | |
| Intergenic | 10049135 | 48.00% | 0.93 | |
| 1031006* | 52.20% | 0.06 | ||
| 10958726* | 47.40% | 0.97 | ||
| 11157219* | 46.20% | 1.00 | ||
| 11694463* | 51.70% | 0.12 | ||
| 1612945* | 48.50% | 0.86 | ||
| 17602038* | 47.40% | 0.97 | ||
| 17633211* | 47.40% | 0.97 | ||
| 17633258* | 45.80% | 1.00 | ||
| 17706299* | 46.90% | 0.99 | ||
| 17706334* | 49.50% | 0.65 | ||
| 1782134* | 47.10% | 0.98 | ||
| 1782141* | 47.00% | 0.98 | ||
| 1782144* | 47.90% | 0.93 | ||
| 1782145* | 47.90% | 0.93 | ||
| 1782159* | 52.80% | 0.03 | ||
| 1782182* | 47.10% | 0.98 | ||
| 2276560 | 45.90% | 1.00 | ||
| 2798983* | 48.60% | 0.84 | ||
| 2836823* | 54.40% | 0.00 | ||
| 4142603* | 48.20% | 0.90 | ||
| 4245150* | 48.70% | 0.83 | ||
| 4859365* | 53.60% | 0.01 |
Figure 4.
ROC curve representing the predictive accuracy of the Bayesian network in Figure 3 on the fitted values. The curve emanating from the origin has an area under the ROC curve (AURCO) of 50%, which represents random classification. The red ROC curve corresponds to the Bayesian network (AUROC = 75.0%), the 95% confidence interval of which is bounded by the green curves.
An interesting result revealed by the topology of the network in Figure 3 is the role of the SNP, rs16969968 (labeled CHRNA5_2 in the network), which is connected to the phenotype through CHRNA5 (rs578776 labeled as CHRNA5_1). This finding is of particular note given that several recent studies have shown the associations of SNPs in the CHRNA5-CHRNA3-CHRNB4 cluster to both nicotine dependence and lung cancer (Amos et al., 2008; Berrettini et al., 2008; Bierut et al., 2008; Hung et al., 2008; Thorgeirsson et al., 2008). Further, an in-depth study in the COGEND population of the complete family of 16 nicotinic receptor subunits identified both rs578776 and rs16969968 as statistically significantly associated with nicotine dependence even though the correlation between these SNPs is low (N. L. Saccone et al., in press).
SUMMARY AND FUTURE DIRECTIONS
Nicotine dependence has been shown to have a heritable component, and therefore, large-scale genetic studies hold tremendous promise for revealing the genetic bases for nicotine dependence. Single-SNP association analyses have yielded novel findings that shed light on the biological underpinnings of the trait. However, none of even the most highly associated SNPs alone has been shown to predict nicotine dependence better than at random, which is a result that might be anticipated given the complex nature of the trait. While standard analytical approaches, such as logistic regression, require prohibitively large sample sizes to consider multiple interacting SNPs, the Bayesian network approach has been shown to be well-suited to this type of analysis in the context of existing sample sizes. Indeed, when applied to nicotine dependence, the multivariate Bayesian network analysis demonstrated markedly enhanced prediction on fitted values relative to individual SNPs. This work is an important first step in discovering predictors of nicotine dependence. An accurate predictive model would give the research community a new vantage point from which to consider the causes of this trait, and such a model would have the further advantage of being framed in the language of diagnostic accuracy and risk communication.
The next steps toward the goal of generating a valid prognostic model of nicotine dependence should be to seek to improve the predictive accuracy by building a model drawing upon all SNPs in the genomewide analysis and incorporating environmental exposures. Further, the models generated should be validated in independent populations, rather than upon fitted values. In preference of a retrospective case-control design, the ideal study for such a validation would be prospective to allow for true prediction of nicotine dependence status, as opposed to correct classification. The distinction between prediction and correct classification is smaller in the context of SNPs, because they are fixed over a person’s entire lifetime, but is of great importance when modeling the effect of mutable environmental exposures. Moreover, such an approach would allow for the prospective determination of phenotype.
