Abstract
To address the biological heterogeneity of lung cancer, we studied 199 lung adenocarcinomas by integrating genome-wide data on copy number alterations and gene expression with full annotation for major known somatic mutations in this cancer. This revealed non-random patterns of copy number alterations significantly linked to EGFR and KRAS mutation status and to distinct clinical outcomes, and led to the discovery of a striking association of EGFR mutations with under-expression of DUSP4, a gene within a broad region of frequent single-copy loss on 8p. DUSP4 is involved in negative feedback control of EGFR signaling and we provide functional validation for its role as a growth suppressor in EGFR-mutant lung adenocarcinoma. DUSP4 loss also associates with p16/CDKN2A deletion and defines a distinct clinical subset of lung cancer patients. Another novel observation is that of reciprocal relationship between EGFR and LKB1 mutations. These results highlight the power of integrated genomics to identify candidate driver genes within recurrent broad regions of copy number alteration and to delineate distinct oncogenetic pathways in genetically complex common epithelial cancers.
Introduction
The complexity of the highly aberrant cancer genomes seen in most human carcinomas has presented a formidable analytical challenge that has been the focus of recent efforts in genome-wide microarray profiling of gene expression and genomic copy number alterations (CNAs). However, such analyses performed independently face certain important limitations. Copy number profiling alone is best suited for the delineation of relatively focal high-amplitude events such as small high-level amplicons or narrow homozygous deletions; it provides few leads into larger regions of gains or loss that may span almost entire chromosome arms, changes commonly seen in human carcinomas. Expression profiling alone has provided fewer insights than expected into carcinomas with complex karyotypes because the gene expression changes contributed by passenger genes from regions of CNAs add considerable noise to these datasets. Finally, it is likely that there are certain patterns of CNAs (and associated gene expression changes) that cooperate with specific known (and unknown) mutations. Here, we harnessed the power of an integrated genomic approach to begin to reduce the complexity of lung adenocarcinoma and formulate new hypotheses regarding common cooperating events in this cancer.
The landmark discovery in 2004 that lung adenocarcinomas sensitive to the EGFR tyrosine kinase inhibitors contain somatic mutations in the EGFR kinase domain [reviewed in (Sharma et al. 2007)] represented a remarkable convergence of clinical observations and kinome sequencing efforts. However, many of the mutations described so far in lung adenocarcinomas may represent the “low-hanging fruit” and their cooperating genetic alterations remain largely unknown. It is likely that further advances in treating lung adenocarcinoma will require a deeper understanding of its biology and heterogeneity, beyond what is possible by individual genomic technologies. Although a number of studies have performed extensive DNA copy number profiling (Kendall et al. 2007; Weir et al. 2007; Kwei et al. 2008), gene expression profiling [reviewed in (Meyerson et al. 2004)] or mutation screening (Davies et al. 2005; Marks et al. 2007) to characterize the lung adenocarcinoma genome, these individual approaches are reaching a point of diminishing returns, uncovering low prevalence mutations or amplifications but not clarifying the broader picture of how common mutations interact with common CNAs in this cancer.
We report an initial analysis of the largest integrated genomic dataset of lung adenocarcinoma assembled to date. We demonstrate how major mutated human lung cancer genes such as EGFR and KRAS appear as strong candidates without a priori knowledge, based on the integration of copy number and gene expression data. We further show how the integration of these data with mutational screening for all major known lung cancer genes leads to the identification of additional novel candidate lung cancer genes that may be targets of pathogenic mutations or CNAs. Specifically, we find that EGFR mutations in lung adenocarcinomas are strongly associated with low expression of DUSP4 due to broad single copy losses at 8p. Dual-specificity phosphatases (DUSPs) are known to be transcriptionally up-regulated by mitogen-activated protein kinase (MAPK) signaling as a negative feedback mechanism (Owens and Keyse 2007) and DUSPs and other negative regulators of kinase signaling are emerging as putative tumor suppressors in other cancers (Furukawa et al. 2003; Shaw et al. 2007).
Results
Patterns of mutations in lung adenocarcinomas
Frozen samples of 199 primary lung adenocarcinomas from 199 patients were processed for genomic analyses (See Supplementary Materials and Methods). Basic clinical and pathologic data are summarized in Supplementary Table 1. We used a variety of approaches including Sanger sequencing, mutation-specific PCR assays, and mass-spectrometry-based genotyping, to profile the mutational status of established somatic lung cancer genes, including EGFR, KRAS, BRAF, ERBB2, PIK3CA, LKB1, PTEN, and TP53 (see Supplementary Materials and Methods). Mutations in at least one of these genes were detected in 140/199 cases (70%) (Figure 1A). Mutations in EGFR, KRAS, ERBB2, or BRAF, present collectively in 98/199 cases (49%), were completely mutually exclusive, as expected from published data. Mutations in EGFR and LKB1 may also be largely mutually exclusive, with only 1/43 EGFR-mutant tumors also showing a mutation in LKB1, compared with 27/156 EGFR-wild type tumors (p=0.012). Mutations in TP53 were frequent (27%) and commonly occurred with other mutations. Few cases showed mutations in PTEN (4%) or PIK3CA (2%), in line with prior studies (Samuels et al. 2004; Marks et al. 2007).
Figure 1.
A. Survey of major known mutations in 199 lung adenocarcinomas. EGFR, KRAS, BRAF, and ERBB2 mutations are mutually exclusive. Mutant samples (columns) are sorted by grouped mutations per one-way average-linkage hierarchical clustering on binary data. Black represents samples with single mutations, while red are samples harboring mutations in more than one gene. Sample numbers are indicated at left, 140 cases harbored at least one mutation. Samples without detected mutations in these genes are not shown. B. Recurrent genomic copy number alterations in 199 lung adenocarcinomas. Frequency of gain (red) and loss (blue) is shown by genomic position across the 22 autosomes. Centromeres are identified by dotted vertical lines; acrocentric p-arms and loci lacking array coverage are indicated as gaps. The most prevalent losses are at 8p and 9p21; among gains, 1q, 5p, and 8q are most frequent.
