Abstract
Colorectal cancer (CRC) is the third most common cancer worldwide and has poor prognosis. To identify the oncofetal proteins involved in CRC carcinogenesis, differentially expressed proteins among fetal colorectal tissues, CRC, and the paired tumor-adjacent normal colorectal tissues were investigated by a two-dimensional gel electrophoresis and MALDI-TOF/TOF-based proteomics approach. 42 protein spots were differentially expressed among these tissues, and 22 proteins were identified by MS analysis. Desmin and zinc finger protein 829 were found to be elevated in CRC tissue and fetal colorectal tissue compared with normal colorectal tissue. The elevated expression of desmin in CRC tissue and different developmental stages of fetus colon was confirmed by RT-PCR and Western blot analysis. Immunohistochemical analysis showed that the elevated expression of desmin was correlated with the severity and differentiation of CRC and decreased survival rate of CRC patients. Finally by developing a highly sensitive immunoassay, desmin could be detected in human serum and was significantly elevated in CRC patients compared with healthy volunteers. We propose that desmin be considered a potential oncofetal serum tumor marker for CRC that may have significance in the detection of patients with CRC.
Colorectal carcinoma (CRC)1 is the third most common type of cancer and the fourth most frequent cause of death due to cancer worldwide. Almost one million new cases occur annually, amounting to 492,000 related deaths (1). The lifetime risk of developing the disease is close to 6% (2). It has been shown that up to 90% of the patients can be cured by surgery if the CRC is detected at an early stage. But unfortunately, the disease is often diagnosed only at an advanced stage, and prognosis is accordingly poor. Therefore, the early diagnosis is important for proper control of CRC (2–4). Identification of biomarkers for early detection, prognosis, and response to treatment is an important goal for cancer research by multiplex technologies (5). Proteomics approaches are promising tools for the discovery of new cancer biomarkers and prognostic therapeutic drug targets (6–8).
Proteomics analysis is currently considered to be a powerful tool for global evaluation of protein expression and has been widely applied in analysis of diseases, especially in the fields of cancer research (9, 10). In addition to the better known genetic and epigenetic alterations, there are also factors relating to molecular changes in translation, post-translational modification, and intracellular mislocalization involved in tumor initiation and growth, and these factors cannot be detected either by measuring the amount of RNA or by detecting nucleotide sequence variation (11). Analysis of the cancer proteome can be beneficial to understand the association between protein alterations and malignancy (12). Several previous studies have also involved the preliminary application of proteomics in the identification of the biomarkers for CRC (13–18). Comparison of protein expression profiles between CRC and normal cell lines or tissues has revealed replicable and significant changes in the expression levels of a number of proteins, including some oncoproteins, signal transduction proteins, metabolic enzymes, and so on (13–18). These studies reported slightly different findings and conclusions that few proteins were found to vary in concert, and the discrepancies might be due to their regional variability, tissue heterogeneity, or technical problems such as the varying ability of mass spectrometry to identify a particular protein (11, 19).
At present, some high diagnostic value biomarkers in cancer have the characteristic of embryonic origin. The gene or protein is expressed during the period of fetal development but is seldom expressed in adulthood; during tumorigenesis it is expressed at a high level in some cancers, such as α-fetoprotein and glypican-3 for the diagnosis of hepatocellular carcinoma (20–23) and carcinoembryonic antigen (CEA) for the diagnosis of CRC (24). Tumorigenesis and embryogenesis are postulated to share certain common pathways (25, 26). Some fetal proteins may be implicated in the development and progression of CRC. Along the developmental path from embryonic stages to adulthood, the intestinal tract performs diverse biological processes, which are accompanied by a series of anatomical as well as transcriptional and proteomic changes. Delineation of the molecular events involved in these stages would enrich our knowledge of the development of intestinal epithelia and stroma. Such information may lead to the identification of specific markers for detection of CRC and potential therapeutic targets that may inhibit malignant transformation and progression (27).
In the present study, differentially expressed proteins were profiled from fetal colorectal tissues, paired cancer tissues, and corresponding normal tissues. Desmin, which was significantly up-regulated in CRC and fetus colon tissues compared with normal tissue, was chosen for validation and analysis.
EXPERIMENTAL PROCEDURES
Specimens
Tissue specimens and blood samples were collected from the Six People's Hospital, Shanghai Jiao Tong University in accordance with approved human subject guidelines approved by the Scientific and Ethical Committee of Shanghai Jiao Tong University. All these samples were taken by experienced surgeons and examined by experienced pathologists. For proteomics analysis, fresh CRC and paired tumor-adjacent normal colorectal tissues from 10 patients and colorectal adenoma tissues from 10 patients were obtained during 2006–2007. 24 fetal colorectal specimens with different developmental stages were obtained from the Department of Gynecology and Obstetrics. All of the specimens were obtained from surgical resections, immediately frozen in liquid nitrogen, and then frozen at −80 °C until use. For the validation studies, 152 paraffin-embedded CRC tissue samples, 30 CRC paired adjacent normal tissues, and 36 colorectal adenoma specimens were obtained between the years 1999 and 2003. Blood samples were obtained from 92 patients with CRC before surgical resections. 45 specimens of healthy individuals were donated on a voluntary basis. 25 benign bowel disease control serum samples were obtained from patients with one or more of the following diagnoses (frequency of diagnosis is given in parentheses): colorectal adenoma (nine), diverticulosis (six), colitis (five), and morbus Crohn (five). A summary of clinical information for these patients is shown in Tables I and II. The serum samples of CRC patients and healthy controls were selected based on the strict criteria in the current study (see the supplemental information).
Table I. Clinical features of all human tissue samples.
| Clinical features | Number |
|---|---|
| Normal tissues | |
| Mean age (range) (years) | 55.86 ± 13.62 (45–68) |
| Gender (male/female) | 12/18 |
| Total | 30 |
| Adenoma | |
| Mean age (range) (years) | 64.59 ± 12.70 (48–63) |
| Gender (male/female) | 16/30 |
| Pathological type | |
| Tubular adenoma | 8 (22.2%) |
| Villous adenoma | 16 (44.5%) |
| Mixed adenoma | 12 (33.3%) |
| Total | 36 (100%) |
| CRC | |
| Mean age (range) (years) | 67.03 ± 11.31 (53–72) |
| Gender (male/female) | 88/64 |
| Location | |
| Ascending colon | 44 (28.9%) |
| Transverse colon | 7 (4.6%) |
| Descending colon | 13 (8.6%) |
| Sigmoid colon | 38 (25%) |
| Rectum | 50 (32.9%) |
| UICC staging | |
| I | 14 (9.2%) |
| II | 68 (44.7%) |
| III | 50 (32.9%) |
| IV (liver metastasis) | 20 (13.2%) |
| Differentiation grading | |
| Well | 10 (6.6%) |
| Moderate | 98 (64.5%) |
| Poor | 44 (28.9%) |
| Total | 152 (100%) |
| Fetal colorectal tissue | |
| Mean age (range) (months) | 5.53 ± 1.36 (3–10) |
| Within 4 months | 10 |
| 4–10 months | 2 samples/month × 7 months |
| Total | 24 |
Table II. Clinical features of tested serum samples.
CEA and desmin levels in serum were determined by ELISA, and median values (mean ± S.D.) are given for the indicated groups. Mean age ± S.D. and gender distribution for the individual groups are indicated.
| Samples | Total number | Age | Gender |
CEA | Desmin | |
|---|---|---|---|---|---|---|
| Female | Male | |||||
| years | ng/ml | ng/ml | ||||
| Controls | 70 | 62.18 ± 11.03 | 30 | 40 | 1.71 ± 1.27 | 61.55 ± 31.36 |
| Healthy controls | 45 | 62.74 ± 11.26 | 19 | 26 | 1.55 ± 1.46a | 49.21 ± 25.74b |
| Benign bowel disease | 25 | 61.86 ± 9.92 | 11 | 14 | 1.98 ± 0.80a | 71.41 ± 23.47b |
| CRC (all stages) | 92 | 65.32 ± 10.15 | 40 | 52 | 5.73 ± 10.08a | 105.80 ± 60.30b |
| UICC I | 31 | 65.01 ± 11.32 | 13 | 18 | 2.84 ± 1.21c | 118.32 ± 78.86d |
| UICC II | 22 | 64.12 ± 12.03 | 7 | 15 | 2.96 ± 1.53c | 119.57 ± 82.03d |
| UICC III | 21 | 65.14 ± 9.81 | 9 | 12 | 4.78 ± 2.14c | 106.64 ± 80.97d |
| UICC IV | 18 | 67.01 ± 7.44 | 11 | 7 | 38.65 ± 4.76c | 120.55 ± 84.18d |
a One-way ANOVA analysis, p < 0.01.
b One-way ANOVA analysis, p < 0.01.
c One-way ANOVA analysis, p < 0.01; LSD t test, p < 0.01 (UICC I versus UICC IV).
d One-way ANOVA analysis, p > 0.05.