In addition to identifying predictors of nicotine dependence, developing predictive models of the trait should help refine the phenotype definitions of “being a smoker” and nicotine dependence themselves. Proper phenotype definition is fundamental to any search for the origins of complex traits, and phenotyping has been identified as an issue in need of additional attention and clarification by a number of researchers (Lerman & Swan, 2002; Lessov et al., 2004; M. R. Munafo & Johnstone, 2008; Robert, 2006). The nature of the disorder will become clearer as the molecular bases of the trait subtypes become better understood. Along these lines, Cardon and Bell have envisioned an iterative process of developing an understanding of the genetic bases of complex traits, in which weak associations are successively improved upon by generating hypotheses on the basis of subgroups and assessing them in new cohorts (Cardon & Bell, 2001). Through these efforts, the nicotine-dependence research community will develop a deeper knowledge of the causes of nicotine-dependence phenotypes. This knowledge promises to define etiology-driven definitions of disease (Piper et al., 2006) and to reveal new therapeutic avenues.
Finally, the ability to develop a prognostic model of the complex trait nicotine dependence highlights the feasibility of identifying predictors of another complex trait: response to smoking cessation therapy. Currently, the untargeted assignment of existing therapies suffers from disappointing relapse rates. Through pharmacogenetic targeting, success rates could be enhanced by assigning people who want to quit smoking to the therapy that is most likely to work for them. A small number of studies have highlighted the promise of the approach, but as with nicotine dependence, a lack of replication has plagued these investigations (M. Munafo & Lerman, 2006). An efficient multivariate solution, such as the Bayesian network approach, may help address the challenge of finding translatable pharmacogenetic solutions for the treatment of nicotine dependence.
ACKNOWLEDGEMENTS
This study was supported, in part, by a Cutting-Edge Basic Research Award (CEBRA) from NIH/NIDA (R21DA025168), by a Mentored Clinical Scientist Development Award from NIH/NIDCR (5K08DE016956), and by a grant from the National Cancer Institute (CA89392).
In memory of Theodore Reich, founding Principal Investigator of COGEND, the authors are indebted to his leadership in the establishment and nurturing of COGEND and acknowledge, with great admiration, his seminal scientific contributions to the field.
Footnotes
Declaration of interest: As a financial disclosure, L.J.B. is listed as an inventor on a patent (US 20070258898) held by Perlegen Sciences, Inc., covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. L.J.B. has acted as a consultant for Pfizer, Inc. (in 2008). N.L.S. is the spouse of S.F.S., who is listed as an inventor on the above-named patent. The authors alone are responsible for the content and writing of this paper.
REFERENCES
- Agrawal A, Heath AC, Grant JD, Pergadia ML, Statham DJ, Bucholz KK, et al. Assortative mating for cigarette smoking and for alcohol consumption in female Australian twins and their spouses. Behav Genet. 2006;36(4):553–566. doi: 10.1007/s10519-006-9081-8. [DOI] [PubMed] [Google Scholar]
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, et al. Genomewide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APA. Diagnostic and Statistical Manual of Mental Disorders. Washington, DC: American Psychiatric Association; 1987. [Google Scholar]