Recurrent genomic copy number alterations
Array-based comparative genomic hybridization (aCGH) was performed using Agilent 44k arrays. Frequent gains were seen on chromosome arms 1q, 5p, 7p, 8q, 12q, and 14q, and frequent losses on 3p, 6q, 8p, 9p, 13q, and 17p (Figure 1B). These major CNAs are consistent with those reported in other lung adenocarcinoma datasets (Kendall et al. 2007; Weir et al. 2007). We focus below on 8p losses, one of the most common broad CNAs in lung adenocarcinoma, occurring in approximately ¼ of cases. We also identified focal, recurrent, high-amplitude CNAs defined heuristically as minimal common regions (MCR) of amplification or deletion (see Supplementary Materials and Methods), several of which contain well-described oncogenes or tumor suppressor genes that may be driving the selection for these CNAs (Supplementary Table 2). For instance, focal high level amplification at 14q13 centered on TITF1, recently recognized in approximately 12% of lung adenocarcinomas (Kendall et al. 2007; Weir et al. 2007), was also revealed here. Other MCRs may define new cancer genes and we list the boundaries of these intervals and propose genes of interest within them (Supplementary Table 2).
Non-random patterns of CNAs associated with EGFR/KRAS mutations and survival
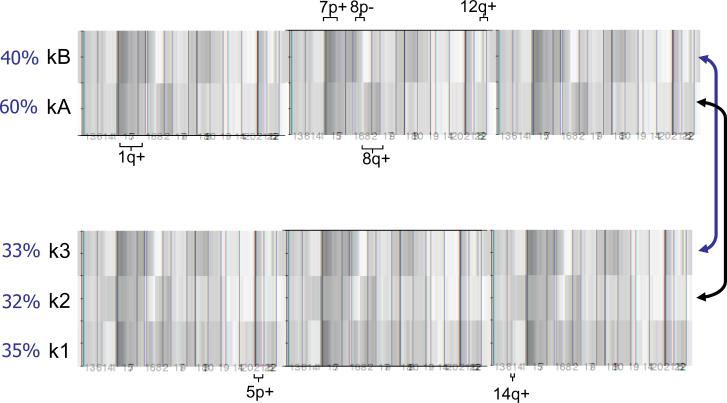
Simple aCGH recurrence plots fail to convey associations between genomic CNAs and hence are not useful in defining distinct pathways of lung adenocarcinoma pathogenesis. We therefore applied an unsupervised clustering algorithm based on non-negative matrix factorization (NMF) to extract recurrent associations between CNAs (see Methods). An analysis of cluster membership (Supplementary Figure 1) showed stable assignments to two or three clusters suggesting the existence of up to three distinct patterns of CNAs within the lung adenocarcinomas in this set. These clusters are shown in aggregate in Figure 2 (and with case-by-case data in Supplemental Figures 2 and 3). Analysis of the two-cluster classification (clusters designated kA and kB) revealed that the kA subgroup, containing about 60% of cases, was distinguished by 1q and 8q gains as well as losses at 5q and 16q. The remaining cases were in the kB subgroup and were characterized by gains of 7p (containing EGFR) and 12q (containing MDM2), and losses at 8p and 10q. In the three-cluster classification (clusters k1-3 respectively), the k2 cluster, similar to the kA cluster, was defined by gains of 1q and 8q. The k3 cluster, similar to the kB cluster, was defined by gains of 7p and 12q. A third cluster, k1, was marked by losses at 5q and 16q and gains at 5p and 14q (containing TITF1).
Figure 2.
Unsupervised clustering of genomic copy number data in 199 lung adenocarcinomas. The NMF method was used to generate K=2 and K=3 clustering, both of which showed stable cluster assignments whereas clustering into more than 3 groups did not yield stable clusters (Supplementary Figure 1). Red is gain and green is loss with the color intensity indicating the level of copy number change; white is no change. Selected notable gains and losses defining individual clusters are indicated. Approximate proportions made up by each cluster are shown as percentages. The two-cluster classification shows that the kA subgroup (n=118) is notable for 1q and 8q gains. The kB subgroup (n=81) is characterized by gains of 7p and 12q, and losses at 8p and 10q. In the three-cluster classification, the k2 cluster (n=64) was similar to the kA cluster (arrow), being defined by gains of 1q and 8q. The k3 cluster (n=65) was similar to the kB cluster (arrow), showing gains of 7p and 12q. The additional cluster k1 (n=70) was marked by gains at 5p and 14q.
To complement the above analysis, we also examined, in a simple pair-wise fashion, six of the CNA associations highlighted by the NMF clustering. This confirmed that many of the associations were statistically significant per moderate copy number thresholds (see Methods). These included associations between EGFR-containing 7p11.2 gains and 8p losses (p=0.007), gains of 1q and 8q (p=0.009), gains of 7p and 12q (p=0.004), losses at 8p and 10q (p<0.001), and gains at 5p and 14q (p=0.02). Except for the latter (Kwei et al. 2008), none of these associations have been noted previously. The co-occurrence of 7p gains and 10q losses was not significant.
The unsupervised clustering of the CNA data also showed a strong correlation with EGFR and KRAS mutation status (Supplementary Table 3). In both the two-cluster and the three-cluster classifications, EGFR mutant tumors were distributed in a highly non-random fashion, with most falling into the kB or k3 cluster, respectively (both p<0.0001), while KRAS-mutant tumors were enriched in the kA and k2 clusters (respectively, p=0.0004 and p<0.0001).
Finally, Kaplan-Meier survival analysis of these NMF clusters showed a significant survival advantage for patients whose tumors were in the EGFR-mutant-rich kB cluster in the 2-cluster separation (p=0.006) (Figure 3). In the 3-cluster separation, the EGFR-mutant-rich k3 cluster showed a survival advantage over the k1 cluster (p=0.03) while patients within the KRAS-mutant-rich k2 cluster were in an intermediate group for clinical outcome. For comparison, Kaplan-Meier survival analysis based on EGFR mutation status alone did not detect statistically significant differences in the present dataset (Supplementary Figure 4). Overall, these analyses indicate that the genetic heterogeneity of lung adenocarcinoma is not random and that coordinated genomic alterations may reflect underlying distinct oncogenic pathways with different clinical outcomes.
Figure 3.
