Abstract
Cardiac sodium channels are responsible for conduction in the normal and diseased heart. We aimed to investigate regional and transmural distribution of sodium channel expression and function in the myocardium. Sodium channel Scn5a mRNA and Nav1.5 protein distribution was investigated in adult and embryonic mouse heart through immunohistochemistry and in situ hybridization. Functional sodium channel availability in subepicardial and subendocardial myocytes was assessed using patch-clamp technique. Adult and embryonic (ED14.5) mouse heart sections showed low expression of Nav1.5 in the HCN4-positive sinoatrial and atrioventricular nodes. In contrast, high expression levels of Nav1.5 were observed in the HCN4-positive and Cx43-negative AV or His bundle, bundle branches and Purkinje fibers. In both ventricles, a transmural gradient was observed, with a low Nav1.5 labeling intensity in the subepicardium as compared to the subendocardium. Similar Scn5a mRNA expression patterns were observed on in situ hybridization of embryonic and adult tissue. Maximal action potential upstroke velocity was significantly lower in subepicardial myocytes (mean ± SEM 309 ± 32 V/s; n = 14) compared to subendocardial myocytes (394 ± 32 V/s; n = 11; P < 0.05), indicating decreased sodium channel availability in subepicardium compared to subendocardium. Scn5a and Nav1.5 show heterogeneous distribution patterns within the cardiac conduction system and across the ventricular wall. This differential distribution of the cardiac sodium channel may have profound consequences for conduction disease phenotypes and arrhythmogenesis in the setting of sodium channel disease.
Electronic supplementary material
The online version of this article (doi:10.1007/s00395-009-0012-8) contains supplementary material, which is available to authorized users.
Keywords: Cardiac sodium channel, Conduction, Cardiac conduction system, Immunohistochemistry, Patch clamp
Introduction
The cardiac voltage-gated sodium channel is responsible for the fast upstroke of the action potential and is essential for proper conduction of the cardiac electrical impulse. Knowledge of its differential cardiac distribution is essential when investigating its role in the normal and diseased heart. During common pathological conditions such as myocardial ischemia and heart failure, decreased sodium current function may cause conduction disturbances and potentially life-threatening arrhythmias [9, 23, 41, 45]. In addition, mutations in the SCN5A gene encoding the pore-forming α-subunit of the cardiac sodium channel (Nav1.5) have been shown to underlie multiple inherited arrhythmia syndromes, including long QT syndrome (type 3, LQT3), Brugada syndrome, conduction disease, sinus node dysfunction and atrial standstill [24]. Multiple biophysical effects of SCN5A mutations are considered to underlie the broad spectrum of disease symptoms observed in inherited sodium channelopathies [35].
To fully appreciate the biophysical effects of the cardiac sodium channel and the observed diversity in phenotypic expression observed in sodium channel disease, knowledge of the regional and transmural distribution of Nav1.5 is essential. While transmural differences in action potential repolarization have previously been studied in detail, information regarding heterogeneous depolarization across the ventricular wall is rather limited [1, 12]. Inhomogeneous sodium channel expression may have profound functional consequences, including transmural heterogeneity in conduction velocity, potentially contributing to arrhythmogenesis [12, 44, 46]. In addition, sodium channel distribution in different regions of the conduction system may influence heart rate and atrioventricular and intraventricular conduction [5, 7, 30, 43].
To date, a number of studies have investigated Nav1.5 distribution in (parts of) the conduction system, but results have been contradictory [17, 33, 47]. Other studies have been performed in enzymatically isolated cardiomyocytes, allowing for only limited interpretation of the relevance of these channels for overall cardiac conduction and function [25, 29, 31]. In the present study, we performed a comprehensive analysis of regional and transmural distribution of Scn5a mRNA and Nav1.5 protein expression in the murine heart, combining immunohistochemistry and in situ hybridization. Our results indicate that Scn5a and Nav1.5 show heterogeneous distribution patterns within the cardiac conduction system and across the ventricular wall. This data thus provides insight into the role of the cardiac sodium channel in normal cardiac electrophysiology and enables increased understanding of the consequences of sodium channel dysfunction for conduction disease phenotypes and arrhythmogenesis.
Methods
Animal experiments were conducted in compliance with the “Guiding Principles in the Use and Care of Animals” published by the National Institutes of Health (NIH publication No. 85–23, revised 1996) and performed in accordance with institutional guidelines for animal use in research.
Antibodies
The following primary antibodies were used: rabbit polyclonal anti-Nav1.5 (1:200, Alomone Laboratories, ASC-005), mouse monoclonal anti-alpha-actinin (1:1000, Sigma), mouse monoclonal anti-desmin (1:200, Monosan, MON-3001), mouse monoclonal anti-Cx43 (1:200, BD Biosciences, 610061), rabbit polyclonal anti-Cx40 (1:250, Chemicon), rabbit polyclonal anti-HCN4 (1:200, Chemicon, AB5808), and rabbit polyclonal anti-cardiac troponin I (anti-cTnI, 1:1000, HytestLtd). Alexa conjugated goat anti-mouse and goat anti-rabbit secondary antibodies were used (1:250, Molecular Probes, Invitrogen). Nuclei were stained using Sytox Green (1:30,000, Molecular Probes).
Immunocytochemistry in transfected HEK293 cells
The human sodium channel α-subunit SCN5A with GFP fused at the carboxyl end was transfected in HEK293 cells using Lipofectamine Reagent (Gibco BRL), according to manufacturer’s instructions. Transfected HEK293 cells were cultured, fixed, and stained with anti-Nav1.5 antibody as described in the “Data supplement”.
Western blot analysis
Total membrane protein fractions were isolated from HEK293 cells transfected with the β1-subunit in the presence or absence of SCN5A. Solubilized membrane protein (50 μg) was blotted to a Hybond transfer membrane (GE Healthcare, Amersham) and incubated with anti-Nav1.5 primary antibody (see “Data supplement”). Protein was visualized using the ECL Plus Western blotting detection kit (GE Healthcare, Amersham).