2-D Gel Electrophoresis
100 mg of tissue sample was ground into powder in liquid nitrogen, homogenized in 1 ml of lysis buffer (7 m urea, 2 m thiourea, 4% CHAPS, 30 mm Tris-HCl, protease inhibitor mixture) on ice, and sonicated (10 × 10-s pulses) on ice. The homogenate was subjected to centrifugation (12,000 rpm) for 1 h at 4 °C. The protein was precipitated with cold acetone at −20 °C for 2 h and dissolved with rehydration buffer (8 m urea, 2 m thiourea, 4% CHAPS, 100 mm DTT, 2% ampholyte). Protein concentrations were determined by the Bradford method (Bio-Rad). IPG strips (18 cm, pH 4–7, non-linear; Bio-Rad) were passively rehydrated using 400 μl of rehydration buffer for 12 h at 17 °C. IEF was performed on an IEF cell (Bio-Rad). The strips were equilibrated in equilibration buffer (25 mm Tris-HCl (pH 8.8), 6 m urea, 20% glycerol, 2% SDS, 130 mm DTT) for 15 min followed by the same buffer containing 200 mm iodoacetamide instead of DTT for another 15 min. 12% SDS-PAGE gels were used for 2-D gel separation. The gels were stained using Coomassie Brilliant Blue R-350 (Merck) according to the supplier's protocol. The protein spots were detected, quantified, and matched using PD-Quest 2D analysis software (Bio-Rad). Each sample was run in triplicate.
In-gel Tryptic Digestion and Protein Identification by Mass Spectrometry
In-gel tryptic digestion and protein identification by mass spectrometry were carried out as described elsewhere (10). Briefly protein spots of interest were excised and destained. In-gel digestion was performed with 0.01 μg/μl trypsin (Promega) for 15 h at 37 °C. The tryptic peptides were extracted from the gel and dried by centrifugal lyophilization. Peptide mixtures were redissolved in 0.5% TFA and analyzed by a 4700 Proteomics Analyzer (Applied Biosystems). Peptide mass maps were acquired in positive reflection mode, averaging 1500 laser shots per MALDI-TOF spectrum and 3000 shots per TOF/TOF spectrum. Assignment of MS/MS data was performed using a MASCOT (version 1.9) search against the Swiss-Prot database (UniProt_SP sprot_84; 230,133 sequences). The search parameters included potential residue mass modification for carbamidomethylation and oxidation, one missed trypsin cleavage, and fragment ion tolerance and peptide tolerance of ±0.2 Da. All of the automatic data analysis and database searching were fulfilled by the GPS ExplorerTM software (version 3.6; Applied Biosystems). The confident identification had a statistically significant (p ≤ 0.05) protein score (based on combined mass and mass/mass spectra) and best ion score (based on mass/mass spectra). Redundancy of proteins that appeared in the database under different names and accession numbers was eliminated. If more than one protein was identified in one spot the single protein member with the highest protein score was singled out from the multiprotein family.
RT-PCR
The primer sequences and the expected sizes of PCR products were as follows: desmin, 5′-CCT GAA GGG CAC TAA CGA TT-3′ (sense) and 5′-CGG AAG TTG AGG GCA GAG T-3′ (antisense) (276 bp); β-actin, 5′-TAT GAC TTA GTT GCG TTA CAC C-3′ (sense) and 5′-CCT TCA CCG TTC CAG TTT-3′ (antisense) (155 bp). Total RNA was extracted using TRIzol reagent (Invitrogen), and RT-PCR was performed with the following conditions: reverse transcription at 48 °C for 30 min and denaturation at 94 °C for 2 min; then amplification for 30 cycles at 94 °C for 0.5 min, annealing at 56.7 °C (desmin) or 50 °C (β-actin) for 0.5 min, and extension at 72 °C for 0.5 min; and then terminal elongation step at 72 °C for 10 min and a final holding stage at 4 °C. The PCR products (5 μl) were analyzed by electrophoresis through 1% agarose gels and visualized by SYBR Gold (Molecular Probes, Eugene, OR) staining.
Western Blotting
Tissue samples were ground in liquid nitrogen and lysed in RIPA lysis buffer (50 mm Tris-HCl (pH 7.4), 0.25% sodium deoxycholate, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, 1 mm NaF, 1 mm Na3V4, 1 mm PMSF). The extracted proteins were separated by SDS-PAGE and immunoblotted using primary antibody against desmin (Abcam, Cambridge, MA). The blots were visualized by enhanced chemiluminescence reagents (Amersham Biosciences). β-Actin was used as an internal control.
Immunohistochemistry
Consecutive paraffin wax-embedded tissue sections (3–5 μm) were dewaxed and rehydrated. Antigen retrieval was performed by pretreatment of the slides in citrate buffer (pH 6.0) in a microwave oven for 12 min. Endogenous peroxidase activity was quenched by incubating the slides in methanol containing 0.6% hydrogen peroxide and washed by water for 4 min. For desmin-specific staining the slides were incubated for 1 h at room temperature with normal goat serum and subsequently incubated at 4 °C overnight with the anti-human desmin antibody (diluted 1:200; Abcam) and rinsed with TBS containing 0.1% bovine serum albumin. Incubation with horseradish peroxidase-linked goat anti-rabbit antibodies for 30 min was followed by reaction with diaminobenzidine and counterstaining with Mayer's hematoxylin. Immunostaining was detected by the Dako Envision System (DakoCytomation GmbH, Hamburg, Germany) according to the manufacturer's instructions.
Saturation and intensity of immunostained stroma was evaluated over five visual representative fields with well preserved carcinoma tissue at a power of ×200 magnification. The immunostaining was evaluated as described in previous studies (11, 28, 29) with some minor modifications. Immunostaining intensity (i) was classified as lack of staining (0), mild staining (1), moderate staining (2), and strong staining (3); the percentage of staining-positive stroma (ii) was semiquantitatively divided into five grades: <5% (0), 6–25% (1), 26–50% (2), 51–75% (3), and >75% (4). The score for each section was measured as (i) × (ii), and the result was defined as negative (−, 0), weakly positive (+, 1–3), positive (++, 4–7), and strongly positive (+++, 8–12). A minimum of five fields for each section was evaluated. Sections were examined separately by two independent investigators without any prior knowledge of each patient's clinical information and outcome. Any discrepancy between the two evaluators was resolved by reevaluation and careful discussion until agreement was reached.
ELISA for Desmin
For detection of desmin in human serum, a sandwich ELISA was developed using streptavidin-coated 96-well microtiter plates (14). 26 μl of human serum sample as calibrator antigen was incubated with 220 μl of antibody reagent containing two different kinds of biotinylated and digoxigenylated anti-desmin antibody (Abcam and Santa Cruz Biotechnology) in 40 mm phosphate buffer (pH 7.4) (0.9% NaCl, 0.1% bovine IgG, 0.022% polymerized rabbit IgG, 1.025% polyethylene glycol 40,000, 1.1% normal rabbit serum, 0.6% Synperonic F68, 0.01% N-methylisothiazolone, 0.1% chloroacetamide). After incubation overnight at room temperature, 100 μl was transferred to a streptavidin-coated microwell plate and incubated for 1 h. Subsequently the plates were washed three times with 0.9% NaCl, 0.1% Tween 20. For the detection of bound antigen-antibody complexes, 100 μl of a monoclonal anti-digoxigenin horseradish peroxidase conjugate (30 milliunits/ml in Universal Conjugate Buffer, Roche Diagnostics GmbH) was added and incubated for 1 h. The excess conjugate was removed by washing the plates three times with 0.9% NaCl, 0.1% Tween 20. The amount of bound conjugate was determined by adding tetramethylbenzidine substrate solution (Roche Diagnostics GmbH) and incubating for 1 h. The reaction was stopped with the addition of 50 μl of sulfuric acid into each wells and gently mixed for 30 s. The optical density was read at 450 nm within 30 min in a microtiter plate reader. The steps of validation of this assay are shown in the supplemental information.