- APA. Diagnostic and Statistical Manual of Mental Disorders. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13(4):368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novel genes identified in a high-density genomewide association study for nicotine dependence. Hum Mol Genet. 2007;16(1):24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Bertelsen S, Fox L, et al. Variants in the nicotinic receptors alter the risk for nicotine dependence. Am J Psychiatry. 2008;165(9):1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms U, Madden PA, Heath AC, Pergadia ML, Shiffman S, Kaprio J. The Nicotine Dependence Syndrome Scale in Finnish smokers. Drug Alcohol Depend. 2007;89(1):42–51. doi: 10.1016/j.drugalcdep.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms U, Silventoinen K, Madden PA, Heath AC, Kaprio J. Genetic architecture of smoking behavior: a study of Finnish adult twins. Twin Res Hum Genet. 2006;9(1):64–72. doi: 10.1375/183242706776403046. [DOI] [PubMed] [Google Scholar]
- Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2(2):91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- Carmelli D, Swan GE, Robinette D, Fabsitz RR. Heritability of substance use in the NAS-NRC Twin Registry. Acta Genet Med Gemellol (Roma) 1990;39(1):91–98. doi: 10.1017/s0001566000005602. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Cigarette smoking among adults—United States: 2003. Morbid Mortal Weekly Report. 2005;52(40):953–956. [PubMed]
- Centers for Disease Control and Prevention. Cigarette smoking among adults—United States: 2005. Morbid Mortal Weekly Report. 2006;52(40):953–956. [PubMed]
- Chakravarti A. Population genetics—making sense out of sequence. Nat Genet. 1999;21 Suppl 1:56–60. doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- Cowell RG, Dawid AP, Lauritzen SL, Spiegelhalter DJ. Probabilistic Networks and Expert Systems. New York: Springer; 1999. [Google Scholar]
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- DHHS. Atlanta, Georgia, USA: Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office of Smoking and Health; The Health Consequences of Smoking: a Report of the Surgeon General. 2004
- Friedman N. Inferring cellular networks using probabilistic graphical models. Science. 2004;303:799–805. doi: 10.1126/science.1094068. [DOI] [PubMed] [Google Scholar]
- Friedman N, Linial M, Nachman I, Pe’er D. Using Bayesian networks to analyze expression data. J Comput Biol. 2000;7(3–4):601–620. doi: 10.1089/106652700750050961. [DOI] [PubMed] [Google Scholar]
- Haberstick BC, Timberlake D, Ehringer MA, Lessem JM, Hopfer CJ, Smolen A, et al. Genes, time to first cigarette, and nicotine dependence in a general population sample of young adults. Addiction. 2007;102(4):655–665. doi: 10.1111/j.1360-0443.2007.01746.x. [DOI] [PubMed] [Google Scholar]
- Hatsukami DK, Stead LF, Gupta PC. Tobacco addiction. Lancet. 2008;371(9629):2027–2038. doi: 10.1016/S0140-6736(08)60871-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heatherton TF, Kozlowski LT, Frecker RC, Fagerström KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerström Tolerance Questionnaire. Br J Addict. 1991;86(9):1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- Heidema AG, Boer JM, Nagelkerke N, Mariman EC, van der ADL, Feskens EJ. The challenge for genetic epidemiologists: how to analyze large numbers of SNPs in relation to complex diseases. BMC Genet. 2006;7:23. doi: 10.1186/1471-2156-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema JM, Corey LA, Kendler KS. A multivariate genetic analysis of the use of tobacco, alcohol, and caffeine in a population-based sample of male and female twins. Drug Alcohol Depend. 1999;57(1):69–78. doi: 10.1016/s0376-8716(99)00053-8. [DOI] [PubMed] [Google Scholar]
- Hoh J, Ott J. Mathematical multilocus approaches to localizing complex human trait genes. Nat Rev Genet. 2003;4(9):701–709. doi: 10.1038/nrg1155. [DOI] [PubMed] [Google Scholar]
- Holtzman NA. The diffusion of new genetic tests for predicting disease. FASEB J. 1992;6(10):2806–2812. doi: 10.1096/fasebj.6.10.1634043. [DOI] [PubMed] [Google Scholar]