Immunohistochemistry on mouse heart sections
Adult male FVB/N mice (age 2–3 months; n = 5 hearts) were anesthetized by an intraperitoneal injection of pentobarbital, after which the heart was excised, snap-frozen in liquid nitrogen and stored at −80°C. Four-chamber view cryosections (7 μm) were mounted on 3-aminopropyltriethoxysilane (AAS) coated glass slides. Mouse embryos at embryonic day 14.5 (ED14.5; n = 4 embryos) were snap-frozen in liquid nitrogen and stored at −80°C, and cryosectioned (10 μm). Sections were permeabilized in 0.2% Triton X-100 in PBS for 1 h, whereas PFA sections were boiled for 10 min in Antigen Unmasking Solution. All sections were blocked in 2% BSA for 30 min and incubated with primary (overnight) and secondary antibody (90 min) in 10% NGS (all at RT). Confocal imaging was performed using a confocal laser scanning microscope (BioRad MRC1024) equipped with a 15 mV Krypton/Argon laser, using the 568 and 488 excitation lines and 605DF32 and 522DF35 emission filters.
In situ hybridization
Embryos (ED14.5; n = 3 embryos) and hearts from adult mice (age 2–3 months; n = 2 hearts) were fixed for 4 h to overnight in freshly prepared 4% paraformaldehyde (PFA) in PBS, dehydrated in a graded ethanol series, paraffin-embedded, sectioned, and mounted on AAS-coated slides. RNA probes complementary to the mRNAs for Scn5a, Cx43, cardiac troponin I (cTnI) and the homeodomain transcription factor Irx5 were labeled with digoxigenin-UTP, hybridized to the sections overnight and immunologically detected (for details, see “Data supplement”).
Action potential measurements in isolated myocytes
Thin slices (approximately 0.5 mm) were removed from the endocardial and epicardial surface of the left ventricular free wall, and single ventricular myocytes from subendocardial and subepicardial origin were enzymatically dissociated as previously described [34]. Action potentials were recorded at 37°C at a stimulation frequency of 2 Hz using the perforated patch-clamp technique. Tyrode’s solution (37°C) contained (in mmol/L): 140 NaCl, 5.4 KCl, 1.8 CaCl2, 1 MgCl2, 5.5 glucose, 5 HEPES; pH 7.4 (NaOH). Patch pipettes (~1.5 MΩ) contained (in mmol/L): 125 K-gluc, 20 KCl, 5 NaCl, 0.22 amphotericin-B, 10 HEPES; pH 7.2 (KOH). Details on action potential generation, acquisition and filtering are described elsewhere [34]. To characterize action potentials, we analyzed resting membrane potential (RMP), action potential amplitude (APA), action potential plateau phase 20 ms after the upstroke, action potential duration at 90% repolarization (APD90), and maximal action potential upstroke velocity (dV/dtmax). dV/dtmax is considered a suitable index for functional sodium current availability. Although it slightly overestimates sodium channel availability at low temperatures (7–10°C), the discrepancies between sodium current and dV/dtmax are reduced at higher temperatures (26–27°C) [37] and are practically absent at physiological temperature [6]. Action potentials were measured from both subepicardial and subendocardial cells of four hearts, and parameters from ten consecutive action potentials were averaged.
Statistical analysis
Data are presented as mean ± SEM. Differences between groups were analyzed by unpaired Student’s t test. The level of statistical significance was set to P < 0.05.
Results
Anti-Nav1.5 antibody selectivity in transfected HEK293 cells
Details on the polyclonal anti-Nav1.5 antibody are shown in Supplemental Fig. 1. When compared to other murine sodium channel α-subunits, no relevant homology with the epitope recognized by this antibody (maximal 9/19 residues identical) was observed (Supplemental Fig. 1a). On Western blot analysis, HEK cells transfected with SCN5A showed a specific band at the approximate expected size (Supplemental Fig. 1b). Immunocytochemistry of HEK293 cells transfected with cDNA for SCN5A with GFP fused at the carboxyl end showed a high degree of overlap between the signal detected by the anti-Nav1.5 antibody and the GFP signal (Supplemental Fig. 1c). Thus, we considered this anti-Nav1.5 antibody selective for the cardiac sodium channel and appropriate for further immunohistochemical analysis.
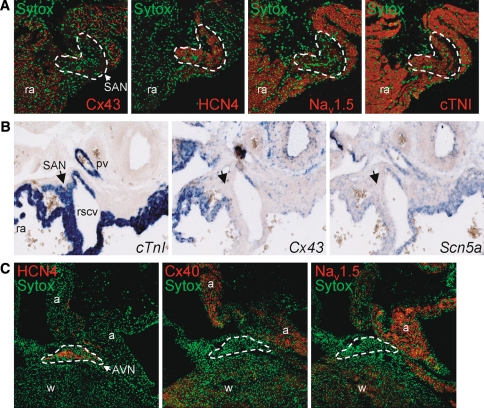
Fig. 1.
a Immunohistochemical analysis of cryosections from adult murine heart showing a detailed view of the sinoatrial node (SAN; marked region). The SAN is clearly demarcated by the presence of HCN4 staining and the absence of Cx43 labeling (middle panel) and displays low levels of Nav1.5 staining. In contrast, abundant Nav1.5 staining is visible in the surrounding right atrial (ra) tissue. b In situ hybridization of embryonic heart (ED14.5) showing mRNA expression of Scn5a, CtnI and Cx43 (in blue). The CtnI-positive and Cx43-negative SAN (denoted by arrow) shows low Scn5a mRNA expression (pv pulmonary vein, rscv right superior caval vein). c Immunohistochemical analysis of cryosections from adult murine heart showing a detailed view of the atrioventricular node (AVN; marked region). The AVN shows intense staining for HCN4, but low expression of both Cx40 and Nav1.5, whereas clear Nav1.5 staining is visible in the atria (a) and working myocardium (w)
Anti-Nav1.5 antibody selectivity in cardiac tissue
To further validate the anti-Nav1.5 antibody, we next studied its expression in the sinoatrial node (SAN), which is generally considered to contain a limited number of Nav1.5 sodium channels [22, 30]. Since double staining of both polyclonal antibodies anti-HCN4 and anti-Nav1.5 was not possible, the distribution of these proteins within the SAN was examined on adjacent serial tissue sections. Nav1.5 staining was found to be very low in the HCN4-positive SAN region, whereas clear anti-Nav1.5 staining was observed in the surrounding atrial myocardium (Fig. 1a). This HCN4-positive region was devoid of the gap junction protein connexin43 (Cx43), as expected for the SAN. These immunohistochemical results were confirmed by in situ hybridization experiments on embryonic mouse tissue. Fig. 1b shows the region of the SAN staining positive for cardiac troponin I (cTnI) mRNA, which displayed limited Cx43 and Scn5a mRNA expression. Furthermore, Nav1.5 staining was also low in the HCN4-positive and Cx40-negative atrioventricular node (AVN; Fig. 1c). These findings further underscore the selectivity and suitability of the anti-Nav1.5 antibody.