CEA Assay
CEA was measured by a commercially available assay (Roche Diagnostics GmbH) according to the manufacturer's instructions.
Stastistical Analysis
Statistical calculations were performed by SPSS statistical software (version 11.0.0; SPSS, Inc.). All data were expressed as mean ± S.D. Comparison between two groups was performed by Wilcoxon two-sample test. Comparisons among multiple groups were performed by one-way ANOVA, LSD t test, or Dunnett t test. Comparisons of ordinal data between two groups were analyzed by rank sum test. Relevance analysis of ordinal data was analyzed by cross-tabulation χ2 test. Survival curves were generated according to the Kaplan-Meier method, and the statistical analysis was performed by log rank test. Multivariate analysis was evaluated by Cox proportional hazard models. Statistical significance was defined as p < 0.05.
RESULTS
Differentially Expressed Proteins among Colorectal Carcinoma, Normal Colorectal Tissue, and Fetal Colorectal Tissue
CRC tissues and adjacent normal colorectal tissues from 10 patients (mean age, 60.30 ± 6.72 years) and fetal colorectal tissues from 10 induced abortion fetuses under 4 months old were analyzed in triplicate by 2-DE. Coomassie staining of 2-D gels detected 685 ± 45, 609 ± 53, and 620 ± 44 protein spots within a pH range from 4 to 7 from fetal colorectal tissue, CRC, and normal colorectal tissue, respectively (Fig. 1A). Statistical analysis of resultant 2-D gels revealed that 42 protein spots were differentially expressed among fetal colorectal tissue, CRC, and normal colorectal tissue (one-way ANOVA; p < 0.05). Among them, only two protein spots were significantly increased in both CRC and fetal colorectal tissue compared with normal colorectal tissue (Fig. 1B).
Fig. 1.
Proteomics analysis of CRC, normal, and fetal colorectal tissues using 2-DE gels. A, comparison of protein profiles among CRC, normal, and fetal colorectal tissues (within 3 months) by 2-DE gels. An average of 685 ± 45, 609 ± 53, and 620 ± 44 spots were visualized for fetal colorectal tissue, CRC, and normal colorectal tissue, respectively. Two representative spots with a similar expression pattern between CRC and the fetal colorectal tissue group but significantly different from the normal colorectal tissue group are indicated. Spot 1 is the zinc finger protein 829; spot 2 at a molecular mass of approximately 60 kDa is desmin. B, cropped 2-DE gels images of the two spots. Desmin (a) and zinc finger protein 829 (b) with a similar expression pattern between CRC and the fetal colorectal tissue group but significantly different from the normal colorectal tissue group were indicated by one-way ANOVA to be statistically significant (p < 0.01). N, normal colorectal tissues; Ca, colorectal cancer; C, fetal colorectal tissues.
Mass Spectrum Identification of Differentially Expressed Proteins
Differentially expressed protein spots were subsequently subjected to MS/MS analysis. 22 proteins were identified from 42 spots (Table III). Differences between the experimental molecular weight/pI and the theoretical values of some proteins occurred due to post-translational modifications such as truncation and/or protein phosphorylation (30–33).
Table III. Proteins identified by MALDI-TOF/TOF.
| Spot no. | Accession no.a | Protein nameb | Gene name | Theoretical molecular massc | Theoretical pIc | S.D.d | Matched peptidese | Unmatched peptidesf | Coverage | Best ion scoreg | Protein scoreh | Peptides identifiedi |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | ||||||||||||
| 1 | P15531 | Nucleoside-diphosphate kinase A | NME1 | 17,309 | 5.83 | 3.21 | 11 | 15 | 64 | 36 | 144 | K↓DRPFFAGLVK↓Y |
| 2 | P62736 | Aortic smooth muscle | ACTA2 | 42,381 | 5.23 | 4.53 | 11 | 14 | 38 | 79 | 183 | K↓SYELPDGQVITIGNER↓F |
| 3 | Q00403 | Transcription initiation factor IIB | GTF2B | 35,324 | 8.67 | 2.25 | 1 | 9 | 3 | 22 | 27 | R↓KAVELDLVPGR↓S |
| 4 | P62937 | Peptidyl-prolyl cis-trans isomerase A | PPIA | 18,229 | 7.68 | 1.62 | 16 | 20 | 65 | 108 | 337 | M↓VNPTVFFDIAVDGEPLGR↓V, K↓SIYGEKFEDENFILK↓H |
| 5 | P16949 | Stathmin | STMN1 | 17,292 | 5.76 | 4.43 | 5 | 10 | 32 | 8 | 28 | K↓TLDFIDVLLLAR↓D |
| 6 | P04406 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 36,201 | 8.57 | 2.36 | 6 | 14 | 26 | 62 | 113 | K↓LISWYDNEFGYSNR↓ |
| 7 | Q06830 | Peroxiredoxin-1 | PRDX1 | 22,324 | 8.27 | 1.48 | 11 | 16 | 49 | 50 | 181 | R↓LVQAFQFTDK↓H |
| 8 | P09651 | Heterogeneous nuclear ribonucleoprotein A1 | HNRNPA1 | 38,936 | 9.26 | 3.12 | 15 | 21 | 34 | 15 | 124 | K↓GGNFGGR↓S |
| 9 | Q13946 | High affinity cAMP-specific 3′,5′-cyclic phosphodiesterase 7A | PDE7A | 55,983 | 7.10 | 1.09 | 6 | 31 | 20 | 92 | 50 | K↓CADICNPCRTWELSK↓Q |
| 10 | Q9NSK0 | Kinesin light chain 4 | KLC4 | 69,054 | 5.82 | 3.60 | 4 | 9 | 10 | 90 | 33 | R↓YYQRALAIYEGQLGPDNPNVAR↓T |
| 11 | P04792 | Heat shock protein β-1 | HSPB1 | 22,826 | 5.98 | 2.79 | 7 | 19 | 32 | 56 | 118 | R↓LFDQAFGLPR↓L |
| 12 | A6NEN9 | Uncharacterized protein LOC158830 | LOC | 21,520 | 10.33 | 2.08 | 6 | 18 | 34 | 22 | 39 | K↓QSGRSDK↓K |
| 13 | P07951 | Tropomyosin β chain | TPM2 | 32,945 | 4.66 | 1.19 | 9 | 13 | 20 | 50 | 68 | R↓IQLVEEELDRAQER↓L |
| 14 | Q3KNS6 | Zinc finger protein 829 | ZNF829 | 51,478 | 4.32 | 3.22 | 4 | 23 | 12 | 14 | 29 | K↓YCSNLNDHQR↓I |
| 15 | P17661 | Desmin | DES | 53,500 | 5.21 | 2.29 | 13 | 30 | 36 | 30 | 193 | R↓TFGGAPGFPLGSPLSSPVFPR↓A |
| 16 | Q9NY65 | Tubulin α-8 chain | TUBA8 | 50,746 | 5.01 | 2.76 | 6 | 9 | 19 | 23 | 46 | R↓QLFHPEQLITGK↓E |
| 17 | P60660 | Myosin light polypeptide 6 | MYL6 | 17,090 | 4.56 | 2.33 | 6 | 15 | 45 | 49 | 133 | K↓EAFQLFDR↓T, K↓NKDQGTYEDYVEGLR↓V |
| 18 | P24844 | Myosin regulatory light polypeptide 9 | MYL9 | 19,871 | 4.80 | 2.06 | 10 | 12 | 39 | 42 | 98 | K↓GNFNYVEFTR↓I |
| 19 | Q01995 | Transgelin | TAGLN | 22,653 | 8.87 | 2.92 | 12 | 19 | 40 | 46 | 77 | R↓GDPNWFMK↓K |
| 20 | P35749 | Myosin-11 | MYH11 | 22,805 | 5.42 | 2.38 | 16 | 21 | 13 | 26 | 158 | K↓LDAFLVLEQLR↓C, K↓SLEADLMQLQEDLAAAER↓A |
| 21 | P63267 | Actin, γ-enteric smooth muscle | ACTG2 | 42,249 | 5.31 | 2.98 | 6 | 17 | 19 | 30 | 73 | K↓SYELPDGQVITIGNER↓F |
| 22 | P07355 | Annexin A2 | ANXA2 | 38,808 | 7.57 | 2.64 | 7 | 31 | 24 | 37 | 55 | R↓RAEDGSVIDYELIDQDAR↓D |
a Accession numbers were derived from the ExPASy database.
