Abstract
In mammals, ataxin-1 (ATXN1) is a member of a family of proteins in which each member contains an AXH domain. Expansion of the polyglutamine tract in ATXN1 causes the neurodegenerative disease spinocerebellar ataxia type 1 (SCA1) with prominent cerebellar pathology. Toward a further characterization of the genetic diversification of the ATXN1/AXH gene family, we identified and characterized members of this gene family in zebrafish, a lower vertebrate with a cerebellum. The zebrafish genome encodes two ATXN1 homologs, atxn1a and atxn1b, and one ATXN1L homolog, atxn1l. Key biochemical features of the human ATXN1 protein not seen in the invertebrate homologs (a nuclear localization sequence and a site of phosphorylation at serine 776) are conserved in the zebrafish homologs, and all three zebrafish Atxn1/Axh proteins behave similarly to their human counterparts in tissue culture cells. Importantly, each of the three homologs is expressed in the zebrafish cerebellum, which in humans is a prominent site of SCA1 pathogenesis. In addition, atxn1a and atxn1b are expressed in the developing zebrafish cerebellum. These data show that in zebrafish, a lower vertebrate, the complexity of the atxn1/axh gene family is more similar to higher vertebrates than invertebrates with a simple central nervous system and suggests a relationship between the diversification of the ATXN1/AXH gene family and the development of a complex central nervous system including a cerebellum.
Keywords: ataxin-1, ataxin-1 like, AXH, SCA1, Danio rerio
Introduction
Ataxin-1 (ATXN1) is a member of a family of proteins that includes the transcriptional repressor HBP1, ATXN1, and ATXN1L. Each of these proteins is characterized by the presence of an ataxin-1/HBP1 (AXH) domain (Mushegian et al., 2005; Bowman et al., 2007; Mizutani et al., 2005). This 120 amino acid region forms an OB-fold (Y. W. Chen et al., 2004) and is implicated in protein-protein interactions (Mizutani et al., 2005; Tsai et al., 2004; Tsuda et al., 2005) as well as protein-oligonucleotide binding (de Chiara et al., 2003). The genomes of Caenorhabditis elegans and Drosophila melanogaster encode small proteins consisting of essentially just an AXH domain, a point consistent with this region having an ability to fold independently (de Chiara et al., 2003).
Of further interest, ATXN1 is the protein mutated in spinocerebellar ataxia type 1 (SCA1), an autosomal-dominant, neurodegenerative disease caused by a polyglutamine expansion in ATXN1 (Orr et al., 1993). SCA1 is one of nine polyglutamine diseases characterized to date (Nakamura et al., 2001; Zoghbi et al., 2000). Clinical symptoms of SCA1 usually present within the third to fourth decade of life and include ataxia, dysphagia, dysarthria and progressive motor dysfunction. Pathologically, SCA1 is characterized by degeneration of specific neuronal populations including the cerebellar Purkinje cells, neurons of the inferior olive, and the deep cerebellar neurons (Subramony et al., 1998).
Recently, an ATXN1 paralog, ataxin-1 like (ATXN1L), was identified (Bowman et al., 2007; Mizutani et al., 2005). Like ATXN1, ATXN1L contains a highly conserved AXH domain (de Chiara et al., 2003; Mizutani et al., 2005). ATXN1L interacts with ATXN1 both in vivo and in vitro and is found to compete with mutant ATXN1 for association in large protein complexes containing capicua (Bowman et al., 2007). In both fly (Mizutani et al., 2005) and mouse models of SCA1(Bowman et al., 2007), ATXN1L overexpression suppressed mutant ATXN1 pathogenesis, suggesting that the identification of disease gene paralogs is an important step towards understanding disease pathogenesis.
Experiments conducted using SCA1 transgenic mouse models indicate there is a developmental aspect to mutant ATXN1-induced disease. In a conditional SCA1 mouse model it was found that timing of mutant ATXN1 expression during cerebellar development affects SCA1 pathology (Serra et al., 2006). Transgenic mice that express a low level of mutant ATXN1 early in development manifest a more severe phenotype compared to transgenic mice with a delayed onset of mutant ATXN1 expression (Burright et al., 1995).
Zebrafish are a well-established model for studying developmental questions. Within 24 hours after fertilization the basic nervous system including a rudimentary cerebellum has formed (Kimmel et al., 1995). In addition, while the overall structure of the adult zebrafish cerebellum is less complex than that of the mammalian cerebellum (Wullimann M.F., 1996), the basic cellular architecture (molecular layer, Purkinje cell layer, and granule cell layer) is conserved between humans and zebrafish (Miyamura et al., 2001). This suggests that the zebrafish may be a relevant model organism for examining the relationship between the ATXN1/AXH gene family and the development and function of the cerebellum.
Here we describe the identification and characterization of two zebrafish ATXN1 homologs, atxn1a and atxn1b, and an ATXN1L homolog, atxn1l. Using a cell culture system, we demonstrate that two key biochemical features of the human ATXN1 protein that are not found in the invertebrate homologs (a nuclear localization sequence and S776 phosphorylation) are conserved in the zebrafish Atxn1 protein family. Finally, we show both that the zebrafish Atxn1/Axh proteins are expressed in adult zebrafish Purkinje cells, and that the zebrafish atxn1 genes are expressed in the developing cerebellum. Thus, the combination of a cerebellum with key cellular components seen in mammals and an atxn1/axh gene family that is highly homologous to that in mammals indicate that zebrafish would be a useful model for studying the function of the ATXN1/AXH gene family in the cerebellum.
Materials and Methods
Animals
Wild type zebrafish were purchased from Segrest Farms and raised in the University of Minnesota Zebrafish Core Facility.
Cloning of Zebrafish ATXN1 Homologs
The human ATXN1 (amino acids 574-687) and ATXN1L (amino acids 469-569) AXH domains were used to perform a BLAT protein search of the whole zebrafish genome (http://genome.ucsc.edu/). A secondary BLAT search was performed using the full length ATXN1 and ATXN1L protein sequence against the AXH domain containing regions. In order to clone the genes by RT-PCR, primers were designed to the highly conserved N and C- terminus regions including the putative start and stop sites (zATXN1a.1 F: 5′-ATGAAGTCAAACCAGGAGCGG-3′, zATXN1a.1 R: 5′-TCACTTTCCAATATTTGATC-3′ zATXN1b.1 F: 5′-ATGAAGTCTAACCAGGAGCGCT-3′, zATXN1b.1 R: 5′-TCAACAACCTGTGCTGGAAC-3′: zATXN1L.1 F: 5′-ATGAAGCCAGTTCACGAGCGC-3′, zATXN1L.1 R: 5′-CTACTTGCCGGCATTCGAGC-3′).