Sodium channel distribution in the AV bundle and His-Purkinje system
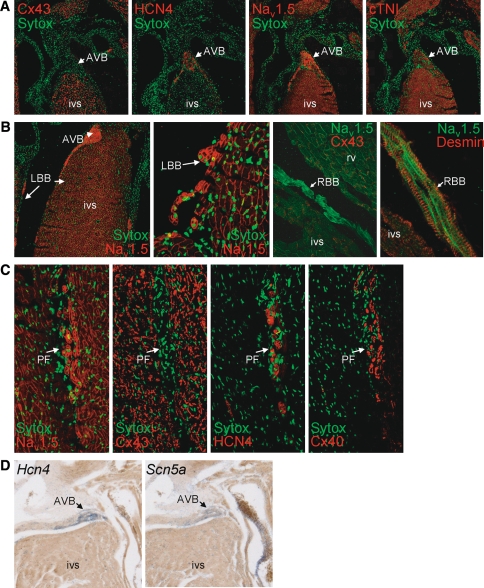
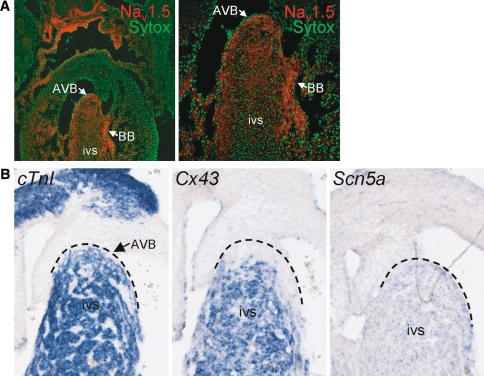
Figure 2 displays the expression and distribution of Nav1.5 in the conduction system in adult mouse ventricular myocardium. The atrioventricular bundle (AVB or His bundle) that displayed the expected presence of HCN4 staining and absence of Cx43 expression showed a high level of Nav1.5 labeling, which was clearly more intense compared to that of ventricular tissue of the septum (Fig. 2a). Myocytes of both tissue types clearly showed staining of the cell contour, indicating membrane labeling of Nav1.5 on the cell surface. The high level of Nav1.5 expression observed in the AVB continued into a thin layer of myocytes running below the subendocardial lining of the septum (Fig. 2b). These subendocardial cells, which constitute the bundle branches and Purkinje fibers of the conduction system, showed intense desmin labeling, were Cx43-negative, Cx40-positive, and HCN4-positive (Fig. 2b, c). These findings were confirmed on Scn5a mRNA level by in situ hybridization experiments in adult hearts (Fig. 2d). A similar distribution pattern in the intraventricular conduction system was observed in embryonic mouse hearts, with high expression levels of both Nav1.5 and Scn5a in the AVB and bundle branches (Fig. 3a, b). Overall, distribution of Nav1.5 and Scn5a labeling intensity within the embryonic and adult heart was as follows: AVB/bundle branches > atrial/ventricular myocardium ⋙ SAN/AVN. The described distribution patterns were consistently observed in all hearts studied.
Fig. 2.
Distribution of Nav1.5 in the atrioventricular bundle (AVB) and His-Purkinje system in adult murine myocardium. a The AVB lacks Cx43 expression, is HCN4-positive, and shows a high Nav1.5 labeling intensity, continuing into (b) the left and right bundle braches, which show intense labeling of Nav1.5 and desmin, but absence of Cx43 (LBB left bundle branch, RBB right bundle branch, rv right ventricle, ivs intraventricular septum). c Serial sections of Purkinje fibers (PF) show high Nav1.5, HCN4, and Cx40 labeling intensity, but no Cx43. d In situ hybridization of adult heart shows similar high expression levels of Hcn4 and Scn5a mRNA in AVB (blue signals) in comparison with the myocardium of the intraventricular septum (ivs)
Fig. 3.
Distribution of Nav1.5 and Scn5a in the conduction system in embryonic hearts (ED14.5). a High expression levels of Nav1.5 protein in the AV bundle (AVB) and bundle branches (BB) in comparison with the ventricular myocardium of the intraventricular septum (ivs); the right panel represents an enlarged section of the left panel. b In situ hybridization shows prominent Scn5a mRNA expression (in blue) but absence of Cx43 mRNA in the AVB (dashed line indicates region of AVB)
Heterogeneous transmural distribution of Nav1.5 and Scn5a in ventricular myocardium
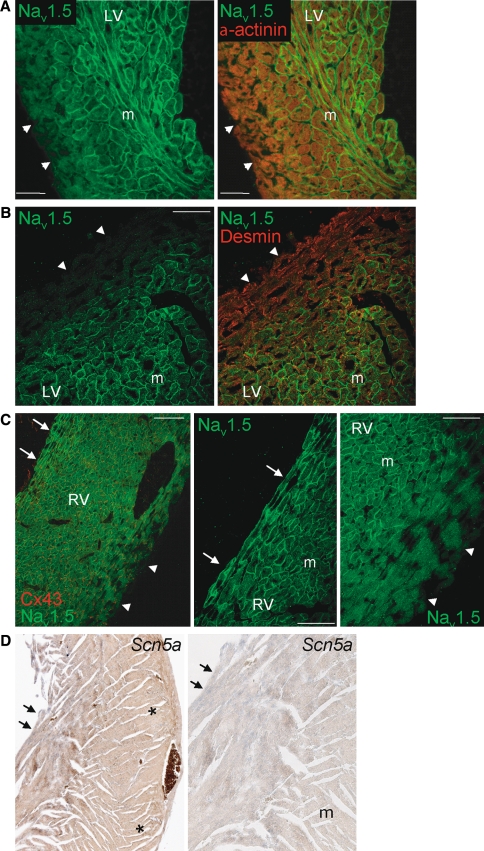
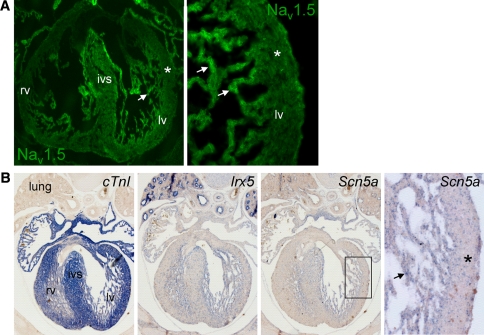
Figure 4a and b shows double labeling of left ventricular myocardium by anti-Nav1.5 with anti-desmin or anti-α-actinin. Although both desmin and α-actinin expression intensity appeared homogenously distributed throughout the ventricular wall, Nav1.5 labeling showed a clear transmural gradient. Nav1.5 expression was greatly reduced in subepicardial (denoted by arrowheads) compared to midmyocardial myocytes. Furthermore, more abundant Nav1.5 expression was observed in subendocardial myocytes (denoted by arrows) as compared to either midmural or subepicardial cells. A similar transmural gradient (subendocardial myocardium > midmural myocardium > subepicardial myocardium) for Nav1.5 was observed in the right ventricle (Fig. 4c). The subepicardial layer with low Nav1.5 expression comprised a larger fraction of the total ventricular wall thickness in the right compared to the left ventricle. Furthermore, a distinct transmural distribution for Nav1.5 was also observed in the right and left ventricle of mouse embryonic tissue (ED14.5; Fig. 5a). Again, the results from immunohistochemical analyses were confirmed by in situ hybridization experiments. Heterogeneous Scn5a mRNA expression with lower expression in the subepicardium compared to subendocardium was observed in both embryonic and adult murine ventricular tissue (Figs. 4d, 5b), in a pattern similar to Irx5, a homeodomain transcription factor with an established transmural gradient (Fig. 5b, right panel) [10, 14]. A similar substantial decrease in Scn5a/Nav1.5 expression levels in the subepicardium compared to subendocardium was consistently observed in each independent heart studied.
Fig. 4.