Impact of aCGH cluster assignment on overall survival. Kaplan-Meier survival showed a survival advantage for patients whose tumors were in the EGFR-mutant-rich kB cluster (designated B) in the 2-cluster separation (p=0.006). In the 3-cluster separation, the EGFR-mutant-rich k3 cluster (designated C) showed a survival advantage over the k1 cluster (designated A) (p=0.03). Outcome differences between the KRAS-mutant-rich k2 cluster (designated B) and the other two clusters were not statistically significant.
Associations between CNAs and expression profiles
As genomic gains are expected to alter the expression of biologically relevant genes, we examined the mRNA expression profiles (based on Affymetrix U133A array hybridizations) of cases defining MCRs of gain in order to identify copy-number driven gene expression changes within or surrounding these MCRs (Supplementary Table 4). As expected, the expression profile of cases defining the MCR of gain at 7p11.2 demonstrated highly significant over-expression of EGFR. Similarly, KRAS was significantly over-expressed in cases defining the MCR of gain at 12p12. Likewise, the expression profile of cases defining the MCR of gain at 12q14 demonstrated significant over-expression of MDM2. For cases defining the MCR of gain at 5p15, SKP2 (Zhu et al. 2004), was significantly over-expressed, while TERT was not. The overexpression of SKP2 (at 5p13) reflects the fact that samples used to delineate MCRs individually have, by definition, broader regions of gain that overlap the MCR. As for TERT, other strong candidate genes within certain MCRs were not found to be significantly over-expressed by transcriptomic analysis, including MYC on 8q24 and TITF1 on 14q13 (Weir et al. 2007; Tanaka et al. 2007; Kendall et al. 2007). It should be emphasized, however, that some transcripts may not be adequately measured by microarrays for technical or biological reasons and therefore these data cannot be used in isolation to exclude driver genes. Finally, the analysis of genes significantly over-expressed in cases defining the MCR of gain at 8q24 highlighted COPS5, previously shown to drive selection for 8q amplicons along with MYC (Adler et al. 2006).
Associations between genomic copy number losses and gene expression profiles were considerably less robust, yielding few if any significant genes in comparable analyses of samples with either 9p21 or 19p13 losses (results not shown). This may be due to the narrower range of expression values seen in the context of genomic losses compared to gene amplifications.
Associations between genomic gains and activating mutations at specific genes
EGFR expression at the mRNA level correlated well with gene copy number by aCGH, as shown in an integrated representation of expression, copy number, and mutation data (Supplementary Figure 5). This analysis also reveals that EGFR-mutation is associated with generally higher levels of EGFR expression among both EGFR-amplified and non-amplified cases (Supplementary Figure 5). A similar analysis of the 12p12 MCR and KRAS mutation status demonstrated a trend for cases defining the MCR to harbor a KRAS mutation, but this did not reach statistical significance (not shown). We did not detect any relationship between LKB1 mutations and (single copy) deletions at 19p13; this may either reflect the less reliable detection of single copy losses compared to multiple copy gains in the midst of admixed non-neoplastic cells, or the reported finding that mutations and deletions at LKB1 generally do not co-occur (Ji et al. 2007). There was no evidence of genomic loss at PTEN in the 8 PTEN-mutant tumors. Finally, among associations between mutations and unrelated loci, we also found a significant co-occurrence of EGFR mutations and p16/CDKN2A deletions (p=0.007), that largely reflects their mutual association with 8p losses, as described below. This finding is also consistent with the reported loss of p14ARF expression in EGFR-mutant lung cancers (Mounawar et al. 2007).
Associations between expression profiles and mutations
Supervised analyses of Affymetrix U133A expression profiles based on EGFR, KRAS, and p53 mutation status were performed (Table 1, Supplementary Table 5). The other five mutated genes (BRAF, ERBB2, PIK3CA, LKB1, PTEN) did not yield robust profiles, but these analyses lacked power given the lower numbers of samples with mutations in these genes. Notably, the number of significantly differentially expressed genes in EGFR-mutant cases was much greater than in KRAS-mutant cases (probe sets significant at FDR <5%: 2571 and 103, respectively). This observation is all the more striking given the marginally stronger statistical power of the KRAS analysis (48 KRAS mutants vs only 43 EGFR mutants). This suggests either that the impact of KRAS mutation on gene expression is less distinctive than that of EGFR mutation, or that EGFR mutations arise in a more restricted and homogeneous cell type than KRAS mutations, or that there is biological or etiologic heterogeneity among KRAS-mutant tumors (Riely et al. 2008). The more distinctive expression profile of EGFR-mutant cases was also supported by an unsupervised clustering analysis (Supplementary Figure 6). We limit ourselves here to three notable observations. First, both the EGFR and KRAS lists contain the respective mutated genes, as measured by multiple probe sets, at or near the top (Table 1), presumably reflecting a consistent level of expression required for their oncogenic effect plus their amplification in a subset of cases. Secondly, we noted that different members of the DUSP family of MAP kinase phosphatases, known to be transcriptionally induced by MAPK signaling to provide negative feedback regulation of the same, are highly differentially expressed in EGFR- and KRAS-mutant tumors (Table 1). In the latter tumors, DUSP6 was relatively over-expressed, consistent with its previous description in other KRAS-mutant signatures (Sweet-Cordero et al. 2005), as was DUSP4 (Supplementary Table 5). In contrast, DUSP4 was the single most highly significantly under-expressed gene in EGFR-mutant tumors, an unexpected finding given that it is normally up-regulated by MAPK signaling (Owens and Keyse 2007). We follow up this observation further below. The third observation emerging from these two lists is the over-representation of genes from specific chromosomal regions, notably under-expressed genes from 8p (including DUSP4) on the EGFR-mutant list and over-expressed genes from 1q on the KRAS-mutant list (Table 1). These reflect the non-random associations of 8p loss and 1q gain with EGFR and KRAS mutations, respectively (see above). Finally, the expression profile of p53 mutation was also robust but its discussion is beyond the scope of this report (Supplementary Table 5).
Table 1.