RNA was prepared with Trizol reagent (Invitrogen) from 24 hpf (hrs post fertilization) embryos. A reverse transcriptase reaction was performed using Superscript III 1st Strand Synthesis Super-Mix (Invitrogen) and a gene specific primer (zATXN1a.1 R, zATXN1b.1 R, or zATXN1L.1 R) according to the manufacturer's instructions. PCR with Pfx Polymerase (Invitrogen) was carried out using the resulting cDNA and the zebrafish primers described above. PCR products were cloned into the pCR-Blunt II-TOPO vector (Invitrogen) and sequenced. To epitope tag the proteins, the zebrafish transcripts were subcloned into the EcoRI sites of the triple-FLAG vector SCII-CDF-EF-3XFLAG (gift from Nik Somia, University of Minnesota). Zebrafish transcripts were translated (http://ca.expasy.org/tools/#translate), and all ClustalW protein alignments were performed using the SDSC Biology Workbench program (http://workbench.sdsc.edu/).
Transfection and Immunoflourescence - Tissue Culture Cells
COS-1 cells were plated onto coverslips and transfected 24 hrs later using Lipofectamine PLUS (Invitrogen) according to the manufacturer's instructions. The following plasmids were used for transfection: CSII-CDF-EF-3XFlag atxn1a, atxn1b, or atxn1l, N-terminal p3XFlag ATXN1L, and pEGFP ATXN1 [30Q](Kaytor et al., 2005). Twenty-four hrs post transfection, the cells were fixed in 3.7% formaldehyde for 10 min, washed twice in 1X PBS, permeabilized in cold acetone for two mins and washed again in 1X PBS. The cells were stained for one hr with the following antibodies: anti-P-S776 ATXN1 (Emamian et al., 2003), M5 (αFLAG, Sigma) or both at a 1:200 dilution. Following three washes with 1XPBS, cells were incubated for one hr with the secondary antibodies (anti- rabbit Alexa Flour 448 and anti- mouse Alexa Flour 546 (Molecular Probes)) and mounted onto slides using glycerol-gelatin (Sigma) supplemented with n-propyl gallate. Images were collected using an Olympus FluoView 1000 confocal microscope.
Immunohistochemistry, RT-PCR and Western Blot - Adult Zebrafish Brains
Adult zebrafish were anesthetized using 4mg/mL Tricaine (Sigma) and decapitated with a razor blade. For immunohistochemistry, zebrafish heads were fixed overnight in 10% buffered formalin, washed in running water for 5-10 min, and placed in Formical 2000 (Decal Chemical Corporation) overnight for decalcification. The specimens were washed in running water for 5-10 min and processed for routine paraffin embedding. 11750 (1:4000), an anti-ATXN1 antibody (Servadio et al., 1995), and AB1778 (1:400), an anti-Calbindin antibody (Chemicon), were used.
For RT-PCR analysis, five cerebella were removed from the decapitated zebrafish heads and pooled. RNA was prepared using Trizol reagent (Invitrogen) following the manufacturer's instructions. As a control, RNA was prepared from COS-1 cells transfected with CSII-CDF-EF-3XFlag atxn1a, atxn1b, or atxn1l. RNA was harvested from the cells 24 hrs post transfection using the Qiagen RNeasy MiniKits (Qiagen). The reverse transcriptase reactions were performed as described above using gene specific primers (zATXN1a.1 R, zATXN1b.1 R, or zATXN1L.1 R) according to the manufacturer's instructions. PCR with Platinum Taq Polymerase (Invitrogen) was carried out using the resulting cDNA and the following primer pairs: zATXN1a.2 F: 5′-AGCTGAAGATTGATTCCAGC-3′ and zATXN1a.2 R: 5′-TGGCTCTTCTTGATGGAACCG-3′, zATXN1b.2 F: 5-TCAACAACCTGTGCTGGAAC-3 and zATXN1b.2 R: 5′-CAAAGCCCAGGTGTGTGTTG-3′, or zATXN1L,2 F: TTCCCTGATACCACCTCAGT-3′ and zATXN1L.2 R: 5′-CCGAATAAACTGCATCTTGA-3. PCR products were analyzed on a 2% agarose gel.
For western blot analysis, two adult zebrafish brains were pooled together and homogenized in lysis buffer (.25M Tris-Cl, pH 7.5) containing 1X protease inhibitors (Roche Biochemicals) and phosphatase inhibitor cocktails 1 and 2 (Sigma). Homogenates were then prepared by three rounds of freezing in liquid nitrogen followed by thawing at 37° C. Lysates were centrifuged at 3000 rpm for 10 min at 4°C, and protein concentration was determined using a Bradford protein assay (Bio-Rad). Cerebellar protein was also prepared from the cerebellum of FVB and ATXN1 knockout mice (Matilla et al., 1998). As a control, protein lysate was prepared from COS-1 cells transfected with CDF-EF-3X-Flag atxn1a, atxn1b, or atxn1l. 100ug of zebrafish protein, 40ug of mouse cerebellar protein, and 3ug of COS-1 lysates were loaded onto a 4-12% Bis-Tris polyacrylamide gel (Invitrogen). After electrophoresis, the proteins were transferred to a nitrocellulose membrane (Protran) and blocked with 5% milk in 1X PBS-0.1% Tween-20. The membrane was probed with anti-P-S776 ATXN1 (Emamian et al., 2003) at 1:1000 overnight in blocking solution and washed three times with 1X PBST. The membrane was then probed with an ECL-anti-rabbit IgG- HRP secondary antibody (GE Healthcare) at 1:2500, and the western was developed using the SuperSignal West Pico Chemiluminescent Kit (Thermo-Scientific).
Whole Mount In Situ Hybridization-Zebrafish Embryos
The following primers were used to amplify regions of atxn1a and atxn1b for probe preparation: zATXN1a.3F: ATACGCTGGCTACATTTCTC, zATXN1a.3R: AGATGGACCAAAACGTCTGT, zATXN1b.3F: GGCAGAGTACCCTCCAACTA, and zATXN1b.3R: TTGACTGTGGGAAACATTTG. SP6 and T7 promoters were added using a PCR approach. DIG labeled probes were prepared using in vitro transcription and the SP6 and T7 polymerases (Roche).