Transmural distribution of Nav1.5 in the ventricular myocardium. Low Nav1.5 labeling intensity in ventricular epicardium (arrowheads) compared to the midmyocardial layer (m) in (a) the apex and (b) left ventricular free wall of the adult mouse heart. For comparison, α-actinin and desmin show a homogeneous transmural labeling. cLeft panel shows low Nav1.5 expression in subepicardium (arrowheads) and high Nav1.5 expression in subendocardium (arrows) of the right ventricle. For comparison, Cx43 shows a homogeneous transmural distribution. Also shown are details of subendocardial surface (middle panel) and subepicardial region (right panel) of the right ventricle. d In situ hybridization of adult ventricular myocardium shows clear Scn5a staining (in blue) in the subendocardium (arrows) and low Scn5a expression in the subepicardium (asterisks). The right panel depicts a magnified section of the left panel indicating higher Scn5a expression in the subendocardium (arrows) compared to midmural (m)
Fig. 5.
a Overview (left panel) and detail (right panel) of embryonic mouse heart (ED14.5) indicating low Nav1.5 labeling intensity in ventricular subepicardium (asterisks) and abundant Nav1.5 in the trabeculated subendocardium. b In situ hybridization of ventricular myocardium in embryonic heart (ED14.5; overview and magnified boxed section in the right panel) displaying a transmural gradient in Scn5a mRNA expression with lower expression in the subepicardium (asterisks) compared to the trabeculated subendocardium (arrows). For comparison, the homeodomain transcription factor Irx5 shows a similar transmural distribution pattern (lv left ventricle, rv right ventricle, ivs intraventricular septum)
Transmural gradient in functional sodium channel availability
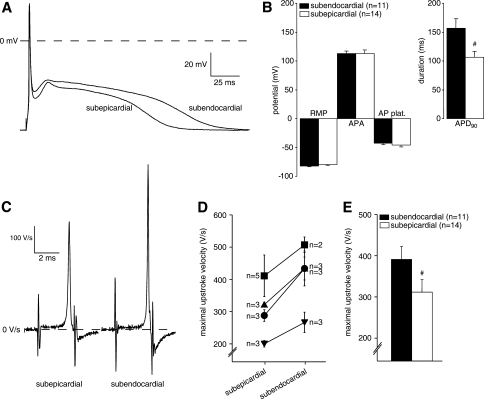
To investigate the functional electrophysiological consequences of the observed differences in Scn5a mRNA and Nav1.5 protein expression levels, we studied the action potential characteristics of subepicardial and subendocardial ventricular myocytes of four hearts. In particular, we focused on the dV/dtmax, which is a suitable index of sodium channel availability. Figure 6a shows representative action potentials recorded from a subepicardial and subendocardial cell of one heart. Subepicardial myocytes displayed considerable shorter action potentials compared to subendocardial cells, while action potential amplitude, plateau phase and resting membrane potential (important for voltage dependence of sodium channel inactivation) were not different (Fig. 6b). On average, the action potential duration at 90% repolarization (APD90) was 109 ± 11 ms in subepicardial myocytes (n = 14) and 153 ± 16 in subendocardial myocytes (n = 11; P < 0.05). Figure 6c shows the first time-derivative of the action potential (dV/dtmax), reflecting the maximal action potential upstroke velocity of the cells depicted in Fig. 6a. In each heart, action potential upstroke velocity was consistently lower in subepicardial myocytes compared to subendocardial cells (Fig. 6d). Consequently, average action potential velocity was significant lower in subepicardial cells (309 ± 32 V/s, n = 14) compared to subendocardial cells (394 ± 32 V/s, n = 11; Fig. 6e), indicating decreased functional sodium channel availability in the subepicardium compared to subendocardium.
Fig. 6.
a Representative examples of action potentials of a subepicardial and subendocardial myocyte from one heart, measured at a stimulation frequency of 2 Hz. b Averaged action potential characteristics (RMP resting membrane potential, APA action potential amplitude, AP plat. action potential plateau phase 20 ms after the upstroke, APD90 action potential duration at 90% repolarization, #P < 0.05 versus subendocardial). c Representative examples of the first time-derivative of the action potential (dV/dtmax), reflecting the maximal action potential upstroke velocity, measured from a subepicardial and subendocardial myocyte from one heart at a stimulation frequency of 2 Hz. d Averaged subepicardial and subendocardial action potential upstroke velocities of each heart measured (n = 4 hearts); numbers of mycoytes measured from each heart are indicated. e Average values for action potential upstroke velocities from all subepicardial and subendocardial myocytes (#P < 0.05 versus subendocardial)
Discussion
In this study, we provide a comprehensive investigation of sodium channel distribution throughout the murine heart. Our results demonstrate a unique differential distribution pattern of Scn5a mRNA and sodium channel protein Nav1.5 expression in various parts of the conduction system. Furthermore, we observed a transmural gradient in Scn5a mRNA expression, Nav1.5 protein expression and sodium channel availability across the ventricular wall.