b For several proteins, a few isoforms were identified in the same individual.
c Theoretical molecular mass (kDa) and pI from the ExPASy database.
d S.D. of protein abundance rations of one certain spot.
e The number of peaks that match to the tryptic peptides.
f The number of peaks that do not match to the tryptic peptides.
g Best ion scores (based on mass/mass spectrums) were from MALDI-TOF/TOF identification. The proteins with a statistically significant protein score and best ion score were considered successfully identified.
h Protein scores (based on combined mass and mass/mass spectrums) were from MALDI-TOF/TOF identification. The proteins with a statistically significant protein score and best ion score were considered successfully identified.
i Each spot corresponding to one certain protein had at least one of the shown peptides identified.
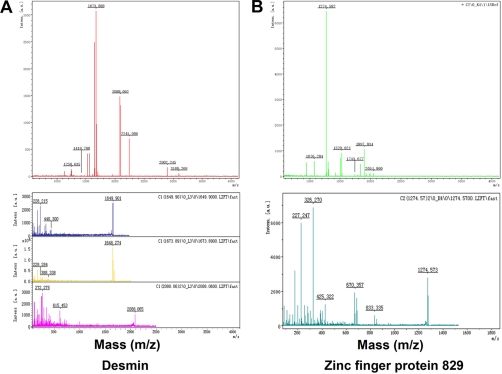
The two protein spots that were significantly increased in both CRC and fetal colorectal tissue compared with normal colorectal tissue were identified as desmin (Fig. 1B, a) and zinc finger protein 829 (Fig. 1B, b). The predicted molecular masses/pI values for desmin and zinc finger protein 829 were 53.5 kDa/5.21 and 51.5 kDa/4.32, respectively, fitting well to the position of the corresponding spot on the 2-DE gel. MS/MS analysis showed that desmin was identified with a high MASCOT score of 193 and 36% sequence coverage (Fig. 2A), but the zinc finger protein 829 was identified with a lower MASCOT score of 29 and 12% sequence coverage (Fig. 2B). From the result of MS/MS analysis, desmin was the more confident identification and became the subsequent focus of this study.
Fig. 2.
Maps of MALDI-TOF/TOF-MS for desmin (A) and zinc finger protein 829 (B) identification. Intens., intensity.
Validation of Desmin by Semiquantitative RT-PCR and Western Blotting Analysis
The differential expression of desmin in CRC was verified by RT-PCR analysis and Western blot analysis at the mRNA and protein levels. RT-PCR analysis showed that similar expression at the mRNA level of desmin was observed in CRC (0.73 ± 0.15 au) and fetal colorectal tissues (0.83 ± 0.17 au), but expression at the mRNA level was significantly different in normal tissues (0.26 ± 0.11 au; p < 0.01) (Fig. 3A). Western blot analysis was performed using anti-desmin antibody, and a similar expression level was also observed in CRC (0.57 ± 0.06 au) and fetal colorectal tissues (0.61 ± 0.14 au), but the expression level was significantly different in normal tissues (0.22 ± 0.04 au; p < 0.01) (Fig. 3B). These results confirmed that higher levels of desmin were expressed in CRC and fetal colorectal tissues compared with normal colorectal tissues at both mRNA and protein levels. The highest mRNA (1.03 ± 0.17 au) and protein levels (0.85 ± 0.21 au) of desmin were found in carcinoma with liver metastasis tissues. The expression of desmin in colorectal adenoma was also increased (or showed an increasing trend; 0.44 ± 0.17 au for mRNA levels and 0.38 ± 0.11 au for protein levels; p < 0.01). The results showed that the increasing trend paralleled the increasing severity of colorectal injury (Fig. 3, A and B).
Fig. 3.
Confirmation of desmin overexpression in CRC and fetal colorectal tissue (within 3 months). A and B, RT-PCR (A) and Western blot (B) indicated similar expression in the mRNA and protein levels of desmin between CRC and fetal colorectal tissues but a significant difference compared with normal tissues (one-way ANOVA, p < 0.01). Meanwhile RT-PCR (A) and Western blot (B) revealed that the expression of desmin at the mRNA and protein levels among four groups including normal colorectal tissues, adenoma tissues, carcinoma tissues, and liver metastasis tissues showed a remarkable increasing trend that followed the progression of carcinogenesis from normal colorectal tissue to adenoma, to carcinoma, and to carcinoma with liver metastasis. N, normal colorectal tissue; AD, adenoma; Ca, carcinoma; F, fetal colorectal tissue; LM, carcinoma with liver metastasis.
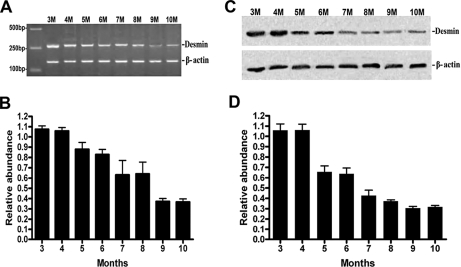
To confirm the oncofetal characteristic of desmin, fetus colorectal tissues from 3 to 10 months were examined by RT-PCR and Western blotting. The expression of desmin at both mRNA and protein levels revealed a decreasing trend from 3 to 10 months (Fig. 4, A and B).
Fig. 4.
Investigation of desmin expression in the different developmental stages (from 3 to 10 months) of fetal colorectal tissue and confirmation of its oncofetal characteristic. A and B, RT-PCR revealed the expression of desmin at mRNA levels in the different developmental stages of colorectal tissues from 3 to 10 months with a remarkable decreasing trend that followed the development of fetus. C and D, Western blot revealed the expression of desmin at protein levels in the different developmental stages of colorectal tissues from 3 to 10 months with a remarkable decreasing trend that followed the development of fetus. M, months.
Analysis of Desmin Expression by Immunohistochemistry
To further investigate the oncogenic properties of desmin in CRC, paraffin-embedded tissues were stained using anti-human desmin antibody. Staining for desmin was mainly located in the cytoplasm of CRC cells, suggesting that the CRC cells were responsible for the overexpression of desmin (Fig. 5A). In 30 normal specimens, no staining and weakly positive staining were detected in 20 and 80% of samples, respectively. No positive or strongly positive staining was detected in normal specimens. In 36 adenoma specimens, no staining, weakly positive staining, and positive staining were detected in 11.1, 69.5, and 19.4% of samples, respectively. In 152 CRC specimens, weakly positive staining, positive staining, and strongly positive staining were detected in 9.9, 74.3, and 15.8% of samples, respectively. In 20 liver metastasis specimens, weakly positive staining, positive staining, and strongly positive staining were detected in 5, 25, and 70% of samples, respectively. The semiquantitative scoring of immunoreactivity of normal colon, adenoma, CRC, and liver metastasis specimens was 1.80 ± 0.82, 2.62 ± 1.64, 5.78 ± 2.00, and 9.45 ± 2.63, respectively. Adenoma, CRC, and liver metastasis specimens expressed more desmin than normal colon (Table IV; p < 0.05).
Fig. 5.