EGFR and KRAS expression profiles. Top 25 genes shown; complete lists provided in Supplementary Table 5.
| Genes differentially expressed in EGFR mutant versus wild type tumors | Genes differentially expressed in KRAS mutant versus wild type tumors | ||||||
|---|---|---|---|---|---|---|---|
| Symbol | Cytoband | SWS | P-value | Symbol | Cytoband | SWS | P-value |
| DUSP4* | 8p12-p11 | -6.7 | 2.4E-11 | MCL1* | 1q21 | 5.1 | 3.3E-07 |
| EGFR* | 7p12 | 6.4 | 1.4E-10 | IGF2BP3* | 7p11 | -5.0 | 5.2E-07 |
| PRDM4* | 12q23-q24.1 | -6.4 | 1.8E-10 | KRAS* | 12p12.1 | 5.0 | 7.1E-07 |
| TXNRD1 | 12q23-q24.1 | -6.0 | 1.5E-09 | PRNPIP | 1p32 | -4.9 | 8.0E-07 |
| GTF2E2 | 8p21-p12 | -6.0 | 1.6E-09 | FTSJ2 | 7p22 | -4.9 | 8.3E-07 |
| GPR177 | 1p31.3 | 6.0 | 1.8E-09 | SNRPD3 | 22q11.23 | -4.8 | 2.0E-06 |
| MGC13098* | 7p13 | 6.0 | 2.0E-09 | KIAA1033 | 12q24.11 | 4.7 | 2.8E-06 |
| GALNT10* | 5q33.2 | 5.9 | 3.5E-09 | RHOB | 2p24 | 4.7 | 2.9E-06 |
| LRRC31 | 3q26.2 | 5.8 | 5.0E-09 | MED18 | 1p35.3 | -4.6 | 3.6E-06 |
| KIAA0319L | 1p34.2 | 5.8 | 7.9E-09 | PAC 886K2 | 1p36.11 | -4.5 | 7.3E-06 |
| MKL2 | 16p13.12 | 5.8 | 8.8E-09 | LANCL2 | 7q31.1-q31.33 | -4.5 | 8.1E-06 |
| RAD51L1 | 14q23-q24.2 | 5.7 | 1.2E-08 | USP2 | 11q23.3 | -4.5 | 8.4E-06 |
| KIAA0495 | 1p36.32 | 5.7 | 1.3E-08 | FOXRED2 | 22q12.3 | -4.4 | 1.3E-05 |
| RHCE | 1p36.11 | 5.7 | 1.4E-08 | RPL13A | 19q13.3 | 4.3 | 1.5E-05 |
| HIP1 | 7q11.23 | 5.6 | 1.7E-08 | SMPDL3A | 6q22.31 | 4.3 | 1.6E-05 |
| GPR172B | 17p13.2 | 5.6 | 2.2E-08 | STK24 | 13q31.2-q32.3 | 4.3 | 1.6E-05 |
| TNFRSF10B | 8p22-p21 | -5.6 | 2.5E-08 | MAFF | 22q13.1 | 4.3 | 1.7E-05 |
| C16orf58* | 16p11.2 | 5.5 | 3.2E-08 | NXPH4 | 12q13.3 | -4.3 | 1.8E-05 |
| RNF40 | 16p11.2-p11.1 | 5.5 | 3.2E-08 | C7orf30 | 7p15.3 | -4.3 | 2.0E-05 |
| MYST1 | 16p11.2 | 5.5 | 4.0E-08 | LOC81691 | 16p12.2 | -4.3 | 2.0E-05 |
| ADCY9 | 16p13.3 | 5.4 | 6.1E-08 | SNHG3-RCC1 | 1p36.1 | -4.3 | 2.0E-05 |
| LRRC47 | 1p36.32 | 5.4 | 6.8E-08 | TRMT5 | 14q23.1 | -4.3 | 2.0E-05 |
| KYNU | 2q22.2 | -5.4 | 7.5E-08 | PPIE | 1p32 | -4.2 | 2.1E-05 |
| RFK | 9q21.13 | -5.4 | 8.2E-08 | IVNS1ABP | 1q25.1-q31.1 | 4.2 | 2.2E-05 |
| HLA-DMA | 6p21.3 | 5.3 | 9.7E-08 | DUSP6 | 12q22-q23 | 4.2 | 2.3E-05 |
Note: for genes that were represented on these lists of the top 25 genes by more than one Affymetrix probe set (indicated by *), only the top scoring probe set is shown; SWS = stratified Wilcoxon statistic with the sign of the statistic indicating either over- or under-expression; two-sided p-values calculated from a normal distribution; PAC886K2 indicates genomic clone containing EST corresponding to Affymetrix probe set 213685_at.
Loss of DUSP4 at 8p in EGFR mutant tumors
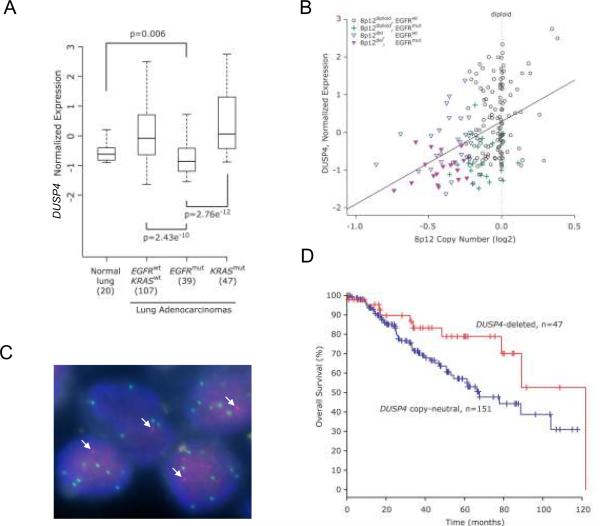
As demonstrated above, DUSP4, a dual specificity MAP kinase phosphatase (a.k.a. MKP-2), was almost uniformly under-expressed in EGFR mutated cases relative to lung adenocarcinomas lacking EGFR mutations (Table 1). A more detailed comparison of DUSP4 transcript levels showed that expression was significantly lower in EGFR-mutant lung adenocarcinomas than in normal lung, KRAS-mutant tumors, and tumors lacking both mutations (Figure 4A). A substantial proportion of EGFR mutant tumors demonstrated single copy genomic loss at 8p12 that included DUSP4. This proportion ranged from 35% to 67% of EGFR-mutant cases depending on the stringency of the sample-specific thresholds used to consider DUSP4 deleted. Conversely, a substantial proportion of DUSP4-deleted tumors contained EGFR mutations, ranging from 41% to 56%, again depending on the aforementioned stringency. Regardless of the threshold used to score deletions, the association of DUSP4 genomic loss and EGFR mutations was highly significant (p<0.0001).