Low Scn5a mRNA and Nav1.5 protein expression in sinoatrial and atrioventricular nodes
Analysis of the murine conduction system revealed very low Scn5a mRNA and Nav1.5 protein expression in the sinoatrial node (SAN) and atrioventricular node (AVN). Previously, low Nav1.5 expression levels in the central atrioventricular node versus high levels of Nav1.5 in the peripherally located transitional cells have been observed in rat and rabbit, and thus may be considered species independent [33, 47]. The low abundance of Nav1.5 in the SAN is in accordance with the relatively low expression of Scn5a mRNA observed in the adult mouse SAN region measured by quantitative RT-PCR [30], and the previously reported low degree of Nav1.5 staining in the SAN of the embryonic and adult mouse heart [22, 28]. We have now confirmed the presence of low Scn5a mRNA expression in the SAN using in situ hybridization, further validating these observations. Functionally, the brain-type sodium channel, and not the cardiac-type, appears essential for heart rate regulation, at least in the mouse [28]. In addition, cardiac-type sodium channels have not been found in the center of the SAN of the human heart, but only in the periphery of the SAN, where they may be essential for proper conduction to the surrounding atrial tissue [16]. Based on these observations, the clinical phenotype of sick sinus syndrome or sinus dysfunction described in some SCN5A mutations may be explained by peripheral SAN dysfunction and impaired impulse propagation to the atria [5, 7, 42]. On the other hand, the persistent inward sodium current and altered channel inactivation kinetics resulting from the SCN5A mutation 1795insD have been proposed to underlie the observed bradycardia and sinoatrial pauses [42]. Furthermore, the SCN5A mutation E161K, associated with sick sinus syndrome, was predicted to cause a reduction in sinus rate by slowing of the diastolic depolarization rate and upstroke velocity of the sinus node action potential [39]. Thus, although the expression level of sodium channels in the SAN appears limited, they may still play a role in sinus node function and heart rate regulation, perhaps even more so in the setting of sodium channel dysfunction.
Abundant Scn5a and Nav1.5 expression in the intraventricular conduction system
We observed abundant expression of Scn5a and Nav1.5 in the atrioventricular bundle (AVB or His bundle), bundle branches and Purkinje fibers, predicting a substantial role for Nav1.5 in fast propagation through the intraventricular conduction system. In previous studies by others, both high and low intensities of Nav1.5 staining have been reported in these areas [17, 47]. This discrepancy in observations may be attributed to differences in tissue handling protocols used, which may have influenced tissue binding properties of the antibody. To address this controversy, we have used in situ hybridization of Scn5a mRNA as an independent tool for assessing the distribution pattern of the cardiac sodium channel in the intraventricular conduction system. The observed pattern of Scn5a mRNA distribution matches that of Nav1.5 protein distribution, thereby ultimately proving the presence of abundant cardiac sodium channel expression in the AVB (or His bundle), bundle branches and Purkinje fibers. Since the cardiac sodium channel is hardly expressed in the atrioventricular node (AVN), the abundance of Nav1.5 in the AVB (or His bundle), bundle branches and Purkinje fibers is likely to be responsible for the atrioventricular and intraventricular dysfunction observed in patients with sodium channel dysfunction due to SCN5A mutations [7, 43].
Functional transmural gradient in cardiac sodium channel distribution
Another finding in the current study is the observation of a transmural gradient in Scn5a, Nav1.5, and functional sodium channel availability. The presence and ionic mechanism of transmural differences in action potential repolarization have been investigated in detail, but studies of transmural differences in depolarization are limited. Transmural action potential heterogeneity of the left ventricle is well known, with subepicardial and midmyocardial action potentials displaying prominent phase-1 repolarization, while subendocardial action potentials do not [1]. In addition, midmyocardial cells have longer action potentials compared to subepicardial and subendocardial cells, while subepicardial action potentials are the shortest [1]. Various membrane channels contribute to these transmural differences in the time course of action potential repolarization, including the transient outward K+ current (Ito1) [27], the slow delayed rectifier K+ current (IKs) [26], and the sustained component of the sodium current (INa(sus)) [48]. Some investigators have reported reduced sodium channel density in isolated myocytes from the subepicardium as compared to the mid- and subendocardium [3, 40], whereas other studies showed either no difference [12] or rather an increased sodium current density in subepicardium versus subendocardium [21]. Similarly, inconsistent results on sodium gating properties of subendocardial versus subepicardial myocytes have been described. In the dog, subepicardial myocytes have a more negative half-inactivation voltage [12], which consequently will lead to a reduced sodium current availability at resting membrane potentials. In contrast, studies in rats revealed no differences in inactivation [3, 21], but one report showed a more negative half-activation potential in subendocardial compared to subepicardial myocytes [21]. These discrepancies may be attributed to differences in experimental protocol, myocyte isolation process and species studied. Our current immunohistochemistry results from whole mouse heart sections show the existence of a transmural gradient in Nav1.5 distribution in both the left and right ventricle in situ. Importantly, Scn5a mRNA expression displayed a similar inhomogeneous distribution across the ventricular wall on in situ hybridization. Reduced Scn5a mRNA expression has also been demonstrated in epicardial compared to endocardial tissue in non-diseased human hearts, clearly suggesting that a transmural gradient in sodium channel availability is also present in human myocardium [18]. The functional relevance of the heterogeneous transmural Scn5a/Nav1.5 distribution was subsequently confirmed by our demonstration of reduced functional sodium channel availability in the subepicardial compared to subendocardial cells from patch-clamp analysis in isolated myocytes. Although we did not measure sodium current characteristics in the current study, dV/dtmax is considered a suitable index for functional sodium current availability [6, 37], and these functional data are in complete accordance with our immunohistochemistry and in situ hybridization results.