Zebrafish embryos (28hpf) were fixed in 4% Paraformaldehyde overnight, dehydrated using 100% methanol, and stored at -20°C. Whole mount in situ hybdridization was performed as follows. Embryos were rehydrated in a methanol series and washed four times in 1XPBT (1X PBS/0.1% Tween 20). Embryos were digested for five mins in PBT containing proteinase K (10ug/ml) and refixed in 4% Paraformaldehyde for 20 minutes. Following five PBT washes, the embryos were prehybridized for four hrs at 65°C in hybridization mix (50% formamide, 5XSSC, 50ug/ml Heparin, 5mM EDTA, 0.5mg/ml rRNA, 0.1% Tween 20, and Citric Acid to adjust pH to 6.0). Probe hybridization was carried out overnight at 65°C in hybridization mix. Following hybridization, embryos were washed at 65°C as follows: 75% Hyb/25% 2XSSC, 50% Hyb/50% 2XSSC, 25% Hyb/75% 2XSSC (15 min each), and 0.2XSSC two times (30 min each). The following washes were then performed at room temperature: 75% 0.2XSSC/25% PBT, 50% 0.2XSSC/50% PBT, 25% 0.2XSSC/75% PBT, and PBT (5 min each). Finally, the embryos were incubated overnight at 4°C with an anti-DIG-AP antibody (1:4000) (Roche) in PBT containing 2% lamb serum, washed seven times with PBT and developed using the NBT/BCIP system (Promega).
Northern Blot Analysis
Atxn1a and atxn1b probes were prepared by PCR using the primers described above and labeled using the Rediprime Labeling system (GE Healthcare). RNA was harvested from COS-1 cells transfected with CSII-CDF-EF-3XFlag atxn1a, atxn1b, or atxn1l, and a northern blot was performed using the Northern Max-Gly system (Ambion). After transfer, the membrane was UV cross-linked and probed with either atxn1a or atxn1b at 65°C for three hrs.
Results
Identification of the Zebrafish ATXN1/AXH Homologs
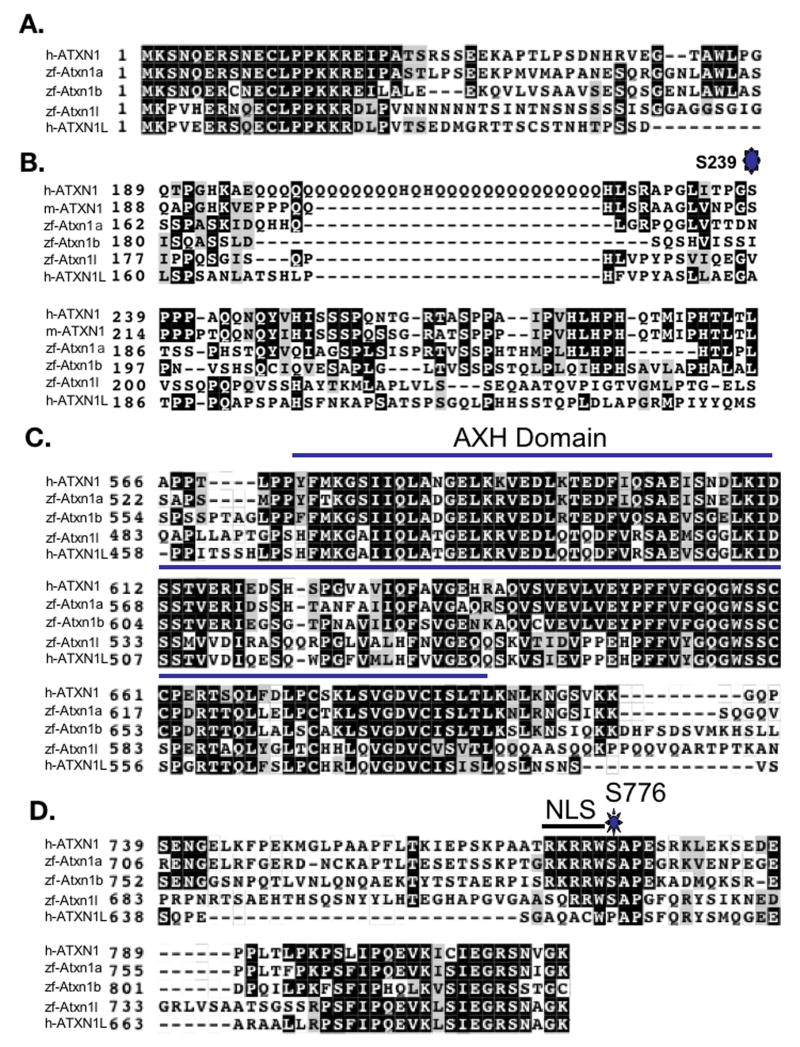
The human ATXN1 protein family members (ATXN1 and ATXN1L) contain a highly conserved AXH domain (Bowman et al., 2007; de Chiara et al., 2003; Mizutani et al., 2005). To identify the zebrafish ATXN1 homologs, we performed a protein BLAT search (http://genome.ucsc.edu) of the zebrafish genome using either the human ATXN1 or ATXN1L AXH domain protein sequences. Genes encoding three putative zebrafish AXH domains were identified with greater than 60% identity to the human ATXN1 AXH domain located on zebrafish chromosomes 19, 16, and 7 respectively. Following further sequence analysis, the complete coding sequences of these three AXH-encoding zebrafish genes were cloned. Based on both protein conservation (Figure 1) and whole genome alignments (Human Chained Alignment: http://genome.ucsc.edu), genes encoding two ATXN1 homologs were identified on chromosomes 19 and 16 in the zebrafish genome (atxn1a and atxn1b) and one encoding an ATXN1L homolog was identified on chromosome 7 (atxn1l). Like their human homologs, all three zebrafish genes consist of two coding exons.
Figure 1. Comparison of the ATXN1/AXH Protein Family Between Zebrafish and Humans.