Immunohistochemical analysis of desmin expression in CRC tissues. A, staining against desmin showed expression of desmin to increase gradually with the progression of carcinogenesis from normal colorectal tissue to adenoma, to carcinoma, and to carcinoma with liver metastasis. The staining of immunoreactivity was expressed as a product of the intensity and the proportion of cells staining positive. (a1, normal colorectal tissue, weakly positive; b1, adenoma, moderately positive; c1, CRC, strongly positive; d1, CRC with liver metastasis, the strongest immunoreactivity. a2–d2, enlargements of representative portions in a1–d1. a3–d3, enlargements of representative portions in a2–d2.) B, Kaplan-Meier curves and statistics showed the correlation between desmin expression and decreased survival (log rank test, p < 0.05).
Table IV. Desmin immunoreactivity in normal mucus, adenoma, CRC, and liver metastasis.
N, normal mucus; Ad, adenoma; Ca, CRC; Lm, liver metastasis; −, negative; +, weakly positive; ++, positive; +++, strongly positive.
| Cases | − | + | ++ | +++a | Average scoreb | |
|---|---|---|---|---|---|---|
| N | 30 | 20% (6/30) | 80% (24/30) | 0 | 0 | 1.80 ± 0.82 |
| Ad | 36 | 11.1% (4/36) | 69.5% (25/36) | 19.4% (7/36) | 0 | 2.69 ± 1.64 |
| C | 152 | 0 | 9.9% (15/152) | 74.3% (113/152) | 15.8% (24/152) | 5.78 ± 2.00 |
| Lm | 20 | 0 | 5% (1/20) | 25% (5/20) | 70% (14/20) | 9.45 ± 2.63 |
a Rank sum test, p < 0.01.
b One-way ANOVA analysis, p < 0.01.
In a total of 152 carcinoma samples, the percentages of well differentiated, moderately differentiated, and poorly differentiated were 6.6, 64.5, and 28.9, respectively, according to histodifferentiation criteria. The percentages of UICC stage I, UICC stage II, UICC stage III, and UICC stage IV were 9.2, 44.7, 32.9, and 13.2%, respectively, according to the UICC staging criteria. The number of CRC specimens located in ascending colon, transverse colon, descending colon, sigmoid colon, and rectum were 44, 7, 13, 38, and 50, respectively. The expression of desmin was significantly increased in the poor differentiation group (p < 0.01) (Table V), but no significant change was observed at different UICC stages or different locations of CRC (p > 0.05; Tables VI and VII).
Table V. Relevance of differentiation grading to desmin immunoreactivity.
−, negative; +, weakly positive; ++, positive; +++, strongly positive.
| Differentiation | Cases | − | + | ++ | +++a | Average scoreb |
|---|---|---|---|---|---|---|
| Well | 10 | 0 | 40% (4/10) | 50% (5/10) | 10% (1/10) | 3.70 ± 2.26 |
| Moderate | 98 | 0 | 7.1% (7/98) | 90.8% (89/98) | 2.1% (2/98) | 6.06 ± 1.50 |
| Poor | 44 | 0 | 11.3% (5/44) | 43.2% (19/44) | 45.5% (20/44) | 8.15 ± 3.11 |
a Cross-χ2 test, p < 0.01.
b One-way ANOVA analysis, p < 0.01; LSD t test, p < 0.01 (well versus moderate, well versus poor, and moderate versus poor).
Table VI. Relevance of UICC staging to desmin immunoreactivity.
−, negative; +, weakly positive; ++, positive; +++, strongly positive.
| UICC staging | Cases | − | + | ++ | +++a | Average scoreb |
|---|---|---|---|---|---|---|
| I | 14 | 0 | 7.2% (1/14) | 71.4% (10/14) | 21.4% (3/14) | 6.57 ± 2.47 |
| II | 68 | 0 | 10.3% (7/68) | 73.5% (50/68) | 16.2% (11/68) | 6.16 ± 2.28 |
| III | 50 | 0 | 10% (5/50) | 76% (38/50) | 14% (7/50) | 6.10 ± 2.21 |
| IV | 20 | 0 | 10% (2/20) | 75% (15/20) | 15% (3/20) | 6.35 ± 2.32 |
a Cross-χ2 test, p > 0.05.
b One-way ANOVA analysis, p > 0.05.
Table VII. Relevance of the location of CRC to desmin immunoreactivity.
AC, ascending colon; TC, transverse colon; DC, descending colon; SC, sigmoid colon; RC, rectum; −, negative; +, weakly positive; ++, positive; +++, strongly positive.
| Location | Cases | − | + | ++ | +++a | Average scoreb |
|---|---|---|---|---|---|---|
| AC | 44 | 0 | 6.8% (3/44) | 61.4% (27/44) | 31.8% (14/44) | 6.89 ± 2.51 |
| TC | 7 | 0 | 14.2% (1/7) | 42.9% (3/7) | 42.9% (3/7) | 7.42 ± 3.51 |
| DC | 13 | 0 | 15.3% (2/13) | 46.2% (6/13) | 38.5% (5/13) | 7.46 ± 2.96 |
| SC | 38 | 0 | 8.3% (4/48) | 58.4% (28/48) | 33.3% (16/48) | 7.10 ± 2.67 |
| RC | 50 | 0 | 10% (5/50) | 78% (39/50) | 12% (6/50) | 6.34 ± 2.18 |
a Cross-χ2 test, p > 0.05.
b One-way ANOVA analysis, p > 0.05.
Survival analysis was also conducted in the 152 CRC cases. The survival curves were generated according to the Kaplan-Meier method, and survival analysis suggested that immunoreactivity of desmin was more likely to present with poor outcome of patients with CRC (log rank test, p < 0.01; Fig. 4B). The 5-year survival rates were 72.19, 39.15, and 20.63% for weakly positive, positive, and strongly positive staining samples, respectively. To determine whether the prognostic value of desmin immunoreactivity was independent of other risk factors associated with the clinical outcome of CRC, multivariate analysis was performed using the Cox proportional hazard model. The risk variables examined included desmin immunoreactivity (weakly/moderately versus strongly positive), the location of CRC, histodifferentiation (well/moderately versus poorly differentiated), and UICC staging (stage I/II versus III). These factors are generally known to significantly affect the outcome of CRC. Desmin immunoreactivity was independently statistically significant as a prognostic factor in survival of patients of CRC (p < 0.05).
Detection of Desmin in Human Serum
After having established overexpression of desmin in CRC tissue, we assessed the potential release of this protein into the periphery and therefore its potential value as a serologic biomarker for the disease. For this purpose, a highly sensitive immunoassay for desmin was established. The serum levels of desmin in patients with CRC (n = 92), in patients with benign bowel disease (n = 25), and in healthy controls (n = 45) were assessed. The mean desmin serum level was 105.80 ± 60.30, 71.41 ± 23.47, and 49.21 ± 25.74 ng/ml in patients with CRC, patients with benign bowel disease, and healthy subjects, respectively (Table II). The difference between healthy subjects and patients with CRC was significant (Wilcoxon two-sample test, p < 0.0001). The same was true for the difference between patients with benign bowel disease and CRC (Wilcoxon two-sample test, p = 0.0084) (Fig. 6A). We also determined the levels of the established tumor maker CEA using a commercial immunoassay. The serum levels for CEA were 1.55 ± 1.46 and 5.73 ± 10.08 ng/ml for healthy subjects and CRC patients, respectively (Wilcoxon two-sample test, p < 0.0001). For benign bowel disease, the mean CEA level was 1.98 ± 0.80 ng/ml (Wilcoxon two-sample test, p = 0.0001) (Fig. 6B). Serum CEA levels were stage-dependent, being much higher in UICC stage IV (38.65 ± 4.67 ng/ml) than in stage I (2.84 ± 1.21 ng/ml) (p < 0.001). In contrast, desmin was not stage-dependent; the difference between UICC stage I and stage IV was not statistically significant.
Fig. 6.