Figure 4.
A. Comparison of DUSP4 levels in normal non-neoplastic lung tissue samples, and lung adenocarcinomas with different EGFR and KRAS mutation status. DUSP4 expression is significantly lower in EGFR-mutant lung adenocarcinomas than in normal lung tissue or other subsets of lung adenocarcinoma. P-values are from two-sided Wilcoxon test. B. DUSP4 copy number versus expression, annotated for EGFR mutation status. Samples with alterations are as indicated in the legend. The correlation of copy number to expression for the DUSP4 locus is highly significant (p<0.0001 and R=0.43, Pearson's product moment correlation). EGFR mutant samples generally had lower DUSP4 expression for a given DUSP4 copy number, compared to samples lacking EGFR mutation. C. DUSP4 / EGFR dual color FISH. DUSP4 signals are red, EGFR in green. Arrows indicate the single DUSP4 signal in each nucleus; there are multiple EGFR signals per nucleus in this EGFR-mutant lung adenocarcinoma. D. DUSP4 loss defines a favorable clinical subset of lung adenocarcinoma. The p=0.042 for difference between survival curves; p=0.031 from univariate Cox proportional hazards regression; hazard ratio of 2.06 (95% confidence interval: 1.02-4.20). Data are from 198 samples with complete data for both variables.
Integration of gene expression with copy number and EGFR mutation status showed that, while low DUSP4 transcript levels correlated well with DUSP4 genomic loss, this relationship was most evident among EGFR-mutant tumors (Figure 4B). We also confirmed by dual-color fluorescence in situ hybridization the co-occurrence of DUSP4 single-copy loss and EGFR amplification in the same tumor cells in EGFR-mutant cases that had both findings by aCGH (Figure 4C). Notably, DUSP4 does not reside in a narrow MCR of loss and therefore would not have emerged as a strong candidate based on algorithms using copy number data alone, but is the top candidate based on the expression profile of EGFR mutant tumors (Table 1), highlighting the value of an integrated genomics approach. Finally, we also screened DUSP4 for somatic mutations. A total of 101 lung adenocarcinoma samples (99 tumor DNAs and cell lines H1299 and H522) were sequenced but no somatic mutations of DUSP4 were identified.
Given the significant co-occurrence of EGFR mutations and p16/CDKN2A deletions (p=0.007) described above, we examined their relationship with DUSP4 deletions. This showed that p16/CDKN2A deletions and DUSP4 deletions are strongly correlated (p=0.0001) (Supplementary Figure 7). Indeed, among DUSP4-diploid tumors, EGFR mutations and p16/CDKN2A deletions were not associated, indicating that their co-occurrence is largely secondary to their mutual association with DUSP4 deletions.
A distinct clinical subset of lung adenocarcinoma defined by DUSP4 loss
Patients with tumors harboring DUSP4 deletion had a better overall survival than patients whose tumors did not show this alteration (p=0.042 for difference between survival curves; p=0.031 from univariate Cox proportional hazards regression; hazard ratio of 2.06) (Figure 4D). Although DUSP4 loss associates with EGFR mutations, it is notable that EGFR mutation status alone had only a sub-significant effect on overall survival in the present set of patients (p=0.18 for difference between survival curves; p=0.159 from univariate Cox proportional hazards regression; hazard ratio of 1.66) (Supplementary Figure 4).
Preferential suppression of EGFR-mutant lung adenocarcinoma growth by DUSP4
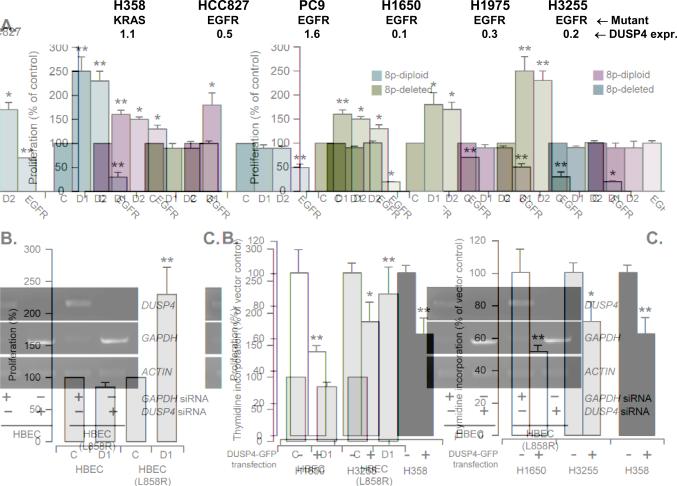
This candidacy of DUSP4 as the prime driver gene for 8p loss in EGFR mutant tumors is biologically plausible given its known transcriptional up-regulation by, and negative feedback regulation of, MAPK signaling (Owens and Keyse 2007). By microarray analysis, DUSP4 expression is decreased following inhibition of mutant EGFR in the H1975 lung adenocarcinoma cell line (Kobayashi et al. 2006). We have also found that DUSP4 is transcriptionally up-regulated by mutant EGFR signaling in HBECs (Supplementary Figure 8). Thus, we hypothesized that the oncogenicity of mutant EGFR may be enhanced by loss or attenuation of the negative autoregulatory loop normally provided by DUSP4. We therefore examined the impact of DUSP4 on the growth of lung adenocarcinoma cells. First, we used RNA interference by short interfering RNAs (siRNA) to study the effects of reducing DUSP4 levels in 6 lung adenocarcinoma cell lines with available data on DUSP4 genomic copy number [data from ref. (Garnis et al. 2006); Affymetrix SNP array data, K. Michel & R. Thomas, unpublished; Agilent 244K aCGH data, J. Bean & W. Pao, unpublished] and DUSP4 transcript levels relative to HBECs (Figure 5A). In the 3 lung adenocarcinoma cell lines with moderate to high DUSP4 expression, all diploid for 8p, namely PC9, HCC827, and H358, DUSP4 knockdown enhanced growth significantly at 48 hours (Figure 5A). In contrast, in the 3 lung adenocarcinoma cell lines with already low DUSP4 expression, associated in 2/3 lines with 8p single copy deletion (H1650 and H3255), DUSP4 knockdown had essentially no effect. To confirm these findings in an isogenic background, we examined the effect of DUSP4 knockdown in HBECs with or without EGFR L858R (Figure 5B). This showed that reducing DUSP4 levels enhances growth in the presence of EGFR L858R but not in the parental line.