- Hughes JR, Stead LF, Lancaster T. Antidepressants for smoking cessation. Cochrane Database Syst Rev. 2007;(1) doi: 10.1002/14651858.CD000031.pub3. CD000031. [DOI] [PubMed] [Google Scholar]
- Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, et al. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302(5644):449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- John U, Meyer C, Hapke U, Rumpf HJ, Schumann A. Nicotine dependence, quit attempts, and quitting among smokers in a regional population sample from a country with a high prevalence of tobacco smoking. Prev Med. 2004;38(3):350–358. doi: 10.1016/j.ypmed.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Khoury MJ, Newill CA, Chase GA. Epidemiologic evaluation of screening for risk factors: application to genetic screening. Am J Pub Health. 1985;75(10):1204–1208. doi: 10.2105/ajph.75.10.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritzen SL, Sheehan NA. Graphical models for genetic analysis. Statist Sci. 2004;18(4):489–514. [Google Scholar]
- Lauritzen SL, Spiegelhalter DJ. Local computations with probabilities on graphical structures and their application to expert systems (with discussion) J Roy Stat Soc B Met. 1988;50:157–224. [Google Scholar]
- Lerman C, Swan GE. Nonreplication of genetic association studies: is DAT all, folks? Nicotine Tob Res. 2002;4(3):247–249. doi: 10.1080/14622200210141220. [DOI] [PubMed] [Google Scholar]
- Lessov-Schlaggar CN, Pergadia ML, Khroyan TV, Swan GE. Genetics of nicotine dependence and pharmacotherapy. Biochem Pharmacol. 2008;75(1):178–195. doi: 10.1016/j.bcp.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessov CN, Martin NG, Statham DJ, Todorov AA, Slutske WS, Bucholz KK, et al. Defining nicotine dependence for genetic research: evidence from Australian twins. Psychol Med. 2004;34(5):865–879. doi: 10.1017/s0033291703001582. [DOI] [PubMed] [Google Scholar]
- Maes HH, Sullivan PF, Bulik CM, Neale MC, Prescott CA, Eaves LJ, et al. A twin study of genetic and environmental influences on tobacco initiation, regular tobacco use, and nicotine dependence. Psychol Med. 2004;34(7):1251–1261. doi: 10.1017/s0033291704002405. [DOI] [PubMed] [Google Scholar]
- Munafo M, Lerman C. Can Pharmacogenetics Help Smokers Quit? Pharmacogenomics. 2006;7(8):1137–1140. doi: 10.2217/14622416.7.8.1137. [DOI] [PubMed] [Google Scholar]
- Munafo MR, Clark TG, Johnstone EC, Murphy MFG, Walton RT. The genetic basis for smoking behavior: a systematic review and meta-analysis. Nicotine Tob Res. 2004;6(4):583–597. doi: 10.1080/14622200410001734030. [DOI] [PubMed] [Google Scholar]
- Munafo MR, Johnstone EC. Genes and cigarette smoking. Addiction. 2008;103(6):893–904. doi: 10.1111/j.1360-0443.2007.02071.x. [DOI] [PubMed] [Google Scholar]
- Murray S. A smouldering epidemic. CMAJ. 2006;174(3):309–310. doi: 10.1503/cmaj.051628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne TJ, Smith PO, McCracken LM, McSherry WC, Antony MM. Assessing nicotine dependence: a comparison of the Fagerström Tolerance Questionnaire (FTQ) with the Fagerström Test for Nicotine Dependence (FTND) in a clinical sample. Addict Behav. 1994;19(3):307–317. doi: 10.1016/0306-4603(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Pergadia ML, Heath AC, Martin NG, Madden PA. Genetic analyses of DSM-IV nicotine withdrawal in adult twins. Psychol Med. 2006;36(7):963–972. doi: 10.1017/S0033291706007495. [DOI] [PubMed] [Google Scholar]
- Piper ME, McCarthy DE, Baker TB. Assessing tobacco dependence: a guide to measure evaluation and selection. Nicotine Tob Res. 2006;8(3):339–351. doi: 10.1080/14622200600672765. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Kendler KS. Genetic and Environmental Influences on Alcohol and Tobacco Dependence among Women. Bethesda, Maryland, USA: National Institutes of Health; 1995. [Google Scholar]