Receiver-operating characteristic curves of CEA and desmin. A and B, serum levels of CEA (A) and desmin (B) among the CRC group, benign bowel disease group, and healthy control group (one-way ANOVA, p < 0.01). C, serum concentrations of CEA and desmin of 92 CRC samples, 45 healthy control samples, and 25 samples from patients with benign bowel diseases were determined by ELISA. Receiver-operating characteristic curves were derived by plotting the relationship between the specificity and the sensitivity at various cutoff levels. The area under the curve was 0.74 ± 0.04 for CEA and 0.77 ± 0.04 for desmin.
The receiver-operating characteristic (ROC) curve and the area under the curve of desmin and CEA were calculated (Fig. 6C). The area under the curve of desmin and CEA was 0.74 ± 0.04 and 0.77 ± 0.04, respectively, and significantly higher than that of the null hypothesis (true area was 0.5; p < 0.01). Thus, the diagnostic accuracy of both markers was in a comparable range. This also suggested that the serum desmin level could serve as a potential diagnostic marker for CRC.
DISCUSSION
The mortality rate of CRC has remained relatively high for many years. Various biomarkers such as CA19-9 and CA242 for CRC diagnosis are available, and CEA is the most commonly used. However, CEA lacks sensitivity as well as specificity for screening an average risk population, and its diagnostic role remains controversial, so CRC is often diagnosed at late stages (34, 35). In contrast, early stage CRC is associated with prolonged survival following surgical resection of the tumor (36). Therefore, the identification of novel tumor markers with higher specificity and sensitivity for early stage diagnosis, prognosis, and treatment has the potential to improve the clinical strategy and outcome of CRC. Despite a number of limitations, a 2-DE and MS-based proteomics strategy provides high throughput simultaneous identification of hundreds of proteins and is still considered a valuable method to screen biomarkers of tumor (5, 37–39). In the present study, we compared the global protein profiles among fetal colorectal tissues, CRC, and matched colorectal tissues using a 2-DE and MS/MS-based approach. This approach makes no assumption about known or unknown molecules, allowing the process to be independent of any presupposed hypotheses (11). A total of 22 differentially expressed proteins were identified among three groups. Most of these proteins were involved in biological processes, which usually play key roles in early development and malignant progression.
To our knowledge, some high diagnostic value biomarkers of cancer have the characteristic of embryonic origin, such as CEA, α-fetoprotein, glypican-3, and nucleophosmin (27). These molecules were highly expressed both in fetal tissues and human tumor tissues but seldom expressed in the normal tissues in adulthood. Indeed it has been widely observed that these genes and proteins are shared between the processes of embryogenesis and tumorigenesis (40). The mechanisms of colorectal development and CRC progression are similar in the processes of mitosis and differentiation, and comparable events are observed during the initiation and progression of CRC (27). According to this theory, we focused on the proteins that are highly expressed in the fetus and CRC tissues compared with normal tissue. By comparing the protein profiles among fetal, cancer, and normal tissues, we identified two proteins, desmin and zinc finger protein 829, with high expression in CRC and fetal colorectal tissue compared with normal colorectal tissue. MS/MS analysis revealed desmin with high confidence and zinc finger protein 829 with a very low confidence. Therefore, desmin became the subsequent focus of the study.
Desmin is a 52-kDa protein with a wide variety of functions. It is a type III intermediate filament in smooth muscle tissue (41). It has been confirmed that desmin is one of the earliest protein markers for muscle tissue in embryogenesis as it is detected in the somites of myoblasts (42). Desmin is present at low levels during embryogenesis but increases as the cell nears terminal differentiation. Recently studies have shown that desmin may be important in mitochondria function (43). There is some evidence that desmin may also connect the sarcomere to the extracellular matrix through desmosomes that could be important in signaling between the extracellular matrix and the sarcomere (44, 45). Now several studies have shown that desmin is a highly sensitive marker for endothelial cell differentiation and tumor invasiveness in several types of cancers, including colon cancer (46), gastrointestinal stromal tumors (47), and embryonal sarcomas (48). Recently proteomics analysis for endometrial carcinoma also discovered the overexpression of desmin in carcinoma tissue (11). Therefore, desmin was further characterized in this study to confirm the potential oncofetal diagnostic and prognostic value for CRC.
2-DE analysis revealed that expression of desmin was significantly higher both in CRC and in fetal colorectal tissue than in normal colorectal tissues. The expression alteration was unambiguously confirmed by RT-PCR and Western blot analysis. In addition, RT-PCR, Western blotting, and immunohistochemistry were further performed to investigate the expression of desmin in different clinical pathological stages. The results showed that the expression of desmin among normal colorectal tissue, adenoma tissue, carcinoma tissue, and liver metastasis tissue exhibited a remarkable increasing trend that paralleled the increasing severity of colorectal tissue injury. To confirm the oncofetal characteristic of desmin, we observed the expression of desmin in the different developmental stages of fetal colorectal tissues. It was noteworthy that expression of desmin in the different fetal developmental stages of colorectal tissues showed a remarkable decreasing trend that followed the development of the fetus, similar to the expression of CEA during fetal development (49, 50).
To further study the potential role of desmin as a biomarker for the prognosis, progression, or other aggressive behaviors of CRC, we examined the correlations between desmin expression and UICC staging, histodifferentiation, location, and survival rate. Our results showed that overexpression of desmin was more likely present in a low degree of differentiated CRC. The survival analysis also showed that overexpression of desmin correlated to low survival rate. No significant correlation was observed between the expression of desmin and UICC staging or location. These results suggested the potential value of the desmin assay in prognosis prediction and clinical treatment of CRC.
After having shown overexpression of desmin in CRC compared with adjacent normal colorectal tissue, we speculated that desmin might be released from tumor to serum, giving rise to elevated desmin levels also in the serum of CRC patients. Therefore, we developed a highly sensitive immunoassay and tested the level of desmin in the serum of CRC patients, healthy volunteers, and patients with benign bowel disease. As a benchmark, we also assessed serum levels of CEA, the established tumor marker for CRC. Mean serum levels of CEA were elevated from 1.55 ± 1.46 ng/ml in healthy volunteers and 1.98 ± 0.80 ng/ml in benign bowel disease to 5.73 ± 10.08 ng/ml in CRC patients. With regard to desmin, we found a strong elevation of desmin in the blood of CRC patients. The mean serum level for desmin was 2.1 times higher in the cohort of CRC patients (105.80 ± 60.30 ng/ml) compared with healthy volunteers (49.21 ± 25.74 ng/ml) and about 1.4 times as high as the mean serum level in the patients with benign bowel diseases (71.41 ± 23.47 ng/ml). Therefore, desmin could prove to be a new, sensitive, and specific marker that assists in the detection of CRC. However, more patient sera are needed to be tested, including pre- and postoperative samples, samples from patients with other cancers, etc., to understand the potential value of this new biomarker.
The ROC curve is a useful method for evaluating clinical usefulness of a biomarker (51) and for comparing the effectiveness between different biomarkers (52). The higher area under the curve represents higher diagnostic ability. The ROC curve analysis showed that the area under the curve of desmin reached 0.77, which means that desmin can be used as a potential serum biomarker. We also assessed serum levels of CEA as a benchmark to compare with desmin. The area under the curve of CEA reached 0.74, which showed a comparable diagnostic performance between CEA and desmin based on our preliminary analysis. CEA, which is the established and the best single tumor marker for CRC, is not recommended for early detection of CRC because of a lack of sensitivity and specificity (53). Since CEA was discovered as a cancer marker in 1965 (49, 50), no biomarker has been established that fulfills the requirements with sufficiently high sensitivity and specificity for early detection of CRC, and to date no novel marker could displace CEA in the detection of the CRC. However, recent proteomics researchers have found two novel biomarkers for CRC, nicotinamide N-methyltransferase (13) and proteasome activator complex subunit (PSME3) (14), which displayed a diagnostic performance comparable to CEA. Therefore, based on our results, desmin could prove to be a potential new, sensitive, and specific marker that assists in the detection of CRC.