Figure 5.
A. DUSP4 knockdown enhances growth of lung adenocarcinoma cell lines. In 3/3 cell lines with moderate to high DUSP4 expression, DUSP4 knockdown by two different siRNAs (D1, D2) resulted in growth enhancement (all p<0.05 except H1975) relative to control siRNA (C). In contrast, in 3/3 EGFR-mutant cell lines with low baseline DUSP4 expression, DUSP4 knockdown had no significant effect. For comparison, EGFR knockdown reduced the growth of 4/5 EGFR-mutant lines. EGFR knockdown did not suppress growth of the KRAS-mutant H358 line. Thymidine incorporation was measured 48-72 hours after transfection. DUSP4 expression was measured by Q-RT-PCR, normalized to TBP expression and expressed as the ratio of the cell line expression to that of HBECs. B. DUSP4 knockdown preferentially enhances growth of human bronchial epithelial cells (HBEC) in the presence of mutant EGFR. To confirm the findings shown in A. in an isogenic background, we examined the effect of DUSP4 knockdown in HBECs stably transfected with EGFR L858R. Reducing DUSP4 levels (using D1 siRNA as in A.) enhances growth compared to control siRNA (C) in the presence of EGFR L858R (p<0.05) but not in the parental line (thymidine incorporation measured 48-72 hours after transfection). DUSP4 knockdown was confirmed by RT-PCR as shown in the adjacent agarose gel image. C. DUSP4 cDNA transfection inhibits cell growth of lung adenocarcinoma cell lines. Thymidine incorporation was measured 24 hours after DUSP4 cDNA transfection. Transfection efficiencies, estimated by FACS analysis of GFP-positive cells after 24 hours of transfection, were 54-60% for H1650 cells, 20-44% for H3255 cells, and 46% for H358 cells. V: vector-transfected cells; D: DUSP4-GFP cDNA transfected cells. For all three figure parts, significant differences relative to control are shown by asterisks if p<0.05 (*) or p<0.01 (**) (t-test, 2 tails, equal variance analysis).
Next, we examined the growth effects of re-expressing or increasing the level of DUSP4 by transfection with a DUSP4-GFP expression plasmid. The expression and appropriate subcellular localization of the fusion protein were confirmed respectively by western blotting using a GFP antibody (Supplementary Figure 9A) and fluorescence microscopy, the latter showing the expected nuclear localization of DUSP4 (Supplementary Figure 9B). In H1650 and H3255, both characterized by EGFR mutation, 8p loss (J. Bean & W. Pao, unpublished data), and low DUSP4 expression, as well as in H358, a KRAS-mutant lung adenocarcinoma line with moderate DUSP4 expression, transient transfection with the DUSP4-GFP expression plasmid resulted in a significant reduction in growth (Figure 5C). Interestingly, attempts to derive corresponding stable transfectants (in H1650) were unsuccessful because of loss of GFP-positive cells after one week of antibiotic selection in the DUSP4-GFP-transfected cultures (but not in the GFP-transfected ones) (Supplementary Figure 10), an observation consistent with the growth inhibition observed in the above transient transfection experiments. Similar difficulties in isolating stable DUSP4 cDNA transfectants have been observed in other settings (Tresini et al. 2007). Overall, these data provide functional validation of the growth suppressive effects of DUSP4 in lung adenocarcinoma lines with activating mutations of kinase signaling pathways.
Discussion
Several levels of data here support the link between DUSP4 loss and EGFR-mutant tumors. Our analysis of the aCGH data showed that lung adenocarcinomas display non-random patterns of co-occurring gains and losses, one of which is characterized by 7p gains (including the EGFR locus) and 8p losses. These tumors frequently showed EGFR mutation (p<10-4), which is not unexpected as mutant EGFR alleles are known to undergo selective amplification (Takano et al. 2005). However, the 8p losses were broad and MCR analysis did not yield a candidate sub-region on this chromosome arm. Previous studies of lung cancers have noted 8p losses, but also failed to narrow the putative target region (Weir et al. 2007). Allelic losses on 8p are well described in other carcinomas, including breast, prostate, and bladder, with most studies finding a complex pattern that cannot be reduced to a single minimally deleted region [reviewed in (Adams et al. 2005)]. Notably, DUSP4 has also been proposed as a driver of 8p losses in breast cancer (Armes et al. 2004). Our integrated genomics strategy showed that DUSP4, at 8p12, was the most consistently under-expressed gene in EGFR mutant cases compared to EGFR wild type cases (nominal p<10-9, two-sided stratified Wilcoxon). By aCGH, using a moderate stringency threshold, the DUSP4 region showed evidence of single-copy genomic loss in approximately 24% of lung adenocarcinomas, including approximately 45% of EGFR mutant cases (p<10-4), while the latter accounted for only 21% of cases in our dataset. No somatic mutations in DUSP4 were detected, suggesting haploinsuffiency as the basic alteration, an oncogenic mechanism recently illustrated by RPS14 loss in the 5q- myelodysplastic syndrome (Ebert et al. 2008). We also show that re-expression of DUSP4 in EGFR-mutant lung adenocarcinoma lines with 8p loss and low endogenous DUSP4 results in reduced growth, and conversely, knockdown of DUSP4 in cell lines with high DUSP4 leads to enhanced growth. Since DUSPs are known to be transcriptionally up-regulated by MAPK signaling as a negative feedback mechanism, the data support the hypothesis that DUSP4 loss cooperates with EGFR mutation to allow full oncogenic activation of the MAPK pathway. Clinically, DUSP4 loss has a significant impact on overall survival, further supporting its biological significance in lung adenocarcinoma.