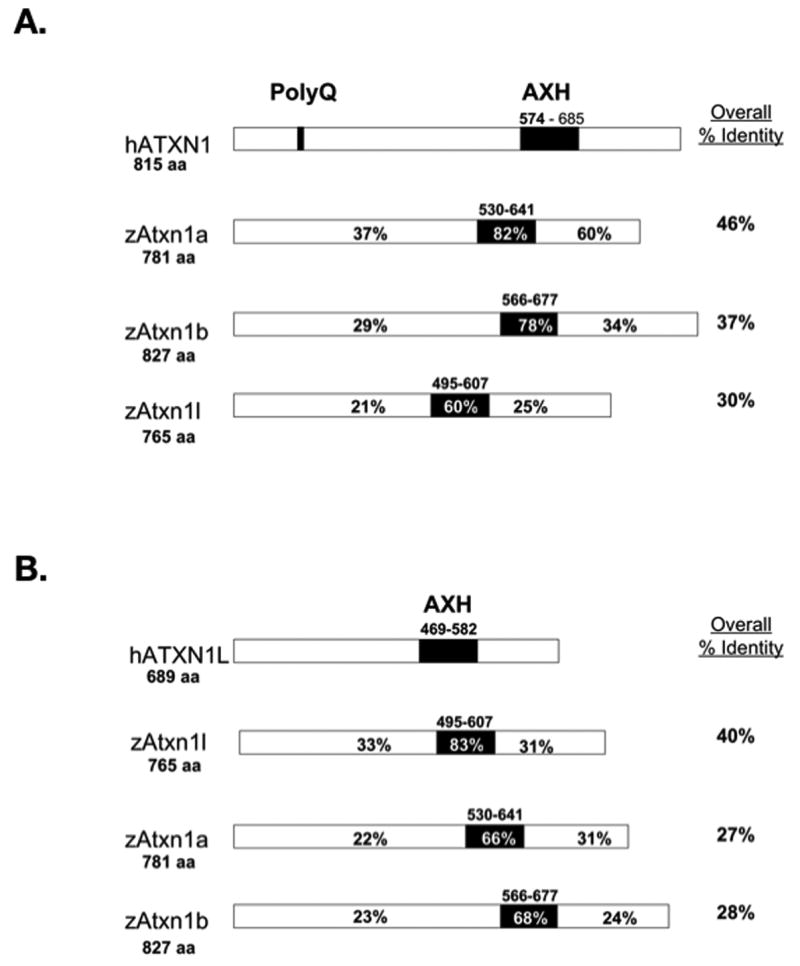
The percent identity of amino acids shared by the zebrafish Atxn1/Axh family members and human ATXN1 (A) or human ATXN1L (B). Percent identity was determined using the ClustalW alignment program. Percent identity is shown for the N-terminus of the protein, the AXH domain (in black), the C-terminus of the protein, and the whole protein. The amino acids defining the AXH domain are shown above the black box. Total amino acid length of each protein is shown at the start of the protein.
Figure 1 compares the three zebrafish Atxn1/Axh proteins to either human ATXN1 (hATXN1) (Figure 1A) or ATXN1L (hATXN1L) (Figure 1B). Overall, zebrafish Atxn1a (zAtxn1a) and Atxn1b (zAtxn1b) were 46% and 37% identical to human ATXN1 (Figure 1A), while zebrafish Atxn1l (zAtxn1l) was 40% identical to human ATXN1L (Figure 1B). The AXH domain was the region of highest conservation within all three proteins. The zebrafish Atxn1a and Atxn1b AXH domains were 82% and 78% identical to the human ATXN1 AXH domain (Figure 1A), while the zebrafish Atxn1l domain was 83% identical to the human ATXN1L AXH domain (Figure 1B).
Conservation of Protein Features Across the ATXN1/AXH Protein Family Members
A more focused look at the alignment of the three zebrafish Atxn1/Axh proteins with both human ATXN1 and ATXN1L showed three regions of high conservation (Figure 2): The N-terminal 21 amino acids (Figure 2A), the AXH Domain (Figure 2C), and the 21 amino acids at the C-terminus (Figure 2D). Notably, like murine ATXN1 and ATXN1L, the three zebrafish homologs do not contain a polyglutamine stretch (Figure 2B).
Figure 2. Three Regions of High Homology Are Identified Between the Human and Zebrafish ATXN1/AXH Proteins.
A ClustalW alignment was performed to identify regions of conservation between human ATXN1 (h-ATXN1), zebrafish Atxn1a (zf-Atxn1a), Atxn1b (zf-Atxn1b), Atxn1l (zf-Atxn1l), and human ATXN1L (h-ATXN1L). Three regions of high conservation were identified: the N-terminus of the protein (A), the AXH domain (C), and the C-terminus of the protein (D). A long polyglutamine repeat was only identified in human ATXN1 (mouse ATXN1 (m-ATXN1) was included to illustrate that in other model systems a long polyglutamine repeat is not conserved) (B).
In addition to the polyglutamine stretch and the AXH domain, two other important features of the human ATXN1 protein include the nuclear localization sequence centered at the lysine at position 772 (Klement et al., 1998) and the serine residue at position 776 (H. K. Chen et al., 2003; Emamian et al., 2003). Protein sequence analysis revealed that K772, a key amino acid important for nuclear localization, was conserved in zebrafish Atxn1a and Atxn1b but not in human or zebrafish ATXN1L. S776 was conserved in all three zebrafish Atxn1/Axh family members but not in human or mouse ATXN1L (Figure 3A). More recently, a second phosphorylation site in human ATXN1 at S239 was identified (Vierra-Green et al., 2005). Although the region downstream of the polyglutamine tract containing S239 was less conserved in the zebrafish, both Atxn1a and Atxn1b contained a serine residue that may correspond to human ATXN1 S239 (Figure 2B).
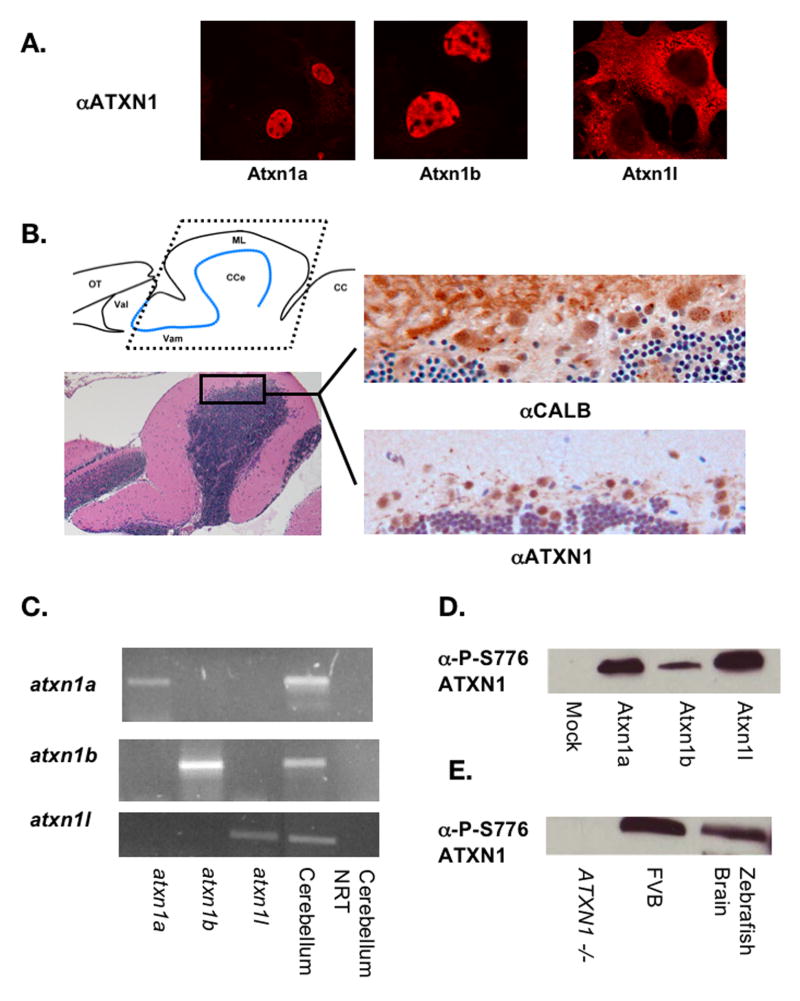
Figure 3. Zebrafish Atxn1/Axh proteins localize to the nucleus and are phosphorylated in tissue culture cells.