In conclusion, a potential oncofetal biomarker, desmin was found and confirmed for diagnosis and predicting prognosis of CRC based on proteomics screening and molecular biology confirmation. The results of our study suggested that desmin has substantial clinical impact, but more patient-centered studies and much larger sample numbers are required for the development of practical applications. Moreover further research is necessary to combine several markers (i.e. CEA, N-methyltransferase, PSME3, desmin, and so on) for the detection of CRC by applying multivariate analysis so that diagnostic performance can be significantly improved. It is anticipated that such a method will ultimately ensure that biomarkers with clinical value make their way into routine clinical practice.
Acknowledgments
We thank Professor M. Michael Wolf (Boston University School of Medicine) for the advice on this study.
Footnotes
* This work was supported by Shanghai Science and Technology Development Fund Grant 05DJ14010), Major Basic Research Program of Shanghai Grant 07DZ19505, and National 973 Basic Research Program of China Grant 2008CB517403.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- CRC
- colorectal cancer
- 2-DE
- two-dimensional polyacrylamide gel electrophoresis
- CEA
- carcinoembryonic antigen
- UICC
- International Union against Cancer
- ROC
- receiver-operating characteristic
- 2-D
- two-dimensional
- ANOVA
- analysis of variance
- au
- arbitrary units
- LSD
- least significant difference.
REFERENCES
- 1.Weitz J., Koch M., Debus J., Höhler T., Galle P. R., Büchler M. W. ( 2005) Colorectal cancer. Lancet 365, 153– 165 [DOI] [PubMed] [Google Scholar]
- 2.Jemal A., Murray T., Ward E., Samuels A., Tiwari R. C., Ghafoor A., Feuer E. J., Thun M. J. ( 2005) Cancer statistics, 2005. CA Cancer J. Clin. 55, 10– 30 [DOI] [PubMed] [Google Scholar]
- 3.Woolf S. H. ( 2000) The best screening test for colorectal cancer–a personal choice. N. Engl. J. Med. 343, 1641– 1643 [DOI] [PubMed] [Google Scholar]
- 4.Walsh J. M., Terdiman J. P. ( 2003) Colorectal cancer screening: scientific review. JAMA 289, 1288– 1296 [DOI] [PubMed] [Google Scholar]
- 5.Celis J. E., Gromov P. ( 2003) Proteomics in translation cancer research: toward an integrated approach. Cancer Cell 3, 9– 15 [DOI] [PubMed] [Google Scholar]
- 6.Alessandro R., Belluco C., Kohn E. C. ( 2005) Proteomic approaches in colon cancer: promising tools for new cancer markers and drug target discovery. Clin. Colorectal Cancer 4, 396– 402 [DOI] [PubMed] [Google Scholar]
- 7.Nedelkov D., Kiernan U. A., Niederkofler E. E., Tubbs K. A., Nelson R. W. ( 2006) Population proteomics: the concept, attributes, and potential for cancer biomarker research. Mol. Cell. Proteomics 5, 1811– 1818 [DOI] [PubMed] [Google Scholar]
- 8.Tyers M., Mann M. ( 2003) From genomics to proteomics. Nature 422, 193– 197 [DOI] [PubMed] [Google Scholar]
- 9.Phizicky E., Bastiaens P. I., Zhu H., Snyder M., Fields S. ( 2003) Protein analysis on a proteomic scale. Nature 422, 208– 215 [DOI] [PubMed] [Google Scholar]
- 10.Sun W., Xing B., Sun Y., Du X., Lu M., Hao C., Lu Z., Mi W., Wu S., Wei H., Gao X., Zhu Y., Jiang Y., Qian X., He F. ( 2007) Proteome analysis of hepatocellular carcinoma by two-dimensional difference gel electrophoresis: novel protein markers in hepatocellular carcinoma tissues. Mol. Cell. Proteomics 6, 1798– 1808 [DOI] [PubMed] [Google Scholar]
- 11.Li Z., Zhao X., Bai S., Wang Z., Chen L., Wei Y., Huang C. ( 2008) Proteomics identification of cyclophilin a as a potential prognostic factor and therapeutic target in endometrial carcinoma. Mol. Cell. Proteomics 7, 1810– 1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petricoin E. F., Zoon K. C., Kohn E. C., Barrett J. C., Liotta L. A. ( 2002) Clinical proteomics: translating benchside promise into bedside reality. Nat. Rev. Drug Discov. 1, 683– 695 [DOI] [PubMed] [Google Scholar]
- 13.Roessler M., Rollinger W., Palme S., Hagmann M. L., Berndt P., Engel A. M., Schneidinger B., Pfeffer M., Andres H., Karl J., Bodenmüller H., Rüschoff J., Henkel T., Rohr G., Rossol S., Rösch W., Langen H., Zolg W., Tacke M. ( 2005) Identification of nicotinamide N-methyltransferase as a novel serum tumor marker for colorectal cancer. Clin. Cancer Res. 11, 6550– 6657 [DOI] [PubMed] [Google Scholar]
- 14.Roessler M., Rollinger W., Mantovani-Endl L., Hagmann M. L., Palme S., Berndt P., Engel A. M., Pfeffer M., Karl J., Bodenmüller H., Rüschoff J., Henkel T., Rohr G., Rossol S., Rösch W., Langen H., Zolg W., Tacke M. ( 2006) Identification of PSME3 as a novel serum tumor marker for colorectal cancer by combining two-dimensional polyacrylamide gel electrophoresis with a strictly mass spectrometry-based approach for data analysis. Mol. Cell. Proteomics 5, 2092– 2101 [DOI] [PubMed] [Google Scholar]
- 15.Alfonso P., Núñez A., Madoz-Gurpide J., Lombardia L., Sánchez L., Casal J. I. ( 2005) Proteomic expression analysis of colorectal cancer by two-dimensional differential gel electrophoresis. Proteomics 5, 2602– 2611 [DOI] [PubMed] [Google Scholar]
- 16.Madoz-Gúrpide J., Cañamero M., Sanchez L., Solano J., Alfonso P., Casal J. I. ( 2007) A proteomics analysis of cell signaling alterations in colorectal cancer. Mol. Cell. Proteomics 6, 2150– 2164 [DOI] [PubMed] [Google Scholar]
- 17.Uemura T., Yerushalmi H. F., Tsaprailis G., Stringer D. E., Pastorian K. E., Hawel L., 3rd, Byus C. V., Gerner E. W. ( 2008) Identification and characterization of a diamine exporter in colon epithelial cells. J. Biol. Chem. 283, 26428– 26435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bi X., Lin Q., Foo T. W., Joshi S., You T., Shen H. M., Ong C. N., Cheah P. Y., Eu K. W., Hew C. L. ( 2006) Proteomic analysis of colorectal cancer reveals alterations in metabolic pathways: mechanism of tumorigenesis. Mol. Cell. Proteomics 5, 1119– 1130 [DOI] [PubMed] [Google Scholar]
- 19.Wang Z., Jiang L., Huang C., Li Z., Chen L., Gou L., Chen P., Tong A., Tang M., Gao F., Shen J., Zhang Y., Bai J., Zhou M., Miao D., Chen Q. ( 2008) Comparative proteomics approach to screening of potential diagnostic and therapeutic targets for oral squamous cell carcinoma. Mol. Cell. Proteomics 7, 1639– 1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillespie J. R., Uversky V. N. ( 2000) Structure and function of alpha-fetoprotein: a biophysical overview. Biochim. Biophys. Acta 1480, 41– 56 [DOI] [PubMed] [Google Scholar]
- 21.Alexander P. ( 1972) Foetal “antigens” in cancer. Nature 235, 137– 140 [DOI] [PubMed] [Google Scholar]
- 22.Mizejewski G. J. ( 2004) Biological roles of alpha-fetoprotein during pregnancy and perinatal development. Exp. Biol. Med. 229, 439– 463 [DOI] [PubMed] [Google Scholar]
- 23.Song H. H., Filmus J. ( 2002) The role of glypicans in mammalian development. Biochim. Biophys. Acta 1573, 241– 246 [DOI] [PubMed] [Google Scholar]
- 24.Sarandakou A., Protonotariou E., Rizos D. ( 2007) Tumor markers in biological fluids associated with pregnancy. Crit. Rev. Clin. Lab. Sci. 44, 151– 178 [DOI] [PubMed] [Google Scholar]