MAPK pathway activation by signaling through growth factor receptors is modulated by negative feedback inhibition at the receptor (O'Reilly et al. 2006), at the level of RAS (e.g. by sprouty proteins) (Shaw et al. 2007), and at the level of ERK (by DUSPs). DUSP4 functions in the nucleus to dephosphorylate and thereby inactivate the ERK, JNK, and p38 MAP kinases (Owens and Keyse 2007). In addition to its role in regulating MAPK-mediated mitogenic signals, DUSP4 has also been implicated in the control of replicative senescence and of p53-mediated apoptosis (Tresini et al. 2007; Shen et al. 2006). DUSP4 is among several DUSPs well-described as transcriptional targets of MAPK signaling (Schulze et al. 2004; Amit et al. 2007). Suppression of mutant EGFR signaling in the H1975 lung adenocarcinoma cell line causes down-regulation of DUSP4 (Kobayashi et al. 2006). More broadly, there is a growing recognition that disruption of negative feedback control of MAPK signaling is an important component of oncogenic kinase signaling (Amit et al. 2007). Finally, our finding that EGFR mutations and p16/CDKN2A deletions are linked through their mutual association with DUSP4 losses suggests that the deregulated mitogenic signaling that occurs in the context of combined DUSP4 loss and EGFR mutation (or possibly other mutations activating the MAPK pathway) may drive selection for loss of the locus encoding p16CDKN2A and p14ARF, known mediators of senescence or apoptosis in response to inappropriate mitogenic signals (Lowe and Sherr 2003; Michaloglou et al. 2005). This recalls the relationship between p16CDKN2A deletions and BRAF and EGFR mutants in melanoma and glioma, respectively (Michaloglou et al. 2005; Ohgaki and Kleihues 2007). Finally, as mutant EGFR and p16 bypass together fail to fully transform HBECs (Sato et al. 2006), the addition of DUSP4 knockdown to such experiments will be of interest.
Although the association of EGFR mutation status with DUSP4 genomic loss and under-expression was striking, the genomic losses on 8p were consistently broad, suggesting the possibility of more than one driver gene. We note two other regions of interest on 8p, the TNFRSF10 TRAIL receptor gene cluster at 8p21.3, similarly implicated by our data on the basis of decreased expression in EGFR-mutant tumors (Table 1) and previously proposed as a tumor suppressor locus in lung cancers (Lee et al. 1999), and a slightly more telomeric gene on 8p21.3, DOK2, encoding an adaptor protein that suppresses KRAS activation. Evidence for DOK2 as another lung adenocarcinoma tumor suppressor on 8p, based on a mouse knockout model that leads to the development of lung adenocarcinomas and also in part on the present integrated dataset, is presented elsewhere (Niki et al. 2007). All 27 stringently detected DUSP4-deleted tumors (including the 15 EGFR-mutant cases) also showed loss of the TNFRSF10 gene cluster and DOK2. Five additional cases showed losses of these two gene loci without DUSP4 deletion; interestingly, none of these 5 samples showed EGFR mutations and 4 were instead KRAS-mutant. This difference between DUSP4/DOK2-codeleted and DOK2-deleted/DUSP4-intact cases was significant (p=0.046), suggesting that lung adenocarcinomas harbor two types of 8p deletions, with those that extend more centromerically to include DUSP4 being more strongly associated with EGFR mutations. Indeed, a mapping of 8p losses in relation to EGFR or KRAS mutation status strengthens the notion of mutation type-specific patterns of 8p loss (Figure 6). The observation that 8p losses in lung adenocarcinomas are consistently broad may reflect a selective advantage provided by the reduced function of more than one gene (here two different negative regulators of kinase signaling), a multiple driver gene model that may also hold for other regions of broad gains or losses.
Figure 6.
8p losses stratified by mutation group. The frequency of deletion on 8p in the region from 20 to 32 megabases (blue, left-axis) is plotted by chromosomal position, along with the position and direction of transcription for three gene loci of interest: DUSP4, DOK2, and the TNF receptor cluster. The frequency of mutation in EGFR and KRAS (right-axis) is plotted by chromosome 8p position, for samples showing 8p losses at a given point in 8p. This highlights the higher frequency of EGFR mutations among cases with 8p losses overall and shows that this association is strongest in a region encompassing DUSP4. In contrast, KRAS mutations are less common in 8p-deleted cases and when present, appear to preferentially affect cases whose 8p losses include DOK2 and the TNF receptor cluster.
Several other previously unrecognized or underappreciated associations emerged from this integrated genomic analysis. We found a significant inverse relationship between mutations in EGFR and LKB1, strengthening observations from others (Matsumoto et al. 2007; Ding et al. 2008). LKB1 cooperates with mutant KRAS in mouse lung tumorigenesis (Ji et al. 2007). Other associations that may point to distinct oncogenic pathways include coamplification of 1q21-23 and 8q24 (including MYC) (p=0.009), and coordinate gains at 5p15 (including TERT) and 14q13 (including TITF1) (p=0.02).
We have described here an initial analysis of the largest integrated genomic study of lung adenocarcinoma assembled to date. The identification of DUSP4 as a novel growth suppressor in lung adenocarcinoma exemplifies the type of emergent observation made possible by the integration of multiple levels of genomic data in large, well annotated datasets. The present associations between mutation status, copy number changes and expression data strengthen the notion that there exist at least 2 or 3 distinct, recurrent oncogenic pathways that drive lung adenocarcinoma. Similar insights are now emerging in other cancers based on the systematic integration of multiple levels of genomic data (The Cancer Genome Atlas Research Network 2008; Parsons et al. 2008).
Methods
See Supplementary information for complete methods.
Supplementary Material
Acknowledgements
We thank Dr Agnes Viale and the MSKCC Genomics Core Laboratory personnel for generation of microarray data; Dr Laetitia Borsu for assistance with Sequenom assays; Drs Hakim Djaballah and Gabriela E. Sanchez of the MSKCC HTS Core Facility for providing GFP siRNAs; Louis Vargas, Yonghong Xiao, and the MSKCC Pathology Core personnel for technical assistance; Drs William Travis, Andre Moreira, and David Klimstra for providing pathologic diagnoses; Drs John Minna and Adi Gazdar for providing the human bronchial epithelial cell (HBEC) lines; Drs Alice T. Hawley, Marasu Niki, and Pier Paolo Pandolfi for assistance with reagents and related studies; and Dr Harold Varmus for support (to R.S.). Barry S. Taylor is a graduate student in the Department of Physiology and Biophysics, Weill Cornell Graduate School of Medical Sciences. We acknowledge the support of the National Cancer Institute (U01-CA84999 to W.G., P01-CA129243 to M.L.), the Doris Duke Charitable Foundation (to W.P.), the Long Island League to Abolish Cancer (to W.P.), the Labrecque Foundation (to W.G.), and the Lung Cancer Research Foundation (to M.L.). The MSKCC Sequenom facility is supported by the Anbinder Fund.