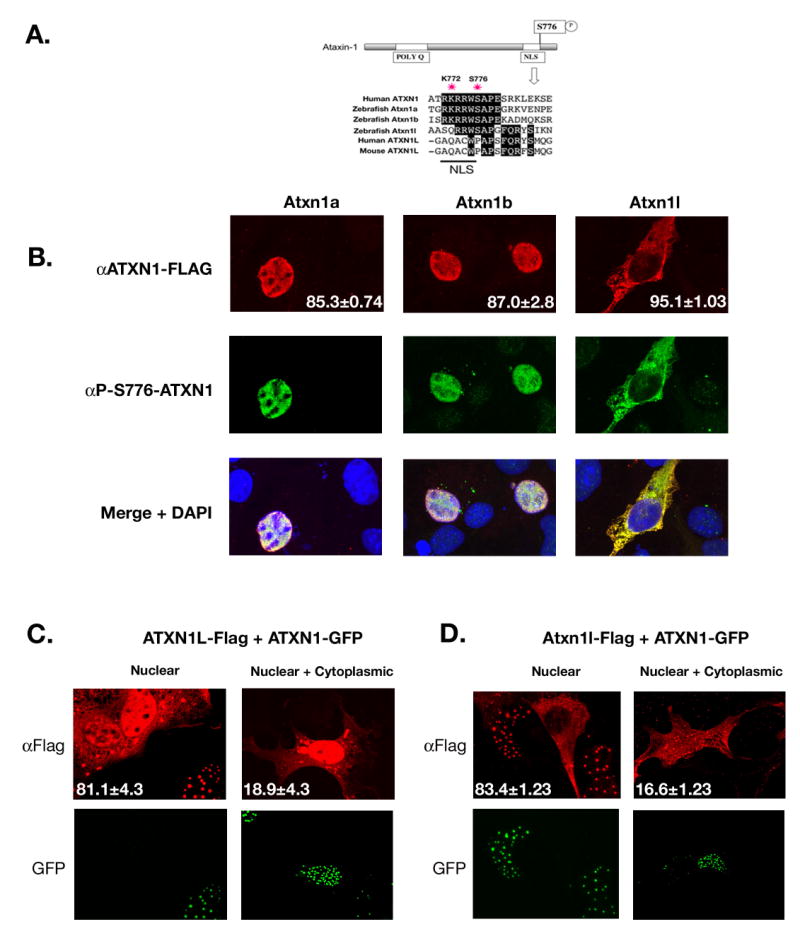
(A).ClustalW alignment was performed to show conservation of both the nuclear localization sequence and S776 residue among the zebrafish Atxn1/Axh proteins and human ATXN1 but not with mouse or human ATXN1L. (B). COS-1 cells were transfected with Flag-tagged zebrafish atxn1a, atxn1b, or atxn1l. Twenty-four hours post transfection these cells were stained using an α-FLAG or α-P-S776 ATXN1 antibody. Zebrafish Atxn1a and Atxn1b localized to the nucleus of the cell while Atxn1l was mostly cytoplasmic. All three proteins were recognized by the phospho-specific ATXN1 antibody. (Each transfection was done in triplicate and 400 cells per experiment were scored for protein localization. The percent of cells with nuclear (Atxn1a and Atxn1b) or cytoplasmic (Atxn1l) localization is noted in the αFlag panel). (C). Flag-tagged human ATXN1L was cotransfected with GFP-tagged human ATXN1 into COS-1 cells and stained with an α-FLAG antibody. Human ATXN1L alone localized to both the nucleus and cytoplasm of the cell (as seen by the two cells not transfected with ATXN1 in the nuclear panel) but when coexpressed with ATXN1, became predominantly nuclear. In a small percentage of cells, cytoplasmic and nuclear localization of ATXN1L was observed in the presence of ATXN1. (This transfection was performed in triplicate, and 100 cotransfected cells per experiment were scored for nuclear or nuclear and cytoplasmic localization of ATXN1L. The percentages are indicated in each representative image.) (D). Flag-tagged zebrafish atxn1l was cotransfected into COS-1 cells with GFP-tagged human ATXN1. When expressed with human ATXN1, zebrafish Atxn1l became predominantly nuclear in contrast to the mostly cytoplasmic location of zebrafish Atxn1l when expressed alone (A). In a small percentage of cells, zebrafish Atxn1l was visualized in both the cytoplasm and nucleus in the presence of human ATXN1. (This transfection was performed in triplicate and 100 cotransfected cells per experiment were scored for Atxn1l localization. Percentage nuclear or nuclear and cytoplasmic is noted in each representative image.)
To examine whether the zebrafish Atxn1/Axh proteins behave similarly to their human counterparts in the cell, COS-1 cells were transfected with Flag-tagged zebrafish atxn1a, atxn1b or atxn1l (Figure 3B). As predicted by sequence analysis, like human ATXN1 (Klement et al., 1998) both zebrafish Atxn1a and Atxn1b localized to the nucleus of the cell while Atxn1l remained mostly cytoplasmic. In addition, staining with an antibody specific for phosphorylation at S776 (Emamian et al., 2003) revealed that in COS-1 cells all three zebrafish proteins were phosphorylated.