- 25.Murray M. J., Lessey B. A. ( 1999) Embryo implantation and tumor metastasis: common pathways of invasion and angiogenesis. Semin. Reprod. Endocrinol. 17, 275– 290 [DOI] [PubMed] [Google Scholar]
- 26.Hanahan D., Weinberg R. A. ( 2000) The hallmarks of cancer. Cell 100, 57– 70 [DOI] [PubMed] [Google Scholar]
- 27.Lee N. P., Leung K. W., Cheung N., Lam B. Y., Xu M. Z., Sham P. C., Lau G. K., Poon R. T., Fan S. T., Luk J. M. ( 2008) Comparative proteomic analysis of mouse livers from embryo to adult reveals an association with progression of hepatocellular carcinoma. Proteomics 8, 2136– 2149 [DOI] [PubMed] [Google Scholar]
- 28.Kreisberg J. I., Malik S. N., Prihoda T. J., Bedolla R. G., Troyer D. A., Kreisberg S., Ghosh P. M. ( 2004) Phosphorylation of Akt (Ser473) is an excellent predictor of poor clinical outcome in prostate cancer. Cancer Res. 64, 5232– 5236 [DOI] [PubMed] [Google Scholar]
- 29.Feng Y. Z., Shiozawa T., Miyamoto T., Kashima H., Kurai M., Suzuki A., Ying-Song J., Konishi I. ( 2007) Overexpression of hedgehog signaling molecules and its involvement in the proliferation of endometrial carcinoma cells. Clin. Cancer Res. 13, 1389– 1398 [DOI] [PubMed] [Google Scholar]
- 30.Zhu K., Zhao J., Lubman D. M., Miller F. R., Barder T. J. ( 2005) Protein pI shifts due to posttranslational modifications in the separation and characterization of proteins. Anal. Chem. 77, 2745– 2755 [DOI] [PubMed] [Google Scholar]
- 31.Cook L. A., Schey K. L., Wilcox M. D., Dingus J., Ettling R., Nelson T., Knapp D. R., Hildebrandt J. D. ( 2006) Proteomic analysis of bovine brain G protein γ subunit processing heterogeneity. Mol. Cell. Proteomics 5, 671– 685 [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez-Piñeiro A. M., de la Cadena M. P., López-Saco A., Rodríguez-Berrocal F. J. ( 2006) Differential expression of serum clusterin isoforms in colorectal cancer. Mol. Cell. Proteomics 5, 1647– 1657 [DOI] [PubMed] [Google Scholar]
- 33.Zheng X., Hong L., Shi L., Guo J., Sun Z., Zhou J. ( 2008) Proteomics analysis of host cells infected with infectious bursal disease virus. Mol. Cell. Proteomics 7, 612– 625 [DOI] [PubMed] [Google Scholar]
- 34.American Society of Clinical Oncology ( 1996) Clinical practice guidelines for the use of tumor markers in breast and colorectal cancer. Adopted on May 17, 1996 by the American Society of Clinical Oncology. J. Clin. Oncol. 14, 2843– 2877 [DOI] [PubMed] [Google Scholar]
- 35.Bast R. C., Jr., Ravdin P., Hayes D. F., Bates S., Fritsche H., Jr., Jessup J. M., Kemeny N., Locker G. Y., Mennel R. G., Somerfield M. R.; American Society of Clinical Oncology Tumor Markers Expert Panel ( 2001) 2000 update of recommendations for the use of tumor markers in breast and colorectal cancer: clinical practice guidelines of the American Society of Clinical Oncology. J. Clin. Oncol. 19, 1865– 1878 [DOI] [PubMed] [Google Scholar]
- 36.Bardeesy N., DePinho R. A. ( 2002) Pancreatic cancer biology and genetics. Nat. Rev. Cancer. 2, 897– 909 [DOI] [PubMed] [Google Scholar]
- 37.Nishigaki R., Osaki M., Hiratsuka M., Toda T., Murakami K., Jeang K. T., Ito H., Inoue T., Oshimura M. ( 2005) Proteomic identification of differentially-expressed genes in human gastric carcinomas. Proteomics 5, 3205– 3213 [DOI] [PubMed] [Google Scholar]
- 38.Kawada N. ( 2006) Cancer serum proteomics in gastroenterology. Gastroenterology 130, 1917– 1919 [DOI] [PubMed] [Google Scholar]
- 39.Rabilloud T. ( 2002) Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs up the mountains. Proteomics 2, 3– 10 [PubMed] [Google Scholar]
- 40.Hendrix M. J., Seftor E. A., Seftor R. E., Kasemeier-Kulesa J., Kulesa P. M., Postovit L. M. ( 2007) Reprogramming metastatic tumour cells with embryonic microenvironments. Nat. Rev. Cancer 7, 246– 255 [DOI] [PubMed] [Google Scholar]
- 41.Li Z., Mericskay M., Agbulut O., Butler-Browne G., Carlsson L., Thornell L. E., Babinet C., Paulin D. ( 1997) Desmin is essential for the tensile strength and integrity of myofibrils but not for myogenic commitment, differentiation, and fusion of skeletal muscle. J. Cell Biol. 139, 129– 144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bär H., Strelkov S. V., Sjöberg G., Aebi U., Herrmann H. ( 2004) The biology of desmin filaments: how do mutations affect their structure, assembly, and organisation? J. Struct. Biol. 148, 137– 152 [DOI] [PubMed] [Google Scholar]
- 43.Goldfarb L. G., Vicart P., Goebel H. H., Dalakas M. C. ( 2004) Desmin myopathy. Brain 127, 723– 734 [DOI] [PubMed] [Google Scholar]
- 44.Paulin D., Li Z. ( 2004) Desmin: a major intermediate filament protein essential for the structural integrity and function of muscle. Exp. Cell Res. 301, 1– 7 [DOI] [PubMed] [Google Scholar]
- 45.Shah S. B., Davis J., Weisleder N., Kostavassili I., McCulloch A. D., Ralston E., Capetanaki Y., Lieber R. L. ( 2004) Structural and functional roles of desmin in mouse skeletal muscle during passive deformation. Biophys. J. 86, 2993– 3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang W., Li X., Rao S., Wang L., Du L., Li C., Wu C., Wang H., Wang Y., Yang B. ( 2008) Constructing disease-specific gene networks using pair-wise relevance metric: application to colon cancer identifies interleukin 8, desmin and enolase 1 as the central elements. BMC Syst. Biol. 2, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liegl B., Hornick J. L., Antonescu C. R., Corless C. L., Fletcher C. D. ( 2009) Rhabdomyosarcomatous differentiation in gastrointestinal stromal tumors after tyrosine kinase inhibitor therapy: a novel form of tumor progression. Am. J. Surg. Pathol. 33, 218– 226 [DOI] [PubMed] [Google Scholar]
- 48.Dias P., Kumar P., Marsden H. B., Morris-Jones P. H., Birch J., Swindell R., Kumar S. ( 1987) Evaluation of desmin as a diagnostic and prognostic marker of childhood rhabdomyosarcomas and embryonal sarcomas. Br. J. Cancer 56, 361– 365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gold P., Freedman S. O. ( 1965) Specific carcinoembryonic antigens of the human digestive system. J. Exp. Med. 122, 467– 481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gold P., Freedman S. O. ( 1965) Demonstration of tumor-specific antigens in human colonic carcinomata by immunological tolerance and absorption techniques. J. Exp. Med. 121, 439– 462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perkins N. J., Schisterman E. F. ( 2006) The inconsistency of “optimal” cutpoints obtained using two criteria based on the receiver operating characteristic curve. Am. J. Epidemiol. 163, 670– 675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schisterman E. F., Faraggi D., Reiser B. ( 2004) Adjusting the generalized ROC curve for covariates. Stat. Med. 23, 3319– 3331 [DOI] [PubMed] [Google Scholar]
- 53.Duffy M. J. ( 2001) Carcinoembryonic antigen as a marker for colorectal cancer: is it clinically useful? Clin. Chem. 47, 624– 630 [PubMed] [Google Scholar]