Implications of transmural heterogeneity in sodium channel distribution and function
The functional implications of transmural heterogeneity in Nav1.5 expression may be substantial. In the subepicardium, decreased inward sodium current will on the one hand slow conduction, and on the other hand allow for a greater impact of the repolarizing potassium currents, resulting in action potential shortening and subsequent transmural dispersion in repolarization. In the setting of myocardial ischemia, a gradient of Nav1.5 expression predisposes to inhomogeneous conduction slowing, greatly increasing the risk of life-threatening arrhythmias [9]. In patients with Brugada syndrome, the typical ECG pattern of ST-segment elevation in the right precordial leads has been attributed to a transmural heterogeneity of repolarization in the right ventricle [2], where the transient outward K+ current (Ito1) responsible for phase-1 action potential repolarization is more prominent compared to the left ventricle [15]. In loss-of-function SCN5A mutations, present in approximately 15–30% of Brugada syndrome patients [36, 38], sodium current reduction combined with the large transient outward current (Ito) in the right ventricular subepicardium is thought to give rise to a prominent early repolarization phase and an even larger transmural repolarization gradient [2]. Brugada syndrome is also thought to arise from right ventricular conduction delay due to localized conduction slowing [4, 13]. The inhomogeneous Nav1.5 distribution across the ventricular wall observed in our immunohistochemistry experiments may contribute to both mechanisms. Since the subepicardial layer comprises a relatively large fraction of the total right ventricular wall (as compared to the left ventricle), the effects of further sodium channel reduction is expected to be most notable in the right ventricle. Thus, in the current study, we have identified transmural heterogeneity in cardiac sodium channel expression as a novel additional pathophysiological player in the Brugada syndrome.
Cardiac distribution of Scn5a mRNA and Nav1.5 protein expression in the mouse embryo
Cardiac expression and function of certain ion channels may be differentially regulated during murine heart development [17, 20, 32]. In our study, the observed distribution patterns of Scn5a mRNA and Nav1.5 protein expression were similar in adult and embryonic murine hearts (ED14.5). Interestingly, the spatio-temporal expression pattern of Scn5a is similar to that of Cx40, and together they are likely responsible for fast conduction through the AVB, bundle branches and Purkinje fibers [11, 19]. Cx40, at least in embryonic hearts, displays a transmural gradient across the ventricular wall in a pattern similar to Scn5a, and its expression is known to be regulated by the T-box transcriptional activator Tbx5 [8, 19]. Sodium channel expression in the SAN is controlled by the T-box transcriptional repressor Tbx3, since Nav1.5 is ectopically expressed in the SAN in Tbx3-deficient mice [22]. Therefore, Tbx3 and Tbx5 may act in concert to determine the unique distribution pattern of Scn5a and Nav1.5 within the heart, with Tbx3 repressing expression in the SAN and AVN, and Tbx5 promoting expression in the atria, ventricle, AVB and bundle branches. The homeodomain transcription factor Irx5 also displays a transmural gradient similar to Scn5a and has been shown to determine the gradient of potassium channel expression and repolarization across the ventricular wall [10, 14]. Thus, it is tempting to speculate that Tbx5 and/or Irx5 are also responsible for the heterogeneous transmural distribution of the cardiac sodium channel.
Conclusion
We have presented an integrative assessment of the unique distribution pattern of the cardiac sodium channel within the murine heart. In particular, Nav1.5 was observed to be heterogeneously expressed in the cardiac conduction system, with low abundance in the nodal tissues and high expression in the intraventricular conduction system. In addition, a transmural gradient across the ventricular wall exists, with low Nav1.5 labeling intensity and functional sodium channel availability in the subepicardium compared to subendocardium. Since the sodium channel ensures proper electrical conduction within various regions of the myocardium, knowledge of its differential cardiac distribution is essential when investigating its role in the normal and diseased heart. The current results thus underline the importance of studying the multiple electrophysiological consequences of an SCN5A mutation in its native environment, i.e., in various regions and compartments of the heart and cardiomyocyte.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank Corrie de Gier-de Vries and Tilly Mommersteeg for their expert help in immunohistochemical analysis. This project was supported by The Netherlands Heart Foundation (Grant 2003/B195), The Interuniversity Cardiology Institute of the Netherlands (ICIN Grant 06401) and by a Fondation Leducq Trans-Atlantic Network of Excellence Grant (05 CVD 01, Preventing Sudden Death). Connie R. Bezzina (Grant 2005/T024), Vincent M. Christoffels (Grant 1996/M002), and Maurice J.B. van den Hoff (Grant 1996/M002) are Established Investigators of The Netherlands Heart Foundation.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00395-009-0012-8) contains supplementary material, which is available to authorized users.