References
- Adams J, Cuthbert-Heavens D, Bass S, Knowles MA. Infrequent mutation of TRAIL receptor 2 (TRAIL-R2/DR5) in transitional cell carcinoma of the bladder with 8p21 loss of heterozygosity. Cancer Lett. 2005;220:137–144. doi: 10.1016/j.canlet.2004.06.052. [DOI] [PubMed] [Google Scholar]
- Adler AS, Lin M, Horlings H, Nuyten DS, van de Vijver MJ, Chang HY. Genetic regulators of large-scale transcriptional signatures in cancer. Nat Genet. 2006;38:421–430. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, et al. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- Armes JE, Hammet F, de SM, Ciciulla J, Ramus SJ, Soo WK, et al. Candidate tumor-suppressor genes on chromosome arm 8p in early-onset and high-grade breast cancers. Oncogene. 2004;23:5697–5702. doi: 10.1038/sj.onc.1207740. [DOI] [PubMed] [Google Scholar]
- Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Sunamura M, Motoi F, Matsuno S, Horii A. Potential tumor suppressive pathway involving DUSP6/MKP-3 in pancreatic cancer. Am J Pathol. 2003;162:1807–1815. doi: 10.1016/S0002-9440(10)64315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnis C, Lockwood WW, Vucic E, Ge Y, Girard L, Minna JD, et al. High resolution analysis of non-small cell lung cancer cell lines by whole genome tiling path array CGH. Int J Cancer. 2006;118:1556–1564. doi: 10.1002/ijc.21491. [DOI] [PubMed] [Google Scholar]
- Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci U S A. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Shimamura T, Monti S, Steidl U, Hetherington CJ, Lowell AM, et al. Transcriptional profiling identifies cyclin D1 as a critical downstream effector of mutant epidermal growth factor receptor signaling. Cancer Res. 2006;66:11389–11398. doi: 10.1158/0008-5472.CAN-06-2318. [DOI] [PubMed] [Google Scholar]
- Kwei KA, Kim YH, Girard L, Kao J, Pacyna-Gengelbach M, Salari K, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27:3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Shin MS, Kim HS, Lee HK, Park WS, Kim SY, et al. Alterations of the DR5/TRAIL receptor 2 gene in non-small cell lung cancers. Cancer Res. 1999;59:5683–5686. [PubMed] [Google Scholar]
- Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Marks JL, Broderick S, Zhou Q, Chitale DA, Li AR, Zakowski M, et al. Prognostic implications of EGFR and KRAS mutations in resected lung adenocarcinoma. J Thoracic Oncology. 2008;3:111–116. doi: 10.1097/JTO.0b013e318160c607. [DOI] [PubMed] [Google Scholar]
- Marks JL, McLellan MD, Zakowski MF, Lash AE, Kasai Y, Broderick S, et al. Mutational analysis of EGFR and related signaling pathway genes in lung adenocarcinomas identifies a novel somatic kinase domain mutation in FGFR4. PLoS ONE. 2007;2 doi: 10.1371/journal.pone.0000426. e426.: e426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Iwakawa R, Takahashi K, Kohno T, Nakanishi Y, Matsuno Y, et al. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene. 2007;26:5911–5918. doi: 10.1038/sj.onc.1210418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Franklin WA, Kelley MJ. Molecular classification and molecular genetics of human lung cancers. Semin Oncol. 2004;31:4–19. doi: 10.1053/j.seminoncol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Mounawar M, Mukeria A, Le CF, Hung RJ, Renard H, Cortot A, et al. Patterns of EGFR, HER2, TP53, and KRAS mutations of p14arf expression in non-small cell lung cancers in relation to smoking history. Cancer Res. 2007;67:5667–5672. doi: 10.1158/0008-5472.CAN-06-4229. [DOI] [PubMed] [Google Scholar]
- Niki M, Hawley AT, Chitale DA, Morotti A, Janas JA, Brennan C, et al. Identification of DOK family proteins as Lung Tumour Suppressors. Cell. 2007 (submitted) [Google Scholar]
- O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene. 2007;26:3203–3213. doi: 10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely GJ, Kris MG, Rosenbaum D, Marks J, Li AR, Chitale DA, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Sato M, Vaughan MB, Girard L, Peyton M, Lee W, Shames DS, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- Schulze A, Nicke B, Warne PH, Tomlinson S, Downward J. The transcriptional response to Raf activation is almost completely dependent on Mitogen-activated Protein Kinase Kinase activity and shows a major autocrine component. Mol Biol Cell. 2004;15:3450–3463. doi: 10.1091/mbc.E03-11-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- Shaw AT, Meissner A, Dowdle JA, Crowley D, Magendantz M, Ouyang C, et al. Sprouty-2 regulates oncogenic K-ras in lung development and tumorigenesis. Genes Dev. 2007;21:694–707. doi: 10.1101/gad.1526207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WH, Wang J, Wu J, Zhurkin VB, Yin Y. Mitogen-activated protein kinase phosphatase 2: a novel transcription target of p53 in apoptosis. Cancer Res. 2006;66:6033–6039. doi: 10.1158/0008-5472.CAN-05-3878. [DOI] [PubMed] [Google Scholar]
- Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- Takano T, Ohe Y, Sakamoto H, Tsuta K, Matsuno Y, Tateishi U, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:6829–6837. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresini M, Lorenzini A, Torres C, Cristofalo VJ. Modulation of replicative senescence of diploid human cells by nuclear ERK signaling. J Biol Chem. 2007;282:4136–4151. doi: 10.1074/jbc.M604955200. [DOI] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CQ, Blackhall FH, Pintilie M, Iyengar P, Liu N, Ho J, et al. Skp2 gene copy number aberrations are common in non-small cell lung carcinoma, and its overexpression in tumors with ras mutation is a poor prognostic marker. Clin Cancer Res. 2004;10:1984–1991. doi: 10.1158/1078-0432.ccr-03-0470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.