Interestingly, human ATXN1L does not contain a predicted NLS sequence at lysine 772 (Figure 3A); yet this protein is found to colocalize with ATXN1 in the nucleus of various human cell lines and like human ATXN1 is expressed in the nucleus of different neuronal populations (Mizutani et al., 2005). Because human ATXN1 and ATXN1L have a similar cellular pattern of expression and are known to interact (Bowman et al., 2007; Mizutani et al., 2005), we explored whether the subcellular localization of human ATXN1L was linked to the expression of ATXN1. COS-1 cells were cotransfected with FLAG-tagged ATXN1L and GFP-tagged ATXN1 (Figure 3C). Human ATXN1L alone localized to both the nucleus and cytoplasm of the cell (as seen by the two cells which do not express human ATXN1 in Fig. 3C). However, when human ATXN1L was expressed with ATXN1, the protein was almost exclusively nuclear. We tested whether the subcellular localization of zebrafish Atxn1l would also be dependent on ATXN1 coexpression. As shown previously, zebrafish atxn1l transfected by itself was predominately cytoplasmic (Figure 3B). Like human ATXN1L, when zebrafish atxn1l was cotransfected with human ATXN1, the protein localized to the nucleus (Figure 3D).
Zebrafish Atxn1/Axh Proteins are Expressed in the Purkinje Cells of the Adult Zebrafish Cerebellum and Are Phosphorylated In Vivo
To determine the cellular expression pattern of the zebrafish Atxn1/Axh family members in the cerebellum, immunohistochemistry was performed using an anti-ATXN1 antibody (Servadio et al., 1995) that recognizes all three zebrafish proteins (Fig 4A). In adult zebrafish, ATXN1 antibody staining of cells in the Purkinje cell layer of the cerebellum was seen. Calbindin was used as a Purkinje cell marker to confirm the identity of the Atxn1/Axh expressing cells (Figure 4B). As in the mice, calbindin stains both the cell body and dendrites of the Purkinje cells while ATXN1 staining is observed only in the cell body (Klement et al., 2003). Because the ATXN1 antibody recognizes all three zebrafish Atxn1/Axh family members, RT-PCR using cerebellar RNA was performed to identify the expression of each homolog. By this analysis, atxn1, atxn1b, and atxn1l transcripts were detected in the adult zebrafish cerebellum (Figure 4C).
Figure 4. Zebrafish Atxn1/Axh proteins are expressed and phosphorylated in the adult cerebellum.
To demonstrate that the anti-ATXN1 antibody used in these studies cross-reacts with all three zebrafish Atxn1/Axh proteins, COS-1 cells were transfected with Flag-tagged zebrafish atxn1a, atxn1b, or atxn1l. Twenty-four hours post transfection, the cells were stained with the anti-ATXN1 antibody (A). Immunohistochemistry was performed on sections from paraffin embedded zebrafish heads using the anti-ATXN1 antibody tested in panel A and an anti-calbindin antibody as a Purkinje cell marker (B). Expression of the zebrafish Atxn1 proteins was detected in the cerebellar Purkinje cells. Both a diagram of the zebrafish cerebellum (with the Purkinje cell layer highlighted in blue) as well as an H&E section are included to provide an orientation to the zebrafish cerebellum. To determine which ATXN1 homologs are expressed in the cerebellum, RT-PCR analysis was performed on RNA isolated from adult zebrafish cerebellum to detect expression of zebrafish atxn1a, atxn1b, or atxn1l (C). RNA from COS-1 cells transfected with each construct individually was used as a control. To determine if the zebrafish Atxn1/Axh protein family is phosphorylated at S776 in vivo, western blot analysis was performed using an anti-P-S776 ATXN1 antibody. Protein lysates from COS-1 cells transfected with zebrafish atxn1a, atxn1b, or atxn1l (D) and lysates from a zebrafish brain (E) were probed with an anti-P-S776 antibody. Mock transfected cells (D) and wild type (FVB) and ATXN1 -/- mouse brains (E) were included as controls. (OT = Optic Tectum, VaL = Lateral Valvula Cerebelli, VaM = Medial Valvula Cerebelli, CCe = Corpus Cerebelli, ML = Molecular Layer, and CC = Crista Cerebelli)
Because phosphorylation of S776 is an important biochemical feature of the ATXN1 protein, western blot analysis was performed to determine if the zebrafish Atxn1/Axh protein family is phosphorylated at S776 in vivo (Figure 4D and 4E). An anti-P-S776 antibody (Emamian et al., 2003) that cross-reacts with all three zebrafish proteins (Figure 4D) recognized phosphorylated Atxn1/Axh in zebrafish whole brain lysate (Figure 4E). Cerebellar lysates from wild type (FVB) and ATXN1 knockout mice were included as controls. Because all three proteins are large and similar in size (83kDa, 87kDa, 82kDa for Atxn1a, Atxn1b, and Atxn1l respectively), they are not distinguishable on the gel (Figure 4D).
Zebrafish atxn1a and atxn1b are Expressed in the Developing Cerebellum
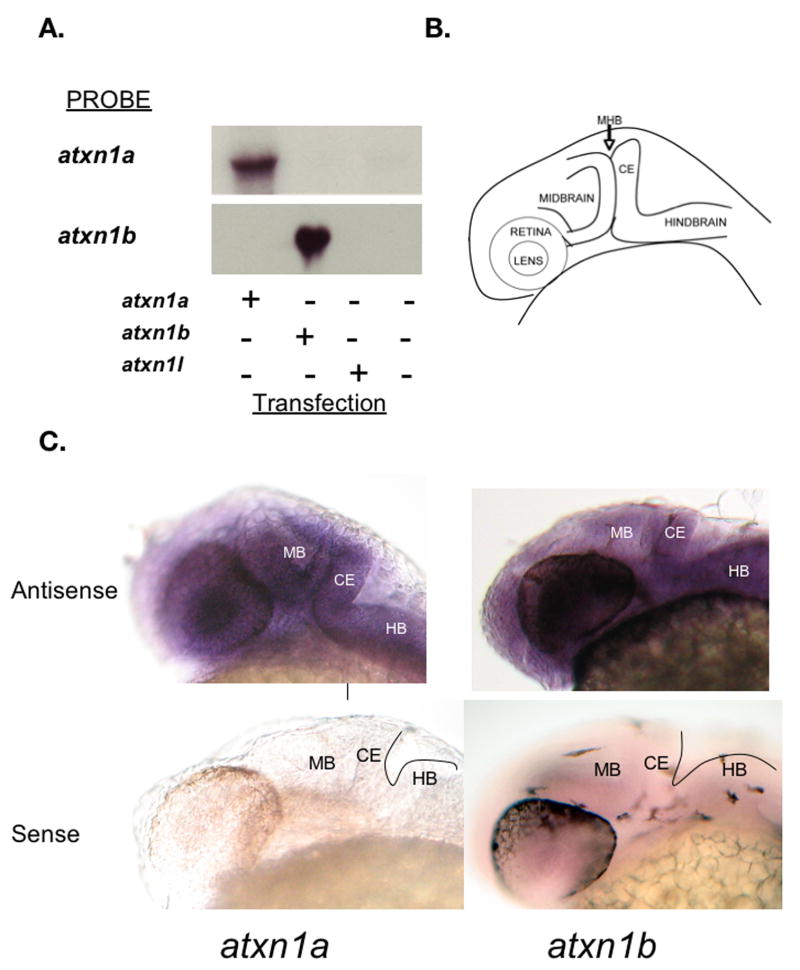
Zebrafish are an important model system for investigating the developmental role of a specific gene. To determine whether the zebrafish atxn1 genes are expressed during zebrafish development whole mount in situ hybridization was performed. At 28 hpf the developing cerebellum can be observed in the zebrafish embryo as diagrammed in Fig 5B. Both atxn1a and atxn1b are expressed in the developing zebrafish brain and specifically in the cerebellar anlage at 28 hpf (Fig 5C). A northern blot was performed to confirm the specificity of the in situ probes for their target atxn1 gene (Fig 5a).