References
- 1.Antzelevitch C, Fish J (2001) Electrical heterogeneity within the ventricular wall. Basic Res Cardiol 96:517–527 [DOI] [PubMed]
- 2.Antzelevitch C, Yan GX, Shimizu W (1999) Transmural dispersion of repolarization and arrhythmogenicity: the Brugada syndrome versus the long QT syndrome. J Electrocardiol 32(Suppl):158–165 [DOI] [PubMed]
- 3.Ashamalla SM, Navarro D, Ward CA (2001) Gradient of sodium current across the left ventricular wall of adult rat hearts. J Physiol 536:439–443 [DOI] [PMC free article] [PubMed]
- 4.Bébarová M, O’Hara T, Geelen JL, Jongbloed RJ, Timmermans C, Arens YH, Rodriguez LM, Rudy Y, Volders PG (2008) Subepicardial phase 0 block and discontinuous transmural conduction underlie right precordial ST-segment elevation by a SCN5A loss-of-function mutation. Am J Physiol Heart Circ Physiol 295:H48–H58 [DOI] [PMC free article] [PubMed]
- 5.Benson DW, Wang DW, Dyment M, Knilans TK, Fish FA, Strieper MJ, Rhodes TH, George AL Jr (2003) Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A). J Clin Invest 112:1019–1028 [DOI] [PMC free article] [PubMed]
- 6.Berecki G, Wilders R, van Ginneken ACG, Verkerk AO (2006) Non-physiological holding potential affects sodium channel repriming kinetics. Biophs J 90:suppl.1:392-Pos (abstract)
- 7.Bezzina CR, Veldkamp MW, van den Berg MP, Postma AV, Rook MB, Viersma JW, van Langen IM, Tan-Sindhunata G, Bink-Boelkens MT, r Hout AH, Mannens MM, Wilde AA (1999) A single Na+ channel mutation causing both long-QT and Brugada syndromes. Circ Res 85:1206–1213 [DOI] [PubMed]
- 8.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG (2001) A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106:709–721 [DOI] [PubMed]
- 9.Cascio WE (2001) Myocardial ischemia: what factors determine arrhythmogenesis? J Cardiovasc Electrophysiol 12:726–729 [DOI] [PubMed]
- 10.Christoffels VM, Keijser AG, Houweling AC, Clout DE, Moorman AF (2000) Patterning the embryonic heart: identification of five mouse Iroquois homeobox genes in the developing heart. Dev Biol 224:263–274 [DOI] [PubMed]
- 11.Coppen SR, Kaba RA, Halliday D, Dupont E, Skepper JN, Elneil S, Severs NJ (2003) Comparison of connexin expression patterns in the developing mouse heart and human foetal heart. Mol Cell Biochem 242:121–127 [DOI] [PubMed]
- 12.Cordeiro JM, Mazza M, Goodrow R, Ulahannan N, Antzelevitch C, Di Diego JM (2008) Functionally distinct sodium channels in ventricular epicardial and endocardial cells contribute to a greater sensitivity of the epicardium to electrical depression. Am J Physiol Heart Circ Physiol 295:H154–H162 [DOI] [PMC free article] [PubMed]
- 13.Coronel R, Casini S, Koopmann TT, Wilms-Schopman FJ, Verkerk AO, de Groot JR, Bhuiyan Z, Bezzina CR, Veldkamp MW, Linnenbank AC, van der Wal AC, Tan HL, Brugada P, Wilde AA, de Bakker JM (2005) Right ventricular fibrosis and conduction delay in a patient with clinical signs of Brugada syndrome: a combined electrophysiological, genetic, histopathologic, and computational study. Circulation 112:2769–2777 [DOI] [PubMed]
- 14.Costantini DL, Arruda EP, Agarwal P, Kim KH, Zhu Y, Zhu W, Lebel M, Cheng CW, Park CY, Pierce SA, Guerchicoff A, Pollevick GD, Chan TY, Kabir MG, Cheng SH, Husain M, Antzelevitch C, Srivastava D, Gross GJ, Hui CC, Backx PH, Bruneau BG (2005) The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell 123:347–358 [DOI] [PMC free article] [PubMed]
- 15.Di Diego JM, Sun Z-Q, Antzelevitch C (1996) Ito and action potential notch are smaller in left vs right canine ventricular epicardium. Am J Physiol 271:H548–H561 [DOI] [PubMed]
- 16.Dobrzynski H, Boyett MR, Anderson RH (2007) New insights into pacemaker activity: promoting understanding of sick sinus syndrome. Circulation 115:1921–1932 [DOI] [PubMed]
- 17.Domínguez JN, de la Rosa A, Navarro F, Franco D, Aránega AE (2008) Tissue distribution and subcellular localization of the cardiac sodium channel during mouse heart development. Cardiovasc Res 78:45–52 [DOI] [PubMed]
- 18.Gaborit N, Le Bouter S, Szuts V, Varro A, Escande D, Nattel S, Demolombe S (2007) Regional and tissue specific transcript signatures of ion channel genes in the non-diseased human heart. J Physiol 582:675–693 [DOI] [PMC free article] [PubMed]
- 19.Gros D, Jarry-Guichard T, Ten Velde I, de Maziere A, van Kempen MJ, Davoust J, Briand JP, Moorman AF, Jongsma HJ (1994) Restricted distribution of connexin40, a gap junctional protein, in mammalian heart. Circ Res 74:839–851 [DOI] [PubMed]
- 20.Harrell MD, Harbi S, Hoffman JF, Zavadil J, Coetzee WA (2007) Large-scale analysis of ion channel gene expression in the mouse heart during perinatal development. Physiol Genomics 28:273–283 [DOI] [PubMed]
- 21.Honen BN, Saint DA (2002) Heterogeneity of the properties of INa in epicardial and endocardial cells of rat ventricle. Clin Exp Pharmacol Physiol 29:161–166 [DOI] [PubMed]
- 22.Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, Ravesloot JH, Moorman AF, Verheijck EE, Christoffels VM (2007) Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev 21:1098–1112 [DOI] [PMC free article] [PubMed]
- 23.Kleber AG, Fast V (1997) Molecular and cellular aspects of re-entrant arrhythmias. Basic Res Cardiol 92(Suppl 1):111–119 [DOI] [PubMed]
- 24.Koopmann TT, Bezzina CR, Wilde AAM (2006) Voltage-gated sodium channels: action players with many faces. Ann Med 38:472–482 [DOI] [PubMed]
- 25.Lei M, Jones SA, Liu J, Lancaster MK, Fung SS, Dobrzynski H, Camelliti P, Maier SK, Noble D, Boyett MR (2004) Requirement of neuronal- and cardiac-type sodium channels for murine sinoatrial node pacemaking. J Physiol 559(Pt 3):835–848 [DOI] [PMC free article] [PubMed]
- 26.Liu DW, Antzelevitch C (1995) Characteristics of the delayed rectifier current (IKr and IKs) in canine ventricular epicardial, midmyocardial, and endocardial myocytes: a weaker IKs contributes to the longer action potential of the M cell. Circ Res 76:351–365 [DOI] [PubMed]