- Rice JP, Saccone NL, Rasmussen E. Definition of the phenotype. Advances in Genetics. 2001;42:69–76. doi: 10.1016/s0065-2660(01)42015-3. [DOI] [PubMed] [Google Scholar]
- Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405(6788):847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- Robert W. Defining and assessing nicotine dependence in humans. In: Gregory Bock JG, editor. Understanding Nicotine and Tobacco Addiction. Hoboken, New Jersey: Wiley; 2006. pp. 36–58. [Google Scholar]
- Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, Pergadia ML, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet Part B Neuropsychiatr Genet. doi: 10.1002/ajmg.b.30828. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P, Abad M, Ramoni MF. Bayesian networks for genomic analysis. In: Dougherty E, editor. EURASIP Book Series on Signal Processing and Communications. New York, NY: Hindawi; 2005. [Google Scholar]
- Sebastiani P, Ramoni MF, Nolan V, Baldwin CT, Steinberg MH. Genetic dissection and prognostic modeling of overt stroke in sickle cell anemia. Nat Genet. 2005;37(4):435–440. doi: 10.1038/ng1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan GE, Carmelli D, Cardon LR. The consumption of tobacco, alcohol, and coffee in Caucasian male twins: a multivariate genetic analysis. J Subst Abuse. 1996;8(1):19–31. doi: 10.1016/s0899-3289(96)90055-3. [DOI] [PubMed] [Google Scholar]
- Swan GE, Carmelli D, Cardon LR. Heavy consumption of cigarettes, alcohol, and coffee in male twins. J Stud Alcohol. 1997;58(2):182–190. doi: 10.15288/jsa.1997.58.182. [DOI] [PubMed] [Google Scholar]
- Swan GE, Carmelli D, Rosenman RH, Fabsitz RR, Christian JC. Smoking and alcohol consumption in adult male twins: genetic heritability and shared environmental influences. J Subst Abuse. 1990;2(1):39–50. doi: 10.1016/s0899-3289(05)80044-6. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer, and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True WR, Xian H, Scherrer JF, Madden PA, Bucholz KK, Heath AC, et al. Common genetic vulnerability for nicotine and alcohol dependence in men. Arch Gen Psychiatry. 1999;56(7):655–661. doi: 10.1001/archpsyc.56.7.655. [DOI] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE. Molecular genetics of nicotine dependence and abstinence: whole genome association using 520,000 SNPs. BMC Genet. 2007;8:10. doi: 10.1186/1471-2156-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbergh DJ, Bennett CJ, Grant MD, Strasser AA, O’Connor R, Stauffer RL, et al. Smoking status and the human dopamine transporter variable number of tandem repeats (VNTR) polymorphism: failure to replicate and finding that never-smokers may be different. Nicotine Tob Res. 2002;4(3):333–340. doi: 10.1080/14622200210142689. [DOI] [PubMed] [Google Scholar]
- Vink JM, Willemsen G, Boomsma DI. Heritability of smoking initiation and nicotine dependence. Behav Genet. 2005;35(4):397–406. doi: 10.1007/s10519-004-1327-8. [DOI] [PubMed] [Google Scholar]
- Xian H, Scherrer JF, Eisen SA, Lyons MJ, Tsuang M, True WR, et al. Nicotine dependence subtypes: association with smoking history, diagnostic criteria, and psychiatric disorders in 5,440 regular smokers from the Vietnam Era Twin Registry. Addict Behav. 2007;32(1):137–147. doi: 10.1016/j.addbeh.2006.03.031. [DOI] [PubMed] [Google Scholar]
- Xian H, Scherrer JF, Madden PA, Lyons MJ, Tsuang M, True WR, et al. The heritability of failed smoking cessation and nicotine withdrawal in twins who smoked and attempted to quit. Nicotine Tob Res. 2003;5(2):245–254. [PubMed] [Google Scholar]
- Xian H, Scherrer JF, Madden PA, Lyons MJ, Tsuang M, True WR, et al. Latent class typology of nicotine withdrawal: genetic contributions and association with failed smoking cessation and psychiatric disorders. Psychol Med. 2005;35(3):409–419. doi: 10.1017/s0033291704003289. [DOI] [PubMed] [Google Scholar]