Figure 5. Expression of the atxn1 genes in the developing zebrafish cerebellum.
A northern blot containing RNA from COS-1 cells transfected with zebrafish atxn1a, atxn1b, or atxn1l was probed with either the atxn1a or atxn1b probe used for in situ hybridization to demonstrate that each probe is specific for its target (A). A diagram of a developing zebrafish embryo at 28hpf is shown to illustrate the location of the cerebellar anlage (B). Whole mount in situ hybridization was performed to determine the expression pattern of the zebrafish atxn1 homologs in the developing brain. Both atxn1a and atxn1b are expressed in the cerebellar anlage at 28hpf. The sense probes are shown as a control (C). (MB = Midbrain, CE = Cerebellar Anlage, HB = Hindbrain).
Discussion
The zebrafish genome contains three members of the ATXN1/AXH gene family, two homologs of ATXN1 designated atxn1a (Chr 19) and atxn1b (Chr 16), as well as a homolog of ATXN1L, atxn1l (Chr 7). A whole genome alignment between the zebrafish and human genomes (using the UCSC Genome Browser Human Chained Alignment) revealed that the genomic sequences containing zebrafish atxn1a and atxn1b are most similar to the region of human chromosome 6 containing ATXN1. Likewise, the genomic region containing zebrafish atxn1l is most similar to human chromosome 16 where ATXN1L is located. Identification of an atxn1 duplication in the zebrafish genome is consistent with data supporting the hypothesis that there was a whole genome duplication event that occurred in the ray-finned fish (including zebrafish). In support of the idea that zebrafish atxn1a (on chromosome 19) and atxn1b (on chromosome 16) are the result of a genome duplication event, these chromosomes in zebrafish were previously identified as paralogous chromosome regions (Taylor et al., 2003).
In Drosophila and C. elegans, invertebrates with a simple central nervous system lacking a cerebellum, the ATXN1/AXH gene family consists of a single gene encoding for a small AXH domain-only protein (de Chiara et al., 2003; Tsuda et al., 2005). In higher vertebrates, such as humans and mice, the ATXN1/AXH gene family consists of two members, ATXN1 and ATXN1L. Likewise, the ATXN1 gene structure is more complex, encoding a protein with some important features not found in the invertebrate homologs. In addition to the AXH domain, an important mediator of protein/protein interactions (de Chiara et al., 2003; Mizutani et al., 2005; Tsuda et al., 2005), there is also the nuclear localization sequence located at K772 (Klement et al., 1998) and the phosphorylation of S776 (H. K. Chen et al., 2003; Emamian et al., 2003). These two features of ATXN1 are conserved in the zebrafish homologs. Both Atxn1a and Atxn1b contain the NLS and localize to the nucleus of the cell in tissue culture. An antibody specific for the phosphorylation of S776 recognizes both Atxn1a and Atxn1b in tissue culture and recognizes phosphorylated Atxn1 protein in the adult zebrafish brain. The conservation of these additional ATXN1 features between zebrafish and humans indicates that the functional pathways involving these elements are likely to also be conserved in the zebrafish and demonstrates that as the structure of the central nervous system becomes more complex there is a corresponding increase in the complexity of the ATXN1 gene structure.
The zebrafish genome also encodes a homolog of ATXN1L. Like its human counterpart, the zebrafish atxn1l gene encodes for a protein that contains an AXH domain and when expressed in COS-1 cells is mostly cytoplasmic. Interestingly, when either human or zebrafish ATXN1L were co-expressed with human ATXN1, both proteins became predominantly nuclear. Thus ATXN1 may help to shuttle ATXN1L into the nucleus where they are known to interact in large protein complexes (Lam et al., 2006). One unique feature of zebrafish Atxn1l is the conservation of a key serine residue (S776) phosphorylated in ATXN1. A proline is found at this position both in mouse and human ATXN1L (Figure 3). In tissue culture cells zebrafish Atxn1l was phosphorylated at this serine indicating that the function of zebrafish Atxn1l may overlap more with Atxn1 than do ATXN1 and ATXN1L in humans.
In addition to the conserved organization of the ATXN1/AXH gene family between zebrafish and humans, the zebrafish atxn1/axh genes are expressed in the adult zebrafish cerebellum in a manner similar to that seen with humans. Immunohistochemical analysis showed that the zebrafish Atxn1/Axh protein family is expressed in the adult cerebellum by large, calbindin- positive neurons, suggesting these cells are Purkinje cells. RT-PCR showed that each member of the zebrafish atxn1/axh gene family is expressed in the zebrafish cerebellum. Finally, whole mount in situ hybridization demonstrates that zebrafish atxn1a and atxn1b are expressed in the cerebellar anlage, supporting a role for these gene products in cerebellar development.
Their easily accessible, transparent embryos, rapid developmental time-frame, and conservation of ATXN1/AXH gene complexity and expression patterns indicate that zebrafish will be a useful model for studying the role of the ATXN1/AXH gene family in the development of a complex central nervous system and perhaps SCA1 pathogenesis. Zebrafish have previously been established as a useful model system for studying modifiers of polyglutamine toxicity (Miller et al., 2005). In a similar manner, as understanding of SCA1 pathogenesis progresses and novel therapeutic targets are identified, a zebrafish SCA1 model will provide a useful means for testing both the toxicity and bioactivity of candidate drugs before moving to a larger animal model (Reviewed in (Lieschke et al., 2007; Zon et al., 2005)).