- 27.Liu DW, Gintant GA, Antzelevitch C (1993) Ionic bases for electrophysiological distinctions among epicardial, midmyocardial, and endocardial myocytes from the free wall of the canine left ventricle. Circ Res 72:671–687 [DOI] [PubMed]
- 28.Maier SKG, Westenbroek RE, Yamanushi TT, Dobrzynski H, Boyett MR, Catterall WA, Scheuer T (2003) An unexpected requirement for brain-type sodium channels for control of heart rate in the mouse sinoatrial node. Proc Natl Acad Sci USA 100:3507–3512 [DOI] [PMC free article] [PubMed]
- 29.Maier SKG, Westenbroek RE, McCormick KA, Curtis R, Scheuer T, Catterall WA (2004) Distinct subcellular localization of different sodium channel α and β subunits in single ventricular myocytes from mouse heart. Circulation 109:1421–1427 [DOI] [PubMed]
- 30.Marionneau C, Couette B, Liu J, Li H, Mangoni ME, Nargeot J, Lei M, Escande D, Demolombe S (2005) Specific pattern of ionic channel gene expression associated with pacemaker activity in the mouse heart. J Physiol 562:223–234 [DOI] [PMC free article] [PubMed]
- 31.Mohler PJ, Rivolta I, Napolitano C, LeMaillet G, Lambert S, Priori SG, Bennett V (2004) Nav1.5 E1053 K mutation causing Brugada syndrome blocks binding to ankyrin-G and expression of Nav1.5 on the surface of cardiomyocytes. Proc Natl Acad Sci USA 101:17533–17538 [DOI] [PMC free article] [PubMed]
- 32.Nguemo F, Sasse P, Fleischmann BK, Kamanyi A, Schunkert H, Hescheler J, Reppel M (2008) Modulation of L-type Ca(2+) channel current density and inactivation by beta-adrenergic stimulation during murine cardiac embryogenesis. Basic Res Cardiol (in press). doi:10.1007/s00395-008-0755-7 [DOI] [PubMed]
- 33.Petrecca K, Amellal F, Laird DW, Cohen SA, Shrier A (1997) Sodium channel distribution within the rabbit atrioventricular node as analysed by confocal microscopy. J Physiol 501:263–274 [DOI] [PMC free article] [PubMed]
- 34.Remme CA, Verkerk AO, Nuyens D, van Ginneken AC, van Brunschot S, Belterman CN, Wilders R, van Roon MA, Tan HL, Wilde AA, Carmeliet P, de Bakker JM, Veldkamp MW, Bezzina CR (2006) Overlap syndrome of cardiac sodium channel disease in mice carrying the equivalent mutation of human SCN5A–1795insD. Circulation 114:2584–2594 [DOI] [PubMed]
- 35.Remme CA, Wilde AAM, Bezzina CR (2008) Cardiac sodium channel overlap syndromes: different faces of SCN5A mutations. Trends Cardiovasc Med 18:78–87 [DOI] [PubMed]
- 36.Schulze-Bahr E, Eckardt L, Breithardt G, Seidl K, Wichter T, Wolpert C, Borggrefe M, Haverkamp W (2003) Sodium channel gene (SCN5A) mutations in 44 index patients with Brugada syndrome: different incidences in familial and sporadic disease. Hum Mutat 21:651–652 [DOI] [PubMed]
- 37.Sheets MF, Hanck DA, Fozzard HA (1988) Nonlinear relation between Vmax and INa in canine cardiac Purkinje cells. Circ Res 63:386–398 [DOI] [PubMed]
- 38.Smits JPP, Eckardt L, Probst V, Bezzina CR, Schott JJ, Remme CA, Haverkamp W, Breithardt G, Escande D, Schulze-Bahr E, LeMarec H, Wilde AA (2002) Genotype-phenotype relationship in Brugada syndrome: electrocardiographic features differentiate SCN5A-related patients from non-SCN5A-related patients. J Am Coll Cardiol 40:350–356 [DOI] [PubMed]
- 39.Smits JP, Koopmann TT, Wilders R, Veldkamp MW, Opthof T, Bhuiyan ZA, Mannens MM, Balser JR, Tan HL, Bezzina CR, Wilde AA (2005) A mutation in the human cardiac sodium channel (E161 K) contributes to sick sinus syndrome, conduction disease and Brugada syndrome in two families. J Mol Cell Cardiol 38:969–981 [DOI] [PubMed]
- 40.Szabó G, Szentandrássy N, Biró T, Tóth BI, Czifra G, Magyar J, Bányász T, Varró A, Kovács L, Nánási PP (2005) Asymmetrical distribution of ion channels in canine and human left-ventricular wall: epicardium versus midmyocardium. Eur J Physiol 450:307–316 [DOI] [PubMed]
- 41.Tomaselli GF, Zipes DP (2004) What causes sudden death in heart failure? Circ Res 95:754–763 [DOI] [PubMed]
- 42.Veldkamp MW, Wilders R, Baartscheer A, Zegers JG, Bezzina CR, Wilde AA (2003) Contribution of sodium channel mutations to bradycardia and sinus node dysfunction in LQT3 families. Circ Res 92:976–983 [DOI] [PubMed]
- 43.Wang DW, Viswanathan PC, Balser JR, George AL Jr, Benson DW (2002) Clinical, genetic, and biophysical characterization of SCN5A mutations associated with atrioventricular conduction block. Circulation 105:341–346 [DOI] [PubMed]
- 44.Wiegerinck RF, van Veen TAB, Belterman CN, Schumacher CA, Noorman M, de Bakker JMT, Coronel R (2008) Transmural dispersion of refractoriness and conduction velocity is associated with heterogeneously reduced connexin43 in a rabbit model of heart failure. Heart Rhythm 5:1178–1185 [DOI] [PubMed]
- 45.Wiegerinck RF, de Bakker JMT, Opthof T, de Jonge N, Kirkels H, Wilms-Schopman FJG, Coronel R (2009) The effect of enhanced gap junctional conductance on ventricular conduction in explanted hearts from patients with heart failure. Basic Res Cardiol (in press). doi:10.1007/s00395-008-0771-7 [DOI] [PubMed]
- 46.Wolk R, Cobbe SM, Hicks MN, Kane KA (1999) Functional, structural, and dynamic basis of electrical heterogeneity in healthy and diseased cardiac muscle: implications for arrhythmogenesis and anti-arrhythmic drug therapy. Pharmacol Ther 84:207–231 [DOI] [PubMed]
- 47.Yoo S, Dobrzynski H, Fedorov VV, Xu SZ, Yamanushi TT, Jones SA, Yamamoto M, Nikolski VP, Efimov IR, Boyett MR (2006) Localization of Na+ channel isoforms at the atrioventricular junction and atrioventricular node in the rat. Circulation 114:1360–1371 [DOI] [PubMed]
- 48.Zygmunt AC, Eddlestone GT, Thomas GP, Nesterenko VV, Antzelevitch C (2001) Larger late sodium conductance in M cells contributes to electrical heterogeneity in canine ventricle. Am J Physiol Heart Circ Physiol 281:H689–H697 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.