Acknowledgments
We thank the University of Minnesota Zebrafish Core Facility for handling the zebrafish colony and for helpful advice and Dr. Steve Ekker for helpful discussion. This work was supported by a National Ataxia Foundation Postdoctoral Fellowship (KMC) and National Institute of Health grants NS022920 and NS045667 (HTO).
Work Cited
- Bowman AB, Lam YC, Jafar-Nejad P, Chen HK, Richman R, Samaco RC, Fryer JD, Kahle JJ, Orr HT, Zoghbi HY. Duplication of Atxn1l suppresses SCA1 neuropathology by decreasing incorporation of polyglutamine-expanded ataxin-1 into native complexes. Nat Genet. 2007;39:373–9. doi: 10.1038/ng1977. [DOI] [PubMed] [Google Scholar]
- Burright EN, Clark HB, Servadio A, Matilla T, Feddersen RM, Yunis WS, Duvick LA, Zoghbi HY, Orr HT. SCA1 transgenic mice: a model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell. 1995;82:937–48. doi: 10.1016/0092-8674(95)90273-2. [DOI] [PubMed] [Google Scholar]
- Chen HK, Fernandez-Funez P, Acevedo SF, Lam YC, Kaytor MD, Fernandez MH, Aitken A, Skoulakis EM, Orr HT, Botas J, Zoghbi HY. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113:457–68. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- Chen YW, Allen MD, Veprintsev DB, Lowe J, Bycroft M. The structure of the AXH domain of spinocerebellar ataxin-1. J Biol Chem. 2004;279:3758–65. doi: 10.1074/jbc.M309817200. [DOI] [PubMed] [Google Scholar]
- de Chiara C, Giannini C, Adinolfi S, de Boer J, Guida S, Ramos A, Jodice C, Kioussis D, Pastore A. The AXH module: an independently folded domain common to ataxin-1 and HBP1. FEBS Lett. 2003;551:107–12. doi: 10.1016/s0014-5793(03)00818-4. [DOI] [PubMed] [Google Scholar]
- Emamian ES, Kaytor MD, Duvick LA, Zu T, Tousey SK, Zoghbi HY, Clark HB, Orr HT. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron. 2003;38:375–87. doi: 10.1016/s0896-6273(03)00258-7. [DOI] [PubMed] [Google Scholar]
- Kaytor MD, Byam CE, Tousey SK, Stevens SD, Zoghbi HY, Orr HT. A cell-based screen for modulators of ataxin-1 phosphorylation. Hum Mol Genet. 2005;14:1095–105. doi: 10.1093/hmg/ddi122. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Klement IA, Skinner PJ, Kaytor MD, Yi H, Hersch SM, Clark HB, Zoghbi HY, Orr HT. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- Lam YC, Bowman AB, Jafar-Nejad P, Lim J, Richman R, Fryer JD, Hyun ED, Duvick LA, Orr HT, Botas J, Zoghbi HY. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–47. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–67. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Matilla A, Roberson ED, Banfi S, Morales J, Armstrong DL, Burright EN, Orr HT, Sweatt JD, Zoghbi HY, Matzuk MM. Mice lacking ataxin-1 display learning deficits and decreased hippocampal paired-pulse facilitation. J Neurosci. 1998;18:5508–16. doi: 10.1523/JNEUROSCI.18-14-05508.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VM, Nelson RF, Gouvion CM, Williams A, Rodriguez-Lebron E, Harper SQ, Davidson BL, Rebagliati MR, Paulson HL. CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J Neurosci. 2005;25:9152–61. doi: 10.1523/JNEUROSCI.3001-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamura Y, Nakayasu H. Zonal distribution of Purkinje cells in the zebrafish cerebellum: analysis by means of a specific monoclonal antibody. Cell Tissue Res. 2001;305:299–305. doi: 10.1007/s004410100421. [DOI] [PubMed] [Google Scholar]
- Mizutani A, Wang L, Rajan H, Vig PJ, Alaynick WA, Thaler JP, Tsai CC. Boat, an AXH domain protein, suppresses the cytotoxicity of mutant ataxin-1. Embo J. 2005 doi: 10.1038/sj.emboj.7600785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Jeong SY, Uchihara T, Anno M, Nagashima K, Nagashima T, Ikeda S, Tsuji S, Kanazawa I. SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum Mol Genet. 2001;10:1441–8. doi: 10.1093/hmg/10.14.1441. [DOI] [PubMed] [Google Scholar]
- Orr HT, Chung MY, Banfi S, Kwiatkowski TJ, Jr, Servadio A, Beaudet AL, McCall AE, Duvick LA, Ranum LP, Zoghbi HY. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4:221–6. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- Serra HG, Duvick L, Zu T, Carlson K, Stevens S, Jorgensen N, Lysholm A, Burright E, Zoghbi HY, Clark HB, Andresen JM, Orr HT. RORalpha-mediated Purkinje cell development determines disease severity in adult SCA1 mice. Cell. 2006;127:697–708. doi: 10.1016/j.cell.2006.09.036. [DOI] [PubMed] [Google Scholar]
- Servadio A, Koshy B, Armstrong D, Antalffy B, Orr HT, Zoghbi HY. Expression analysis of the ataxin-1 protein in tissues from normal and spinocerebellar ataxia type 1 individuals. Nat Genet. 1995;10:94–8. doi: 10.1038/ng0595-94. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–90. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CC, Kao HY, Mitzutani A, Banayo E, Rajan H, McKeown M, Evans RM. Ataxin 1, a SCA1 neurodegenerative disorder protein, is functionally linked to the silencing mediator of retinoid and thyroid hormone receptors. Proc Natl Acad Sci U S A. 2004;101:4047–52. doi: 10.1073/pnas.0400615101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H, Jafar-Nejad H, Patel AJ, Sun Y, Chen HK, Rose MF, Venken KJ, Botas J, Orr HT, Bellen HJ, Zoghbi HY. The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell. 2005;122:633–44. doi: 10.1016/j.cell.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Vierra-Green CA, Orr HT, Zoghbi HY, Ferrington DA. Identification of a novel phosphorylation site in ataxin-1. Biochim Biophys Acta. 2005;1744:11–8. doi: 10.1016/j.bbamcr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–47. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]